Abstract
Nine carbapenem-resistant Enterobacteriaceae isolates collected from eight patients in five German hospitals were investigated. Six isolates produced the OXA-48 carbapenemase, and three isolates produced OXA-162, which is a point mutant form of OXA-48. Both carbapenemase genes were located on IncL/M-type conjugative plasmids. Insertion sequence IS1999 (truncated or not by IS1R) was located upstream of the blaOXA-48 and blaOXA-162 genes in all of the isolates. Pulsed-field gel electrophoresis typing indicated the clonal transmission of an OXA-48-producing Klebsiella pneumoniae strain in two hospitals.
TEXT
Carbapenem resistance in Enterobacteriaceae is based on various mechanisms that may involve upregulation of efflux pumps or loss of porins. Most prevalent is the acquisition of carbapenem-hydrolyzing enzymes, or carbapenemases. Some commonly identified carbapenemases are KPC-, NDM-, and OXA-48-type enzymes whose respective genes are located on plasmids that enable their transfer between different enterobacterial species (19). The OXA-48 carbapenemase was first described in Klebsiella pneumoniae epidemic isolates from Turkey and then in several European countries, such as France and Belgium. Recently, it has also been identified in enterobacterial isolates recovered from non-European countries, such as Lebanon, Tunisia, Senegal, Morocco, Israel, and India (2, 5, 9, 10, 12, 18). In addition to K. pneumoniae, OXA-48 has been identified in Escherichia coli, Enterobacter cloacae, Citrobacter freundii, and Providencia rettgeri (2). This enzyme is able to hydrolyze penicillins and carbapenems but possesses poor activity against broad-spectrum cephalosporins. Multidrug resistance in OXA-48-producing strains often results from the coproduction of various resistance mechanisms, in particular, extended-spectrum β-lactamases (ESBLs) and other resistance determinants.
Here we report on the molecular analysis of carbapenem-resistant Enterobacteriaceae isolates that were recovered in Germany between 2008 and 2010 and sent to the Robert Koch Institute, Wernigerode, for further characterization. Nine isolates, including E. coli (n = 2), K. pneumoniae (n = 4), Raoultella ornithinolytica (n = 1), C. freundii (n = 1), and E. cloacae (n = 1), were selected since they gave negative phenotypic test results for the production of metallo-β-lactamases or KPC enzyme production (MBL-Etest [bioMérieux, Nürtingen, Germany] and KPC+MBL Confirm ID Kit [Alere GmbH, Zug, Switzerland]).
In April and May 2008, two E. coli isolates were isolated from a wound swab and secretion from a tracheal cannula (colonization) in two hospitals in Berlin (hospitals A and B). One patient developed sepsis but recovered. The second patient, exhibiting several comorbidity factors, developed sepsis and ventilator-associated pneumonia and was treated with various antibiotics (tigecycline, piperacillin-sulbactam, and meropenem). In addition, one R. ornithinolytica strain recovered from a blood culture and one C. freundii strain recovered from bronchoalveolar lavage fluid were isolated from a 67-year-old patient in hospital A in September 2009.
Between November 2009 and January 2010, four multidrug-resistant K. pneumoniae isolates were sent in from intensive care units of two hospitals (hospitals C and D) located within 40 km of each other in the federal state of North Rhine-Westphalia. The strains had been isolated from urine cultures or tracheal aspirates of four different patients. These patients all presented with underlying diseases (myocardial infarction, congestive heart failure, plasmacytoma), and two patients had previously received meropenem. Additionally, an E. cloacae strain was isolated in 2009 from a drainage swab in hospital E, which is located in southern Germany. None of the patients reported any link with Turkey, one patient (E. coli, hospital B) came from Syria, and another patient (E. cloacae, hospital E) was from Libya.
Antimicrobial susceptibility testing of 10 antibiotics (ampicillin, cefoxitin, cefotaxime, ceftazidime, gentamicin, kanamycin, chloramphenicol, tetracycline, ciprofloxacin, and sulfamethoxazole-trimethoprim) was done by broth microdilution according to the CLSI guidelines (3). MIC determinations for carbapenems (imipenem, meropenem) were performed by Etest (bioMérieux). Occurrence of β-lactamases was detected by PCR amplification and sequencing of ESBL genes (blaTEM, blaSHV, blaCTX-M, and blaOXA) and several carbapenemase genes (blaVIM, blaIMP, blaNDM-1, blaKPC, and blaOXA-48) (6, 13, 14). Identification of qnr-like genes encoding plasmid-mediated quinolone resistance determinants was performed as described previously (13). Transfer of resistance was performed by broth mating assays using a sodium azide-resistant E. coli J53 recipient (4). Plasmid DNA of clinical isolates and transconjugants was isolated using the Qiagen Plasmid Mini Kit (Qiagen, Hilden, Germany). Southern hybridization of the plasmids using digoxigenin (DIG)-labeled, blaOXA-48-specific probes and signal detection using CDP-Star were performed by following the manufacturer's guidelines (Roche Diagnostics Ltd., West Sussex, United Kingdom). In addition, all nine isolates were typed by pulsed-field gel electrophoresis (PFGE) using XbaI-restricted whole genomic DNA.
Both E. coli isolates were resistant to carbapenems but remained susceptible to expanded-spectrum cephalosporins. All other isolates were resistant to cefotaxime and ceftazidime and either resistant (K. pneumoniae isolates) or intermediately susceptible to imipenem and meropenem. Coresistances to fluoroquinolones (seven isolates), aminoglycosides (nine isolates), and sulfamethoxazole-trimethoprim (three isolates) were frequently observed (Table 1).
Table 1.
Phenotypic and genotypic characteristics of OXA-type carbapenemase-producing clinical isolates and transconjugants
| Straina | Hospital | Yr | β-Lactamase(s) | Antimicrobial resistance profile | MIC (mg/liter)e |
PFGE type | |
|---|---|---|---|---|---|---|---|
| IPM | MPM | ||||||
| E. coli 84/08 | A | 2008 | OXA-162, TEM-1 | AMP FOX GEN CMP OTE CIP SXT | 8 | 16 | 1 |
| R. ornithinolytica 215/09b | A | 2009 | OXA-162, TEM-1, OXA-1, SHV-5 | AMP CTX CAZ KAN CMP CIP | 4 | 4 | 2 |
| C. freundii 216/09b | A | 2009 | OXA-162, SHV-5 | AMP FOX CTX CAZ GEN CMP | 2 | 2 | 3 |
| E. coli 131/08 | B | 2008 | OXA-48, TEM-1, OXA-1 | AMP GEN CMP OTE SXT | 32 | 32 | 4 |
| K. pneumoniae | |||||||
| 229/09 | C | 2009 | OXA-48, TEM-1, OXA-9, SHV-11, CTX-M-15 | AMP FOX CTX CAZ GEN KAN AMK CIP | >32 | >32 | 5 |
| 238/09 | C | 2009 | OXA-48, TEM-1, OXA-9, SHV-11, CTX-M-15 | AMP FOX CTX CAZ GEN KAN AMK CIP | >32 | >32 | 5 |
| 239/09 | C | 2009 | OXA-48, TEM-1, OXA-9, SHV-11, CTX-M-15 | AMP FOX CTX CAZ GEN KAN AMK CIP | >32 | >32 | 5 |
| 16/10 | D | 2010 | OXA-48, TEM-1, OXA-9, SHV-11, CTX-M-15 | AMP FOX CTX CAZ GEN KAN AMK CIP | 32 | >32 | 5 |
| E. cloacae 1/10 | E | 2010 | OXA-48, TEM-1, CTX-M-15 | AMP FOX CTX CAZ GEN KAN CMP CIP SXT | 4 | 8 | 6 |
| E. coli J53 | |||||||
| Tc 84/08 | OXA-162, TEM-1 | AMP GEN OTE SXT | 0.25 | 1 | |||
| Tc 131/08 | OXA-48, TEM-1 | AMP GEN CMP | 0.25 | 0.5 | |||
| Tcc | OXA-48 or OXA-162 | AMP | 1 | 1 | |||
| Rcd | ≤0.063 | ≤0.063 | |||||
Tc, transconjugant.
Isolates from the same patient.
Characteristics of transconjugants Tc 215/09, Tc 216/09, Tc 229/09, Tc 238/09, Tc 239/09, Tc 16/10, and Tc 1/10.
Recipient E. coli J53 resistant to sodium azide.
Determined by Etest. AMP, ampicillin; FOX, cefoxitin; CTX, cefotaxime; CAZ, ceftazidime; GEN, gentamicin; KAN, kanamycin; AMK, amikacin; CMP, chloramphenicol; OTE, oxytetracycline, CIP, ciprofloxacin; SXT, sulfamethoxazole-trimethoprim; IPM; imipenem; MPM, meropenem.
PCR and sequencing analysis revealed that the three isolates from hospital A (E. coli, C. freundii, and R. ornithinolytica) harbored the blaOXA-162 gene, whereas the blaOXA-48 gene was detected in E. cloacae isolates and the four K. pneumoniae isolates (Table 1). OXA-162 is a recently identified OXA-48-type variant, differing from OXA-48 by a Thr-to-Ala substitution at position 224 (DBL numbering; 17). Additionally, the blaTEM-1 gene was identified in eight out of the nine isolates, and the blaSHV-11 and blaOXA-9 genes were identified in all of the K. pneumoniae isolates. Furthermore, genes encoding ESBL SHV-5 or CTX-M-15 were found in isolates resistant to ceftazidime and cefotaxime (Table 1). The qnrB1 gene was additionally identified in the E. cloacae isolate.
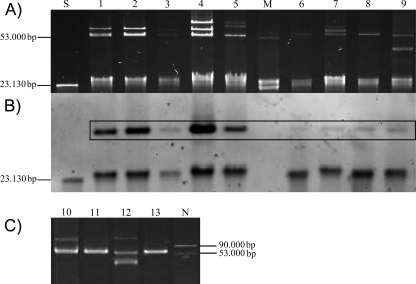
Conjugation assays were successful for all isolates and allowed the identification of blaOXA-162- and blaOXA-48-carrying plasmids with a size of ca. 60 kb in all isolates transferred into E. coli recipients (Fig. 1). No other resistance genes were cotransferred on these plasmids. PCR-based typing targeting genes identified from other blaOXA-48-bearing plasmids as recently described (15) showed that the genes blaOXA-48 and blaOXA-162 identified in the present study corresponded to IncL/M-type plasmids, further reinforcing the hypothesis that the current spread of the blaOXA-48-like genes in different strain backgrounds and different countries is mainly the consequence of the diffusion of an epidemic plasmid. Analysis of the genetic environment located upstream of the blaOXA-48 and blaOXA-162 genes (1, 2) revealed the presence of insertion sequence IS1999 in the four K. pneumoniae isolates, although IS1999 was truncated by insertion sequence IS1R in all of the other isolates, as described previously (2).
Fig 1.
Plasmid preparations from OXA-type carbapenemase-producing clinical strains and transconjugants (Tc). (A) Native plasmid preparation of clinical strains and transconjugants in agarose gel. (B) Southern hybridization of plasmids of clinical strains and transconjugants on nylon membrane with a DIG-labeled blaOXA-48 probe. (C) Native plasmid preparation of clinical strains isolated in 2010 and transconjugants in agarose gel. Lanes: M, plasmid marker E. coli K12J53 V517 (53,000-bp plasmid); N, plasmid marker E. coli K12J53 V517 plus E. coli K12J53 R222 (53,000-bp and 90,000-bp plasmids); S, DIG-labeled Molecular Weight Marker II (Roche Diagnostics Ltd., West Sussex, United Kingdom); 1, E. coli 131/08; 2, Tc 131/08; 3, E. coli 84/08; 4, Tc 84/08; 5, R. ornithinolytica 215/09; 6, Tc 215/09; 7, C. freundii 216/09; 8, Tc 216/09; 9, K. pneumoniae 229/09; 10, E. cloacae 1/10; 11, Tc 1/10; 12, K. pneumoniae 16/10; 13, Tc 16/10. Positive hybridization signals are framed. Hybridization signals of less than 50 kb result from plasmid residues and linear plasmid DNA, respectively.
The antibiotic resistance patterns and β-lactamase contents of the four K. pneumoniae isolates recovered from two different hospitals were identical. Additional sequencing of outer membrane protein genes ompK35 and ompK36, performed as described previously (11), revealed the disruption of ompK36 by an insertion sequence in all four K. pneumoniae, resulting in porin loss and increased carbapenem MICs, as previously described (11). The higher carbapenem MICs observed for the E. coli and E. cloacae clinical isolates than those for the respective transconjugants may likely be attributable to permeability defects in the clinical isolates, related to porin loss or efflux mechanisms. By PFGE typing, identical restriction patterns were observed for all four isolates, indicating the clonal spread of a multidrug-resistant K. pneumoniae strain. No links among the four patients from the two hospitals located 40 km apart could be evidenced.
The present study showed the emergence of OXA-48 and OXA-162 producers among enterobacterial isolates in Germany. Although the spread of OXA-48 producers has been recently identified in different countries in the Mediterranean area and western Europe (2, 8), it is noteworthy that Turkey represents a main reservoir. Considering the high frequency of population exchanges between Germany and Turkey, we speculate that at least some of the isolates currently emerging in Germany could be from Turkey. We identified the novel OXA-162 enzyme, which is a point mutant derivative of OXA-48 and has also been identified recently in Turkey according to the GenBank database (accession numbers HM015773 and GU197550). Identification of the same blaOXA-162-carrying plasmid in R. ornithinolytica and C. freundii isolated from one patient may have resulted from horizontal gene transfer. We further detected loss of the porin OmpK36 in K. pneumoniae as a combined mechanism of carbapenem resistance, as identified in K. pneumoniae 11978 (7, 16).
Here we identified carbapenemases OXA-48 and OXA-162 in different multidrug-resistant Enterobacteriaceae species that coproduce ESBLs and other plasmid-mediated resistance determinants like Qnr. We observed the dissemination of blaOXA-48-like genes by conjugative plasmid transfer, as well as the regional spread of a multidrug-resistant, OXA-48-producing K. pneumoniae clone. Because of limited therapeutic options and higher mortality caused by these carbapenem-resistant Enterobacteriaceae, continuous surveillance and molecular characterization of OXA-48 producers are needed to shed light upon all of the transmission pathways in Germany and over continents. Taking into account the relationships between Germany and many countries located in North Africa and the Middle East, this study underlines the need to detect OXA-48 producers as early as possible.
ACKNOWLEDGMENTS
We thank George A. Jacoby for providing the E. coli J53 recipient strain and Lina Cavaco and Beatriz Guerra for providing Qnr control strains. We extend special thanks to Sybille Müller-Bertling for performing phenotypic and genotypic analyses.
This work was funded by the Ministry of Health, Germany, and the INSERM U914, France.
Footnotes
Published ahead of print 30 January 2012
REFERENCES
- 1. Aubert D, Naas T, Héritier C, Poirel L, Nordmann P. 2006. Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of beta-lactam resistance genes. J. Bacteriol. 188:6506–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carrër A, et al. 2010. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob. Agents Chemother. 54:1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing. CLSI M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 4. Clowes RC, Rowley D. 1954. Some observations on linkage effects in genetic recombination in Escherichia coli K-12. J. Gen. Microbiol. 11:250–260 [DOI] [PubMed] [Google Scholar]
- 5. Goren MG, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2011. Plasmid-encoded OXA-48 carbapenemase in Escherichia coli from Israel. J. Antimicrob. Chemother. 66:672–673 [DOI] [PubMed] [Google Scholar]
- 6. Gröbner S, et al. 2009. Emergence of carbapenem-non-susceptible extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates at the university hospital of Tübingen, Germany. J. Med. Microbiol. 58:912–922 [DOI] [PubMed] [Google Scholar]
- 7. Gülmez D, et al. 2008. Carbapenem-resistant Escherichia coli and Klebsiella pneumoniae isolates from Turkey with OXA-48-like carbapenemases and outer membrane protein loss. Int. J. Antimicrob. Agents 31:523–526 [DOI] [PubMed] [Google Scholar]
- 8. Kalpoe JS, Al Naiemi N, Poirel L, Nordmann P. 2011. Detection of an Ambler class D OXA-48-type β-lactamase in a Klebsiella pneumoniae strain in The Netherlands. J. Med. Microbiol. 60:677–678 [DOI] [PubMed] [Google Scholar]
- 9. Ktari S, et al. 2011. Spread of Klebsiella pneumoniae isolates producing OXA-48 β-lactamase in a Tunisian university hospital. J. Antimicrob. Chemother. 66:1644–1646 [DOI] [PubMed] [Google Scholar]
- 10. Lascols C, et al. 2011. Increasing prevalence and dissemination of NDM-1 metallo-{beta}-lactamase in India: data from the SMART study (2009). J. Antimicrob. Chemother. 66:1992–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee CH, et al. 2007. Collateral damage of flomoxef therapy: in vivo development of porin deficiency and acquisition of blaDHA-1 leading to ertapenem resistance in a clinical isolate of Klebsiella pneumoniae producing CTX-M-3 and SHV-5 beta-lactamases. J. Antimicrob. Chemother. 60:410–413 [DOI] [PubMed] [Google Scholar]
- 12. Moquet O, et al. 2011. Class D OXA-48 carbapenemase in multidrug-resistant enterobacteria, Senegal. Emerg. Infect. Dis. 17:143–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pfeifer Y, Matten J, Rabsch W. 2009. Salmonella enterica serovar Typhi with CTX-M β-lactamase, Germany. Emerg. Infect. Dis. 15:1533–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pfeifer Y, et al. 2011. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother. 66:1998–2001 [DOI] [PubMed] [Google Scholar]
- 15. Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 56:559–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poirel L, Héritier C, Tolün V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poirel L, Naas T, Nordmann P. 2010. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob. Agents Chemother. 54:24–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poirel L, et al. 2011. Cross-border transmission of OXA-48-producing Enterobacter cloacae from Morocco to France. J. Antimicrob. Chemother. 66:1181–1182 [DOI] [PubMed] [Google Scholar]
- 19. Walsh TR. 2010. Emerging carbapenemases: a global perspective. Int. J. Antimicrob. Agents 36(Suppl. 3):S8–S14 [DOI] [PubMed] [Google Scholar]