Abstract
A carbapenemase-resistant Klebsiella pneumoniae strain, clone ST258 producing KPC-3, was fully characterized. The entire plasmid content was investigated, thereby identifying plasmids of the IncFIIk (two of them similar to pKPQIL and pKPN3, respectively), IncX, and ColE types, carrying a formidable set of resistance genes against toxic compounds, metals, and antimicrobial drugs and a novel iron(III) uptake system.
TEXT
Nosocomial infection with Klebsiella pneumoniae has a worldwide distribution. The presence of invasive devices, contamination of respiratory support equipment, immunocompromised status, treatment in an intensive care unit or nursing home, and use of antibiotics are factors that increase the likelihood of nosocomial infection. Acquisition of K. pneumoniae has become a major problem in many hospitals because of resistance to multiple antibiotics, the scarcity of therapeutic options left to treat these patients, and the association of invasive infections with disturbingly high mortality rates (6).
Carbapenem resistance in K. pneumoniae strains can arise by the loss or modification of porins (OmpK35 and OmpK36) associated with the production of expanded-spectrum or AmpC β-lactamases or by the acquisition of resistance genes encoding metallo-β-lactamases (MBLs) and nonmetallo-carbapenemases (KPC, GES, or OXA types) (9, 16, 17, 21).
In the last several years, an extremely drug-resistant K. pneumoniae sequence type, 258 (ST258), producing the KPC carbapenemase, has been detected as a nosocomial pathogen around the world, causing epidemics of national and international proportions. K. pneumoniae KPC producers have been described mainly in Israel but in recent studies have been also identified in South and North America and in several European countries, including Italy (2, 7, 10, 13, 17, 18, 22).
The blaKPC-3 gene has been found as part of the 10-kb Tn3-like element Tn4401 (3). Tn4401 has been found on both IncN and IncFIIk plasmids (8, 11). Recently, the pKpQIL plasmid carrying blaKPC-3 from K. pneumoniae ST258 from Israel has been completely sequenced (11) (GenBank accession number GU595196).
Active surveillance of carbapenem-resistant K. pneumoniae and retrospective case-control studies have been performed during the last 4 years (2007 to 2010) at the 1,200-bed Umberto I teaching hospital of Rome, Italy (14). Previous studies demonstrated that a K. pneumoniae ST37 clone was dominant in this hospital, colonizing many patients but also causing severe outbreaks in the intensive care unit (5). This clone showed ertapenem resistance and decreased susceptibility to meropenem and imipenem due to extended-spectrum β-lactamase (ESBL) production, depletion of the OmpK35 porin, and the presence of a mutated OmpK36 protein designated OmpK36V (5).
No carbapenemase-producing K. pneumoniae strains were identified in our hospital until June 2010, when an Italian patient was transferred from another Italian hospital to the laparoscopic surgery ward for an abdominal drainage. From this site, a K. pneumoniae strain was isolated (strain 55873), showing resistance to all antibiotics, including carbapenems (MICs: ertapenem, >8 μg/ml; imipenem, 8 μg/ml; meropenem, >16 μg/ml; determined by the Vitek2 system, AST-N089 card [bioMérieux, Marcy l'Etoile, France], according to the European Committee on Antimicrobial Susceptibility Testing [EUCAST] guidelines), except gentamicin, colistin, and tigecycline. The multilocus sequence typing for this strain was carried out as previously described (4), and the strain was assigned to ST258 (http://www.pasteur.fr/recherche/genopole/PF8/mlst/).
Combined-disc tests of meropenem (10 μg) with or without 400 μg of phenylboronic acid (PBA) or 10 μl of 0.1 M EDTA on Mueller-Hinton agar II detected nonmetallo-carbapenemase production in this ST258 strain (19). PCR amplification of β-lactamase genes and sequencing of the amplicons demonstrated the presence of the blaKPC-3, blaTEM-1, and blaSHV-11 genes (1, 5, 12, 15).
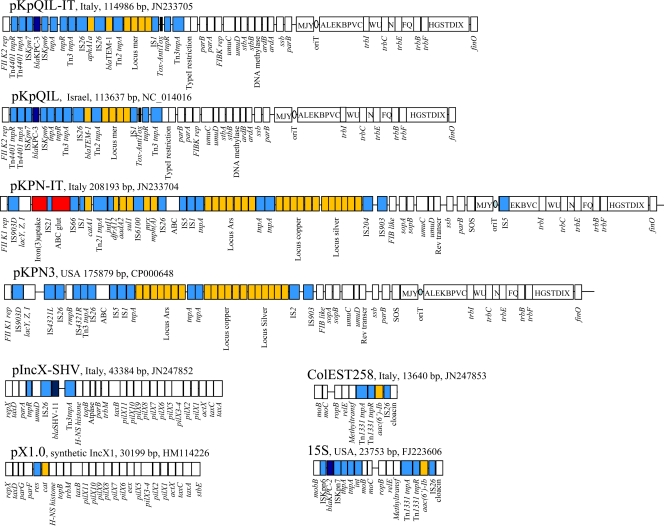
The coding sequences of ompK36 and ompK35 genes were amplified and sequenced for the ST258 strain 55873 (5). The ompK35 gene showed a TGA nonsense mutation at codon 89, resulting in early termination of translation and depletion of this porin (data not shown). The ompK36 gene encoded a novel OmpK36 protein variant, showing the insertion of codons Asp135 and Gly136 in the L3 loop of this protein. This insertion, in analogy with what was previously hypothesized for the CTX-M-15-ST37 clone, may contribute to increasing the MICs of different drugs (5, 9). The complete DNA sequences of plasmids contained in this strain were obtained by applying the 454-Genome Sequencer FLX procedure on a library constructed on total plasmid DNA purified by the Invitrogen PureLink HiPure Plasmid Filter Midiprep kit (Invitrogen, Milan, Italy), according to the manufacturer's procedure. Seventy-two contigs ranging from 49,686 to 353 bp, with at least 15-fold coverage, were obtained using the GS-FLX gAssembler software and compared with GenBank data using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). DNA comparison showed the presence of four major plasmid scaffolds, highly homologous to plasmid pKpQIL (NC_014016), pKPN3 (CP000648), 15S (FJ223606), and IncX1 (HM114226). Following the read-status output and BLAST results, contigs were assembled in four complete plasmid sequences by PCR-based gap closure (Fig. 1). In particular, the assembly of contigs flanked by IS26 elements was individually confirmed by PCR and DNA sequencing of the amplicons.
Fig 1.
Major structural features of plasmids pKpQIL-IT (JN233705), pKPN-IT (JN233704), IncX-SHV (JN247852), and ColEST258 (JN247853). Plasmids sequenced in this study were compared with plasmids pKpQIL (NC_014016), pKPN3 (CP000648), pX1.0 (HM114226), and 15S (FJ223606), respectively. White boxes indicate plasmid scaffold regions. The genes associated with the Tra locus are indicated as capital letters within the respective boxes. Resistance genes are indicated by orange boxes, except for the blaKPC and blaSHV-11 genes, which are indicated by dark blue boxes. Transposon-related genes (tnpA, tnpR, and tnpM), insertion sequences, and the class1 integrase genes are indicated by light blue boxes. The two putative virulence clusters acquired by the pKPN-IT plasmid with respect to pKPN3 are indicated by red boxes.
An IncFIIk-FIBk plasmid (JN233705; IncF replicons were defined as in reference 20) was very similar to plasmid pKpQIL (96% query coverage, 99% maximum nucleotide identity). The unique significant difference was the composite transposon IS26-aphA1-IS26, conferring kanamycin resistance that the pKpQIL-IT plasmid has acquired, while it was absent on the pKpQIL plasmid from Israel (11). The ST258 strain 55873 also contained another IncFIIk-FIB-like plasmid, which we named pKPN-IT (JN233704), which was highly related to plasmid pKPN3 (79% query coverage, 100% maximum nucleotide identity) identified in K. pneumoniae from the United States, conferring resistance to arsenic, copper, and silver. The plasmid pKPN-IT showed the same scaffold and resistance loci as did pKPN3, but it acquired a Fec-like iron(III) dicitrate transport system, a glutathione ABC transport system, a class 1 integron carrying trimethoprim and streptomycin resistance genes (dfrA12, orfF, and aadA2), and the chloramphenicol and macrolide resistance genes [catA1, mph(A)] (Fig. 1). The presence of these two plasmids endowed this ST258 strain with a formidable set of resistance genes against toxic compounds, metals, and antimicrobial drugs. The presence of the iron(III) uptake system is an interesting feature, likely involved in the capacity of the bacterium to acquire iron in the human host. The third plasmid identified in ST258 strain 55873 (IncX-SHV; JN247852) carried the blaSHV-11 gene and was related to the IncX family of plasmids such as the Escherichia coli plasmid pOLA52 (EU370913; 35% query coverage, 99% maximum nucleotide identity) and the synthetic conjugative molecular parasite pX1.0 (HM114226; 32% query coverage, 96% maximum nucleotide identity). Finally, a small ColE-like plasmid (JN247853), named ColEST258 and carrying the aac(6′)-Ib gene as part of a Tn1331 element, was also identified in this strain (Fig. 1). Interestingly, ColEST258 was identical (100% query coverage, 100% maximum nucleotide identity) to the K. pneumoniae plasmid 15S from the United States that contained the Tn4401 transposon carrying the blaKPC-2 gene variant (8).
Our study contributes to the description of the characteristics of K. pneumoniae clones that currently represent a serious potential risk for nosocomial settings.
Nucleotide sequence accession numbers.
GenBank accession numbers are as follows: pKpQIL-IT, JN23705; pKPN-IT, JN233704; IncX-SHV, JN247852; ColEST258, JN247853.
ACKNOWLEDGMENTS
This work was funded by grants to A.C. from the Istituto Superiore di Sanità and the Italian Minister of Health.
Footnotes
Published ahead of print 17 January 2012
REFERENCES
- 1. Arlet G, Rouveau M, Philippon A. 1997. Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum beta-lactamase. FEMS Microbiol. Lett. 152:163–167 [DOI] [PubMed] [Google Scholar]
- 2. Baraniak A, et al. 2009. The emergence of Klebsiella pneumoniae ST258 with KPC-2 in Poland. Antimicrob. Agents Chemother. 53:4565–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cuzon G, et al. 2010. Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. Emerg. Infect. Dis. 16:1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. García-Fernández A, et al. 2010. An ertapenem-resistant ESBL-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob. Agents Chemother. 54:4178–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Giamarellou H, Poulakou G. 2009. Multidrug-resistant Gram-negative infections: what are the treatment options? Drugs 69:1879–1901 [DOI] [PubMed] [Google Scholar]
- 7. Giani T, et al. 2009. Emergence in Italy of Klebsiella pneumoniae sequence type 258 producing KPC-3 carbapenemase. J. Clin. Microbiol. 47:3793–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gootz TD, et al. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob. Agents Chemother. 53:1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD. 2006. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of blaACT-1 β-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrob. Agents Chemother. 50:3396–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kitchel B, et al. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2010. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob. Agents Chemother. 54:4493–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mabilat C, Goussard S, Sougakoff W, Spencer RC, Courvalin P. 1990. Direct sequencing of the amplified structural gene and promoter for the extended-broad-spectrum beta-lactamase TEM-9 (RHH-1) of Klebsiella pneumoniae. Plasmid 23:27–34 [DOI] [PubMed] [Google Scholar]
- 13. Mezzatesta ML, et al. 7 May 2011. Outbreak of KPC-3-producing, and colistin-resistant, Klebsiella pneumoniae infections in two Sicilian hospitals. Clin. Microbiol. Infect. doi:10.1111/j.1469-0691.2011.03572 [DOI] [PubMed] [Google Scholar]
- 14. Orsi GB, et al. 2011. Risk factors and clinical significance of ertapenem-resistant Klebsiella pneumoniae in hospitalised patients. J. Hosp. Infect. 78:54–58 [DOI] [PubMed] [Google Scholar]
- 15. Pérez-Pérez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poirel L, Heritier C, Tolun V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rasheed JK, et al. 2008. Detection of the Klebsiella pneumoniae carbapenemase type 2 carbapenem-hydrolyzing enzyme in clinical isolates of Citrobacter freundii and K. oxytoca carrying a common plasmid. J. Clin. Microbiol. 46:2066–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Samuelsen O, et al. 2009. Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J. Antimicrob. Chemother. 63:654–658 [DOI] [PubMed] [Google Scholar]
- 19. Tsakris A, et al. 2010. A simple phenotypic method for the differentiation of metallo-beta-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J. Antimicrob. Chemother. 65:1664–1671 [DOI] [PubMed] [Google Scholar]
- 20. Villa L, García-Fernández A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65:2518–2529 [DOI] [PubMed] [Google Scholar]
- 21. Weldhagen GF, Prinsloo A. 2004. Molecular detection of GES-2 extended spectrum beta-lactamase producing Pseudomonas aeruginosa in Pretoria, South Africa. Int. J. Antimicrob. Agents 24:35–38 [DOI] [PubMed] [Google Scholar]
- 22. Woodford N, et al. 2008. Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom. J. Antimicrob. Chemother. 62:1261–1264 [DOI] [PubMed] [Google Scholar]