Abstract
During a β-lactam resistance surveillance study, 12 IMP-18-positive Pseudomonas aeruginosa isolates belonging to 9 different pulsed-field gel electrophoresis groups were identified. In nine isolates, a class I integron with a novel gene array was identified that contained blaIMP-18 and blaOXA-224, while in two isolates the class I integron contained blaIMP-18 and blaOXA-2 but in a new arrangement. Our findings show the dissemination of two novel class I integrons in P. aeruginosa from different regions of Puerto Rico.
TEXT
Pseudomonas aeruginosa is an important nosocomial pathogen associated with high morbidity and mortality. Its treatment is, at times, very difficult because of the emergence of antibiotic-resistant isolates, for which the carbapenems are the treatment of choice. The metallo-β-lactamases (MBLs) are an important medical problem, because they can hydrolyze most β-lactam antibiotics, including the carbapenems (2, 13, 18). The MBLs include nine families with their respective variants: IMP, VIM, NDM, SPM-1, GIM-1, SIM-1, AIM-1, KHM-1 and DIM-1 (18, http://www.lahey.org). Although the IMP-type enzymes are more commonly identified in Asia, they have been reported sporadically worldwide (2, 18). The blaIMP genes have been identified mainly in Enterobacteriaceae, P. aeruginosa, and Acinetobacter spp. and are frequently located on class I integrons, which can facilitate their horizontal spread (2, 18). In a recent review of IMP-type MBLs, 59 different class I integrons harboring various IMP gene cassettes were identified (18). The IMP-18 variant was first detected in the continental United States, followed by Mexico, Puerto Rico, and France (3, 6, 7, 14, 15). The objectives of this study were to characterize 12 IMP-18-positive P. aeruginosa isolates and to determine the genetic background of the blaIMP-18 isolates collected during a 6-month PCR-based surveillance study of β-lactam resistance in 17 hospitals from six different geographical regions of Puerto Rico (12).
Twelve of 272 multi-β-lactam-resistant P. aeruginosa isolates were identified as blaIMP positive by PCR. The patients' basic epidemiological information and the susceptibility of the isolates to selected antibiotics, based on microdilution panels (TREK Diagnostic Systems), are shown in Table 1. Seven of the 12 isolates were obtained from a single hospital located in the Puerto Rico Medical Center. No blaIMP-18-positive isolates were detected in the north and west regions. Only colistin demonstrated consistent antimicrobial activity (83%) against the isolates.
Table 1.
Baseline clinical information and antimicrobial susceptibilities to selected antibiotics of IMP-positive P. aeruginosa isolates
| P. aeruginosa isolate | Geographical regiona | Age (yrs) | Genderb | Anatomical site of collectionc | Hospital unitd | Test agente MIC (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FEP | ATM | TZP | IPM | MEM | DORf | CST | ||||||
| M1PA9-12 | San Juan metropolitan | 73 | M | RT | ICU | >16 | >16 | 34/4 | >8 | >8 | >2 | 2 |
| M1PA9-15 | San Juan metropolitan | 66 | M | UT | Gen Ward | >16 | 8 | 16/4 | >8 | >8 | >2 | 2 |
| S1PA9-25 | South | 0.08 | M | RT | ICU | >16 | <2 | 8/4 | >8 | <1 | 2 | 1 |
| C1PA9-07 | Central | 56 | M | UT | ICU | >16 | >16 | <8/4 | >8 | <1 | 2 | 4 |
| E1PA9-8 | East | 92 | M | RT | ICU | >16 | >16 | 32/4 | >8 | >8 | >2 | 1 |
| MC1PA9-13 | PRMC | 27 | F | RT | Gen Ward | >16 | 4 | 16/4 | >8 | >8 | >2 | 4 |
| MC1PA9-25 | PRMC | 63 | M | UT | Gen Ward | >16 | >16 | >64/4 | >8 | >8 | >2 | 2 |
| MC1PA9-33 | PRMC | 58 | M | Blood | Gen Ward | >16 | 16 | >64/4 | >8 | >8 | >2 | 2 |
| MC1PA9-34 | PRMC | 60 | F | Blood | ICU | >16 | 4 | 8/4 | >8 | >8 | >2 | 2 |
| MC1PA9-37 | PRMC | 65 | M | UT | Gen Ward | >16 | 16 | >64/4 | >8 | >8 | >2 | 2 |
| MC1PA9-38 | PRMC | 41 | M | Blood | Gen Ward | >16 | 16 | 64/4 | >8 | >8 | >2 | 2 |
| MC7PA9-5 | PRMC | 89 | M | UT | Gen Ward | >16 | 8 | 64/4 | >8 | >8 | >2 | 2 |
PRMC, Puerto Rico Medical Center.
M, male; F, female.
RT, respiratory tract; UT, urinary tract.
ICU, intensive care unit; Gen Ward, general ward.
Test agents and their Clinical and Laboratory Standards Institute (CLSI) susceptibility breakpoints (in μg/ml): FEP, cefepime (≤8); ATM, aztreonam (≤8); TZP, piperacillin-tazobactam (≥64/4); IPM, imipenem (≤4); MEM, meropenem (≤4); DOR, doripenem (≤2); CST, colistin (≤2).
The doripenem breakpoint value was obtained from the Doribax package insert.
All organisms were screened by PCR using panels of family-specific β-lactamase primers for the detection of the following genes: KPC, IMP and VIM, TEM, SHV, OXA-1, -2, and -9, CTX-M extended-spectrum β-lactamases, and OXA carbapenemases. The bacterial DNA template, primers used, and the PCR conditions were as previously described (6, 8, 9, 15, 16, 17). Eleven isolates demonstrated additional β-lactamases: blaOXA-1 in 9, blaTEM in 7, and blaOXA-2 in two isolates. After DNA sequencing of the PCR amplicon obtained with the primers for blaOXA-1, the results obtained indicated that the amplicon differed from blaOXA-1 by 3 amino acid substitutions, making it a new variant, named blaoxa-224. No additional β-lactamase genes were identified in one isolate (S1PA9-25).
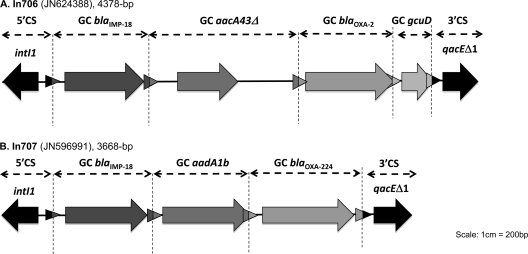
The structure of the variable region of the class I integron containing the IMP-18 gene cassette was determined as previously described by Sánchez-Martínez et al. (14). All PCR amplicons generated were sequenced independently and bidirectionally at least twice. Sequence alignment and analysis were performed online using the BLAST program (www.ncbi.nlm.nih.gov). Two novel class I integrons with the following structures were identified: In706 (blaIMP-18-aadA43Δ-blaOXA-2-gcuD) in two isolates and In707 (blaIMP-18-aadA1b-blaOXA-224) in nine isolates (Fig. 1). The two new integron sequences were submitted to the INTEGRALL database (10; http://integrall.bio.ua.pt/) for integron (In) number assignment and to the GenBank database (see below).
Fig 1.
Schematic representation of two novel class I integrons containing blaIMP-18. Two arrangements of the class 1 integron containing the blaIMP-18 gene were identified by a PCR mapping strategy described by Sánchez-Martínez et al. (14). In both class I integrons, the blaIMP-18 gene is located in the first position next to the 5′ conserved region. (A) In706, the disrupted aacA43Δ gene cassette (GC) was identified at the second position. blaOXA-2 and gcuD of a hypothetical orfD gene of unknown function were identified in the third and fourth positions, respectively. (B) The diagram of In707 shows the blaIMP-18 gene followed by aadA1b and blaOXA-224. Open reading frames (ORFs) attl1 and attC are indicated by arrows with different gray colors. Scale: 1 cm = 200 bp. Illustration courtesy of the INTEGRALL database (http://integrall.bio.ua.pt/).
The sizes of the class I integrons sequenced were 4,378 bp for In706 and 3,668 bp for In707, and their structures were compared to the three previously described P. aeruginosa-containing blaIMP-18 integrons isolated in Mexico (In96 and In169) and the United States (In133) (1, 3, 14). The location of blaIMP-18 in the first position in both integrons is similar to In96 and In169, but different from In133. The blaIMP-18 has been previously associated with blaOXA-2 in the In169 class I integron identified in Mexico, which harbors two copies of the aadA1 gene cassette in the second and fourth positions with the blaOXA-2 gene between them (14). In In706, the blaOXA-2 gene was also identified in the third position as in In169, but with other gene cassettes. At the second position, a variant of the aacA43 gene cassette (GenBank accession number HQ247816) was identified that differed by two amino acids (R60Q and R95K) and 6 nucleotide substitutions. Since it is not known if this variant is functional, the gene cassette was identified as aacA43Δ. A hypothetical orfD gene of unknown function, gcuD, located in the fourth position was previously found associated with a blaIMP-15 variant in a class I integron obtained from two P. aeruginosa isolates from Mexico (GenBank accession numbers GQ856540 and GQ856541) (11, 18). In all cases, gcuD was located near the 3′ conserved sequence (18). To our knowledge, this is the first time that the blaIMP-18 variant has been found associated with blaOXA-224 in a class I integron (In707). blaOXA-224 was recently identified in a class I integron (In662; GenBank accession number JN412067) but was not associated with an MBL gene (10; http://integrall.bio.ua.pt/). In isolate S1PA9-25, blaIMP-18 was located in the first position near the 5′ conserved region; however, the rest of the gene cassette(s), including the 3′ conserved sequence, was not detected.
Pulsed-field gel electrophoresis (PFGE) was performed as previously described (4, 5). A total of 9 distinct PFGE patterns, 2 related and 6 genetically unrelated groups, were identified in the 12 isolates (Fig. 2). For In707, the class I integron was found in 9 isolates (groups 1 to 6), and for In706 it was found in two (groups 8 and 9). Seven of the 12 isolates, harboring either In706 or In707, were obtained from a single Puerto Rico Medical Center hospital and belonged to one related and four unrelated PFGE groups. The presence of a blaIMP-18 class I integron in genetically related and unrelated isolates, either in a single institution or in several geographically unrelated hospitals, suggests that both clonal and horizontal transfer are contributing to its dissemination. However, Southern blot assays and PCR performed on genomic and plasmid DNA with and without treatment with plasmid-safe DNase (Epicentre) to determine the integrons' locations gave inconclusive results (data not shown).
Fig 2.
Genetic relatedness determinations by pulsed-field gel electrophoresis. The In706 class I integron was detected in P. aeruginosa isolates with PFGE groups patterns 8 and 9 from a single hospital, while In707 was identified in groups 1 to 6 from either a single or several hospitals.
The finding of two new class I integrons containing blaIMP-18 are in agreement with previous reports that emphasized the plasticity and multiple arrangement of the class I metallo-β-lactamase integrons (1, 18).
Accession numbers.
The PCR amplicon obtained with the primers for blaOXA-1 indicated that the amplicon differed from blaOXA-1 by 3 amino acid substitutions, making it a new variant, named blaoxa-224. Its sequence was submitted to GenBank and assigned accession number JN596991. The two new integron sequences were submitted to the INTEGRALL database for integron number assignment and to the GenBank database, and the accession numbers JN624388 (In706) and JN596991 (In707) were assigned.
ACKNOWLEDGMENTS
This work was partially funded by Ortho-McNeil Janssen (Johnson & Johnson), Merck Sharp and Dohme, Pfizer Caribbean, the PR Health Department, and NCRR/NIH-RCMI award G12RR03051.
We acknowledge the following members of the Puerto Rico Antibiotic Resistance Study Group: Miguel Colón, Osvaldo Laboy, Carlos F León, Agripino Lugo, Vanessa Olivo, Diana M. Otero, Ramón Ramírez Ronda, Jorge L. Santana, María I. Santé, and Nilda Zapata. We thank the participating hospital bacteriology laboratories personnel for collecting the isolates and epidemiological information, in particular Carmen Báez, Myriam Corazón, Madeline Cruz, Leyda E. Echevarría, Maria Maldonado, Aixa Martínez, María Matos, Miriam Nistal, Nereida Santiago, Linnette Santos, Abigail Torres, and Nayda Vázquez. We are grateful for the support of Ada M. Cortez, Enid J. García, and Johnny Rullán from the PR Department of Health. We appreciate the dedicated technical assistance of Caleb Fernández and the undergraduate students of our laboratory. We also thank Wieslaw J. Kozek for reviewing the manuscript.
Footnotes
Published ahead of print 30 January 2012
REFERENCES
- 1. Borgianni L, et al. 2011. Genetic context and biochemical characterization of the IMP-18 metallo-beta-lactamase identified in a Pseudomonas aeruginosa isolate from the United States. Antimicrob. Agents Chemother. 55:140–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-beta-lactamases: a last frontier for beta-lactams? Lancet Infect. Dis. 11:381–393 [DOI] [PubMed] [Google Scholar]
- 3. Garza-Ramos U, et al. 2008. Metallo-beta-lactamase IMP-18 is located in a class 1 integron (In96) in a clinical isolate of Pseudomonas aeruginosa from Mexico. Int. J. Antimicrob. Agents 31:78–80 [DOI] [PubMed] [Google Scholar]
- 4. Goering RV. 2004. Pulsed-field gel electrophoresis, p 185–196 In Persing DH, et al. (ed), Molecular microbiology: diagnostic principles and practice. ASM Press, Washington, DC [Google Scholar]
- 5. Goering RV. 2010. Pulsed field gel electrophoresis: a review of application and interpretation in the molecular epidemiology of infectious disease. Infect. Genet. Evol. 10:866–875 [DOI] [PubMed] [Google Scholar]
- 6. Hanson ND, Hossain A, Buck L, Moland ES, Thomson KS. 2006. First occurrence of a Pseudomonas aeruginosa isolate in the United States producing an IMP metallo-beta-lactamase, IMP-18. Antimicrob. Agents Chemother. 50:2272–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hocquet D, et al. 2010. Nationwide investigation of extended-spectrum beta-lactamases, metallo-beta-lactamases, and extended-spectrum oxacillinases produced by ceftazidime-resistant Pseudomonas aeruginosa strains in France. Antimicrob. Agents Chemother. 54:3512–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moland ES, et al. 2002. Occurrence of newer beta-lactamases in Klebsiella pneumoniae isolates from 24 U.S. hospitals. Antimicrob. Agents Chemother. 46:3837–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moland ES, et al. 2006. Prevalence of newer beta-lactamases in gram-negative clinical isolates collected in the United States from 2001 to 2002. J. Clin. Microbiol. 44:3318–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moura A, et al. 2009. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25:1096–1098 [DOI] [PubMed] [Google Scholar]
- 11. Partridge SR, Tsafnat G, Coiera E, Iredell JR. 2009. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33:757–784 [DOI] [PubMed] [Google Scholar]
- 12. Robledo IE, Aquino EE, Vazquez GJ. 2011. Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob. Agents Chemother. 55:2968–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sacha P, et al. 2008. Metallo-beta-lactamases of Pseudomonas aeruginosa: a novel mechanism resistance to beta-lactam antibiotics. Folia Histochem. Cytobiol. 46:137–142 [DOI] [PubMed] [Google Scholar]
- 14. Sanchez-Martinez G, et al. 2010. In169, a new class 1 integron that encoded bla(IMP-18) in a multidrug-resistant Pseudomonas aeruginosa isolate from Mexico. Arch. Med. Res. 41:235–239 [DOI] [PubMed] [Google Scholar]
- 15. Wolter DJ, et al. 2009. Surveillance of carbapenem-resistant Pseudomonas aeruginosa isolates from Puerto Rican Medical Center Hospitals: dissemination of KPC and IMP-18 beta-lactamases. Antimicrob. Agents Chemother. 53:1660–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Woodford N, et al. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351–353 [DOI] [PubMed] [Google Scholar]
- 17. Woodford N, Fagan EJ, Ellington MJ. 2006. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J. Antimicrob. Chemother. 57:154–155 [DOI] [PubMed] [Google Scholar]
- 18. Zhao WH, Hu ZQ. 2011. IMP-type metallo-beta-lactamases in Gram-negative bacilli: distribution, phylogeny, and association with integrons. Crit. Rev. Microbiol. 37:214–226 [DOI] [PubMed] [Google Scholar]