Abstract
We performed a genomewide screen for altered sensitivity to antifungal drugs, including clotrimazole and terbinafine, that target ergosterol biosynthesis using a Schizosaccharomyces pombe gene deletion library consisting of 3,004 nonessential haploid deletion mutants. We identified 109 mutants that were hypersensitive and 11 mutants that were resistant to these antifungals. Proteins whose absence rendered cells sensitive to these antifungals were classified into various functional categories, including ergosterol biosynthesis, membrane trafficking, histone acetylation and deacetylation, ubiquitination, signal transduction, ribosome biosynthesis and assembly, regulation of transcription and translation, cell wall organization and biogenesis, mitochondrion function, amino acid metabolism, nucleic acid metabolism, lipid metabolism, meiosis, and other functions. Also, proteins whose absence rendered cells resistant to these antifungals were classified into functional categories including mitochondrion function, ubiquitination, membrane trafficking, cell polarity, chromatin remodeling, and some unknown functions. Furthermore, the 109 sensitive mutants were tested for sensitivity to micafungin, another antifungal drug that inhibits (1,3)-β-d-glucan synthase, and 57 hypersensitive mutants were identified, suggesting that these mutants were defective in cell wall integrity. Altogether, our findings in fission yeast have shed light on molecular pathways associated with the cellular response to ergosterol biosynthesis inhibitors and may provide useful information for developing strategies aimed at sensitizing cells to these drugs.
INTRODUCTION
Fungal diseases, especially opportunistic fungal infections, have increased dramatically over the past 2 decades as a result of the growing population of patients with compromised immune systems due to chemotherapy, the transplantation of solid organs or hematopoietic stem cells, infection with HIV, aggressive treatments of cancer, and autoimmune disorders (16, 35, 40). To date, invasive fungal infections are usually treated with drugs that interfere with the biosynthesis or integrity of ergosterol, which is an essential component of fungal plasma membranes. In fact, three of the five classes of antifungal drugs in clinical use are ergosterol biosynthesis inhibitors, such as azoles and terbinafine. However, certain factors, including host toxicity or the emergence of drug resistance, compromise the efficacy of treatment (2, 7, 10, 29).
We have been studying the ergosterol biosynthesis pathway in the fission yeast Schizosaccharomyces pombe, because this system is amenable to genetic analysis and has many advantages in terms of its similarity to some pathogenic fungi. We previously identified a mutation in the essential gene hmg1+, encoding the sterol biosynthetic enzyme 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase (HMGR), which catalyzes the conversion of HMG-CoA to mevalonate and constitutes the rate-limiting step in the biosynthesis of ergosterol (17).
In order to improve the treatment of fungal disease, it is necessary to identify novel targets by which the inhibitors of these targets act in synergism with existing antifungals, and a combination therapy with existing antifungals would convert the fungistatic antifungal effect into potent fungicidal activities. In this study, we report the first-ever genomewide screen of S. pombe haploid deletion mutants for genes affecting the sensitivity or resistance of antifungal drugs that target ergosterol biosynthesis, including clotrimazole and terbinafine. The screening may lead to the uncovering of novel targets for antifungal drugs that would enhance the efficacy of existing antifungal drugs as well as minimize the probability of the emergence of drug resistance.
MATERIALS AND METHODS
Yeast stains and media.
A genomewide deletion mutant library purchased from BiONEER (South Korea) was employed for the genetic screen. The viable haploid deletions used in this screen were generated by using a method for PCR-based targeted gene deletion with a genetic background of h+ leu1-32 ura4-D18 ade6-M210 or -M216 (25). The haploid deletion library used in this study consists of 3,004 mutants that are estimated to be nonessential for viability in the haploid and represents around 80% of the nonessential genes in S. pombe.
Cells were grown in rich yeast extract with supplements (YES) at 27°C unless otherwise indicated (31). Adenine, histidine, leucine, uracil, and lysine were added at 225 mg/liter. Gene disruptions are abbreviated by the gene preceded by Δ (for example, Δspo9). Proteins are denoted by roman letters, and only the first letter is capitalized (for example, Spo9).
Deletion library screens for altered sensitivity of antifungal drugs.
In this screen, ergosterol biosynthesis inhibitors, namely, clotrimazole and terbinafine, were used. Clotrimazole, a member of the azoles, has the same mechanism of action as that of other azoles that inhibit lanosterol 14-α-demethylase (Erg11), an essential enzyme in the ergosterol biosynthetic pathway (34), and it showed the most potent and stable inhibitory effects on fission yeast cell growth among the azoles tested (clotrimazole, fluconazole, miconazole, and sulconazole) (data not shown). Terbinafine is in the allylamine class of drugs and targets the enzyme squalene epoxidase (Erg1) (34). Both of these inhibitors are acknowledged as first-line agents for the treatment of fungal disease. The sensitivities and resistances of the yeast cells to these antifungals were determined by three different methods, as described below.
(i) Streak assay.
In the first assay, the growth of each yeast strain was assayed by streaking out to form single colonies on plates containing YES or YES plus various concentrations of antifungals, and the plates were incubated at 27°C for 4 days. The colony size of the wild-type cells gradually increased over approximately 5 to 6 days. For the estimation of drug sensitivity or resistance, the colony sizes were assessed after 4 days. To test for drug hypersensitivity, different concentrations of drugs, ranging from 0.01 to 0.03 μg/ml of terbinafine and 0.05 to 0.15 μg/ml of clotrimazole, were used. To test for drug resistance, different concentrations of drugs, ranging from 0.08 to 0.2 μg/ml of terbinafine and 0.2 to 0.3 μg/ml of clotrimazole, were used.
(ii) Microtiter assay.
The MICs for the strains were determined by broth microdilution according to CLSI guidelines (9), with some modifications. Fission yeast cells with an auxotrophic marker such as ura4 fail to grow at a neutral pH (3), and some deletion mutants show hypersensitivity to high temperatures. Given these characteristics of fission yeast cells in the deletion library, the determination of MICs using RPMI 1640 (pH 7.0) and incubation at 35°C, according to CLSI criteria, may not be suitable. In addition, the inoculum density in each well was increased to ∼106 yeast cells/well because we failed to detect cell growth as turbidity at the low density (2.5 × 103 yeast cells/ml) used in the CLSI methodology. Thus, strains were inoculated into 96-well microtiter plates containing 200 μl of YES medium and different concentrations of drugs to yield ∼106 yeast cells/well. A drug-free control was also included. The concentration range of the drugs in microtiter plate wells was 0.015 μg/ml to 8 μg/ml. The plates were incubated at 27°C and examined by use of a microplate reader after 48 h. MICs were determined as the concentrations of the drugs causing a 50% inhibition of growth. The experiments were done in triplicate.
(iii) Spot assay.
The yeast cells were grown to saturation in YES liquid medium at 27°C. The cultures were then resuspended in fresh YES medium to give an optical density (OD) at 660 nm of 0.3, corresponding to about 107 cells/ml, and serially diluted to concentrations of 1 × 10−1 to 1 × 10−4. Five-microliter samples of 10-fold serial dilutions of each yeast cell culture were spotted onto each plate, as described above for the streak assay, and incubated at 27°C for 4 days. Altered sensitivity was assessed manually by analyzing the number and size of the colonies formed on each plate in relation to the control.
Confirmation of clotrimazole- and terbinafine-sensitive or -resistant strains.
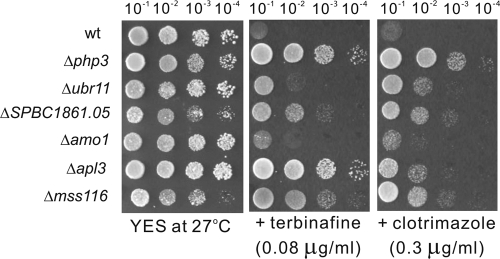
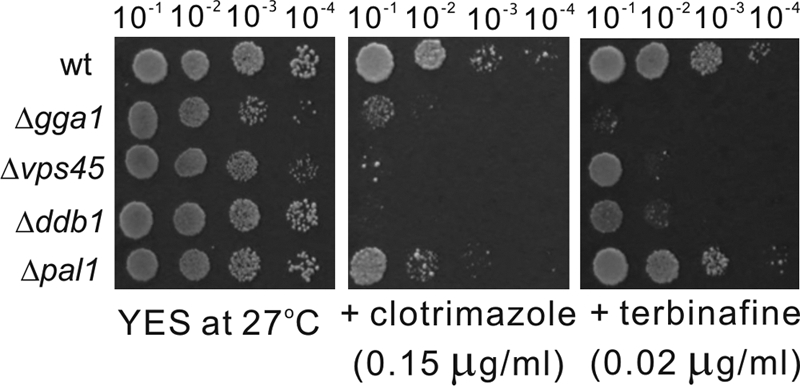
In this study, we used a streak assay for a preliminary screen and a spot assay for a secondary screen, because as a classical experimental approach, they are widely used for the study of yeast cells due to their simplicity and accuracy, respectively. We also used a microtiter assay as an alternative method for detecting the sensitivity of yeast strains to drugs. The results showed that there is a positive correlation between sensitivities detected by the microtiter assay and the spot assay. Thus, mutants with altered sensitivity identified in the preliminary screen were further confirmed by the semiquantitative spot assay (1) and microtiter assay (9), and the sensitivity or resistance of the strains was assessed by the results of the spot assay, as described previously by Kennedy et al. (24), Xia et al. (42), and Alamgir et al. (1). Representative examples of the S. pombe deletion mutants screened for inhibited growth in the presence of clotrimazole or terbinafine are shown in Fig. 1, and representative examples of the S. pombe deletion mutants screened for resistance to these drugs are shown in Fig. 2. Using the wild-type strain as a control, the sensitivity was estimated relative to that of untreated controls, the strains were then classified, and the extent of growth that fell within the corresponding ranges were scored as follows: strongly sensitive (+++), indicating that the mutant cells completely failed to grow on the drug-containing plates, while the wild-type cells grew normally (Fig. 1) (e.g., the clotrimazole-sensitive phenotype of the Δddb1 and Δvps45 strains and the terbinafine-sensitive phenotype of the Δgga1 strain); moderately sensitive (++), indicating that only the first circle on the left of the mutant cells was observed to grow on the drug-containing plates (Fig. 1) (e.g., the terbinafine-sensitive phenotype of the Δddb1 and Δvps45 strains and the clotrimazole-sensitive phenotype of the Δpal strain); and mildly sensitive (+), indicating that colonies were observed to grow on the drug-containing plates but that the numbers of colonies were significantly reduced compared to those of wild-type cells (Fig. 1) (e.g., the terbinafine-sensitive phenotype of the Δpal strain). On the other hand, resistance was also estimated relative to that of untreated controls, the strains were scored, and the extent of growth that fell within the corresponding ranges was scored as follows: strongly resistant (+++), indicating that the mutant cells grew very well under these extreme conditions, while the wild-type cells failed to grow (Fig. 2) (e.g., the terbinafine-resistant phenotype of the Δphp3 and Δapl3 strains); moderately resistant (++), indicating that the first and second circles on the left of the mutant cells grew well (Fig. 2) (e.g., terbinafine-resistant phenotype of the ΔSPBC1861.05 and Δmss116 strains); and mildly resistant (+), indicating that only the first circle on the left of the mutant cells was observed to grow on the drug-containing plates but that the numbers of the colonies were significantly increased compared to those of wild-type cells (Fig. 2) (e.g., the terbinafine-resistant phenotype of the Δubr11 and Δamo1 strains). All of the mutants identified in the screen grew well in YES medium, indicating that the phenotype on clotrimazole- or terbinafine-containing plates is a result of the effect of the drugs (Fig. 1 and 2).
Fig 1.
Representative examples of the S. pombe deletion mutants screened for growth inhibition in the presence of clotrimazole or terbinafine. Wild-type (wt) and deletion mutant cells grown at log phase were spotted onto each plate as indicated and then incubated at 27°C for 4 days.
Fig 2.
Representative examples of the S. pombe deletion mutants screened for resistance to ergosterol biosynthesis inhibitors, namely, clotrimazole and terbinafine. Wild-type (wt) and deletion mutant cells grown at log phase were spotted onto each plate as indicated and then incubated at 27°C for 4 days.
Glucanase sensitivity assay.
Cell wall digestion by β-glucanase (Zymolyase; Seikagakukogyo, Tokyo, Japan) was performed as previously described (8). Briefly, exponentially growing cells at 27°C were suspended at a concentration of 107 cells/ml. The cells were then treated with β-glucanase at a concentration of 100 μg/ml at 27°C. Cell lysis was monitored by measuring the optical density at 660 nm.
Complementation analysis.
Each gene was expressed under the authentic or nmt1 promoter in a multicopy vector. Expression plasmids and vector controls were transformed into the mutant strains by the lithium acetate method and plated onto selective medium. To test for the complementation of the ergosterol biosynthesis inhibitor-sensitive phenotype, transformants were grown to saturation in selective medium, and serial dilutions were spotted onto each plate, as described above for the streak assay, and incubated at 27°C for 4 days.
Determination of ergosterol levels.
Ergosterol extraction was performed essentially as previously described (32, 33), with some modifications. Briefly, cells were grown to saturation in YES at 27°C and washed two times with ultrapure water. One milliliter of chloroform-ethanol (2:1, vol/vol) eluate, including 40 μmol/liter pyrogallol and 10 μmol/liter pyrene as an internal standard, was then added. The sample was mixed for 3 h. Subsequently, the sample was centrifuged at 16,110 × g for 5 min. After centrifugation, the supernatant was evaporated by using a centrifugal vacuum evaporator. The residue was redissolved in 100 μl of methanol.
Ergosterol analysis was carried out with an Alliance high-performance liquid chromatography (HPLC) system (Waters) coupled to an LTQ linear ion trap mass spectrometer with an atmospheric-pressure chemical ionization source (Thermo Scientific). An Inertsil ODS-3 column (with a 30-mm by 2.1-mm internal diameter [i.d.] and a particle size of 2 μm; GL Sciences) was utilized for the separation process. Water and methanol were used as the mobile phases. A 10-μl aliquot was injected onto the column. Analysis was conducted in the positive-ion mode.
Selected reaction monitoring was employed to quantify ergosterol, and selected ion monitoring was employed to detect the internal standard pyrene. The m/z 379.34 ([M + H − H2O]+) ion was selected as the precursor ion of ergosterol, and the m/z 295.24 product ion of ergosterol was monitored. The collision energy was set at 35%. The m/z 203.09 ([M + H]+) ion was monitored to detect pyrene.
The ergosterol content of wild-type cells was taken as 100%, and the ergosterol contents of the other deletion mutants were calculated as a percentage of that of the wild-type cells.
Bioinformatics.
Database searches were performed by using the National Center for Biotechnology Information BLAST network service (www.ncbi.nlm.nih.gov) and the Sanger Center S. pombe database search service (www.sanger.ac.uk).
RESULTS AND DISCUSSION
Identification of clotrimazole- and terbinafine-sensitive S. pombe deletion mutants.
We screened a genomewide library containing 3,004 haploid deletion strains to identify some nonessential genes by which gene deletion caused the yeast cells to become more sensitive than wild-type strains to the antifungal agents that are inhibitors of ergosterol biosynthesis, such as clotrimazole and terbinafine. In this screening, we isolated 109 deletion strains that displayed various levels of sensitivity to the antifungals clotrimazole and terbinafine, and as shown in Table 1, for each gene deletion mutant listed, the systematic name, common gene name (if applicable), as well as a brief description of each gene product (obtained from the Sanger Center website [www.sanger.ac.uk]) are indicated. In cases where the common name of the S. pombe mutant gene is not applicable, for convenience, we named the genes according to their Saccharomyces cerevisiae counterparts. With regard to the effect of clotrimazole, as shown in Table 1, 54 mutants exhibited severe sensitivity, 31 mutants exhibited moderate sensitivity, and 20 mutants exhibited mild sensitivity. Regarding the effect of terbinafine, 5 mutants were classified as being strongly sensitive, 27 mutants were moderately sensitive, and 69 mutants were mildly sensitive. Among the 109 mutants, almost 89% of the mutants showed hypersensitivity to the two inhibitors of ergosterol biosynthesis, suggesting that these genes are involved in various biological processes influenced by an ergosterol deficiency. Nine mutants, namely, gene deletion mutants of dap1+, ada3+, git1+, rnc1+, aah3+, aru1+, aro3+ (SPAC24H6.10c), kti12+ (SPAC30.02c), and irc6+ (SPAC19A8.11c), exhibited hypersensitivity only to clotrimazole and not to terbinafine. On the other hand, gene deletion mutants of gos1+, trt1+, mrs3+(SPAC4G8.08), and SPCC126.13c exhibited hypersensitivity only to terbinafine and not to clotrimazole. It was reported previously that azoles inhibit Erg11 while terbinafine inhibits Erg1 in the ergosterol biosynthesis pathway (34). Our results suggest that the inhibition of Erg11 as well as the inhibition of Erg1 have different outcomes and that intermediate products such as lanosterol may have a distinct physiological function. Alternatively, clotrimazole and terbinafine may affect targets other than their primary targets in the ergosterol biosynthesis pathway. For mammalian cells, it was reported previously that clotrimazole inhibits numerous transport proteins of cellular membranes, such as Na,K-ATPase (5) and gastric H,K-ATPase (41).
Table 1.
S. pombe genes identified in the screen for clotrimazole- and terbinafine-sensitive genesf
| Category and systematic gene name | Common gene name | Gene descriptiona | Spot assay result with: |
MIC (μg/ml)e |
||
|---|---|---|---|---|---|---|
| Clot | Terb | Clot | Terb | |||
| Wild type | 2.0 | 0.25 | ||||
| Ergosterol synthesis | ||||||
| SPAC20G4.07c | sts1 | C-24(28) sterol reductase Sts1 | +++ | +++ | 0.06 | 0.06 |
| SPBC36.06c | spo9 | Farnesyl pyrophosphate synthetase | ++ | +++ | 1.0 | 0.03 |
| SPBC1271.12 | kes1 | Oxysterol binding protein | +++ | +++ | 0.25 | 0.12 |
| SPAC25B8.01 | dap1 | Cytochrome P450 regulator Dap1 | ++ | − | 1.0 | 0.25 |
| Membrane trafficking | ||||||
| SPAC144.06 | apl5 | AP-3 adaptor complex subunit Apl5 | +++ | + | 0.25 | 0.12 |
| SPAC23H3.06 | apl6 | AP-3 adaptor complex subunit Apl6 | +++ | + | 0.25 | 0.12 |
| SPAC30D11.05 | aps3 | AP-3 adaptor complex subunit Aps3 | +++ | + | 0.5 | 0.25 |
| SPBC651.11c | apm3 | AP-3 adaptor complex subunit Apm3 | + | + | 1.0 | 0.25 |
| SPBP16F5.07 | apm1 | AP-1 adaptor complex subunit Apm1 | +++ | + | 0.06 | 0.12 |
| SPAC767.01c | vps1 | Dynamin family protein Vps1 | +++ | + | 0.06 | 0.12 |
| SPAC2G11.03c | vps45 | Vacuolar sorting protein Vps45 | +++ | ++ | 0.12 | 0.015 |
| SPAC1142.07c | vps32 | Vacuolar sorting protein Vps32 | ++ | + | 0.12 | 0.12 |
| SPBC27B12.08 | sip1 | Pof6 interaction protein Sip1 | ++ | + | 0.12 | 0.25 |
| SPAC31A2.13c | sft1c | SNARE Sft1 | +++ | + | 0.12 | 0.06 |
| SPAC1527.02 | sft2 | Golgi transport protein Sft2 | + | ++ | 0.12 | 0.03 |
| SPAC4G8.10 | gos1 | SNARE Gos1 | − | + | 0.5 | 0.12 |
| SPAC869.11 | cat1 | Cationic amino acid transporter Cat1 | +++ | ++ | 1.0 | 0.12 |
| SPCC1795.03 | gms1 | UDP-galactose transporter Gms1 | ++ | + | 0.25 | 0.06 |
| SPAC23C11.14 | zhf1 | Zinc ion transporter Zhf1 | +++ | + | 0.25 | 0.06 |
| SPBC25H2.16c | gga1b | Adaptin | ++ | +++ | 0.25 | 0.03 |
| SPCC794.11c | ent3b | ENTH/VHS domain protein Ent3 | ++ | + | 0.25 | 0.06 |
| SPCC794.03 | uga4b | Amino acid permease | +++ | + | 0.25 | 0.12 |
| SPBC725.10 | NA | TspO homolog | +++ | + | 0.06 | 0.06 |
| Histone acetylation and deacetylation | ||||||
| SPCC24B10.08c | ada2 | SAGA complex subunit Ada2 | +++ | + | 0.06 | 0.12 |
| SPBC28F2.10c | ada3c | SAGA complex subunit Ngg1 | +++ | − | 0.25 | 0.25 |
| SPBC14C8.17c | spt8c | SAGA complex subunit Spt8 | ++ | + | 0.25 | 0.12 |
| SPBC146.09c | lsd1 | Histone demethylase SWIRM1 | ++ | ++ | 0.25 | 0.06 |
| SPAC16C9.05 | cph1 | Clr6 histone deacetylase-associated PHD protein 1 (Cph1) | ++ | ++ | 0.12 | 0.12 |
| SPCC126.13c | NA | Histone deacetylase complex subunit | − | + | 0.25 | 0.06 |
| Ubiquitination | ||||||
| SPAC11G7.02 | pub1 | Ubiquitin-protein ligase E3 | +++ | + | 0.06 | 0.03 |
| SPBC21D10.09c | rkr1b | Ubiquitin-protein ligase E3 | +++ | + | 0.12 | 0.03 |
| SPBC14F5.10c | NA | Ubiquitin-protein ligase E3 | +++ | ++ | 0.5 | 0.06 |
| SPAC13A11.04c | ubp8 | Ubiquitin C-terminal hydrolase | ++ | + | 0.12 | 0.12 |
| SPCC1223.01 | NA | Ubiquitin-protein ligase E3 | +++ | + | 0.12 | 0.06 |
| SPAC17H9.19c | cdt2 | WD repeat protein Cdt2 | ++ | ++ | 0.25 | 0.03 |
| SPAC1687.05 | pli1 | SUMO E3 ligase Pli1 | + | + | 0.5 | 0.12 |
| SPCC970.10c | brl2 | Ubiquitin-protein ligase E3 | ++ | ++ | 0.03 | 0.03 |
| SPAC17H9.10c | ddb1 | Damaged DNA binding protein | +++ | ++ | 0.25 | 0.03 |
| Signal transduction | ||||||
| SPCC1753.02c | git3c | G-protein-coupled receptor Git3 | +++ | + | 0.12 | 0.06 |
| SPAC23H3.13c | gpa2 | Heterotrimeric G protein α-2 subunit | +++ | +++ | 0.12 | 0.06 |
| SPBC32H8.07 | git5 | Heterotrimeric G protein b subunit | +++ | + | 0.25 | 0.06 |
| SPBC215.04 | git11 | Heterotrimeric G protein γ subunit | ++ | + | 0.25 | 0.06 |
| SPBC21C3.20c | git1 | C2 domain protein Git1 | +++ | − | 0.12 | 0.06 |
| SPCC162.10 | ppk33 | Serine/threonine protein kinase | ++ | ++ | 0.25 | 0.06 |
| SPBC106.10 | pka1 | cAMP-dependent protein kinase catalytic subunit Pka1 | +++ | ++ | 0.12 | 0.03 |
| SPAC6F6.01 | cch1 | Calcium channel Cch1 | ++ | ++ | 0.06 | 0.03 |
| SPAC16.01 | rho2 | Rho family GTPase Rho2 | ++ | ++ | 0.25 | 0.06 |
| SPBC15D4.15 | pho2 | 4-Nitrophenylphosphatase | +++ | + | 1.0 | 0.12 |
| SPAC4F10.04 | rrd1b | Protein phosphatase type 2A, intrinsic regulator | ++ | + | 0.12 | 0.03 |
| SPCC757.09c | rnc1 | RNA binding protein that suppresses calcineurin deletion Rnc1 | + | − | 0.25 | 0.25 |
| SPCC297.05c | NA | Diacylglycerol binding protein | +++ | ++ | 0.25 | 0.06 |
| Ribosome biogenesis and assembly | ||||||
| SPAC3A12.10 | rpl2001 | 60S ribosomal protein L20a | +++ | ++ | 0.12 | 0.03 |
| SPAC26A3.07c | rpl1101 | 60S ribosomal protein L11 | ++ | + | 0.5 | 0.12 |
| SPAC3H5.10 | rpl3202 | 60S ribosomal protein L32 | ++ | + | 0.25 | 0.12 |
| SPAC1556.05c | cgr1c | Ribosome biogenesis CGR1 family | ++ | + | 0.5 | 0.12 |
| SPBC16C6.03c | rsa1b | Ribosome assembly protein | + | + | 2.0 | 0.12 |
| SPAC6F6.03c | nog2b | Ribosome export GTPase | +++ | ++ | 0.25 | 0.12 |
| Regulation of transcription and translation | ||||||
| SPBC21B10.13c | yox1 | MBF complex negative regulatory component Yox1 | +++ | + | 0.25 | 0.06 |
| SPBC2F12.11c | rep2 | Transcriptional activator, MBF subunit | + | ++ | 0.25 | 0.03 |
| SPBC19G7.16 | iws1 | Transcription elongation factor complex subunit Iws1 | ++ | ++ | 0.5 | 0.12 |
| SPBC31F10.09c | nut2 | Mediator complex subunit Med10 | ++ | ++ | 0.5 | 0.12 |
| Cellular morphogenesis, cell wall organization, and biogenesis | ||||||
| SPBC947.04d | NA | Cell surface glycoprotein, DIPSY family | +++ | ++ | 0.12 | 0.06 |
| SPAC1F8.06 | fta5c | Cell surface glycoprotein | ++ | + | 0.25 | 0.25 |
| SPCC63.02c | aah3 | Alpha-amylase homolog Aah3 | + | − | 0.5 | 0.25 |
| SPCP1E11.04c | pal1c | Membrane-associated protein | ++ | + | 0.12 | 0.015 |
| SPAC688.11 | end4 | Huntingtin-interacting protein homolog | +++ | + | 0.03 | 0.06 |
| Mitochondrial function | ||||||
| SPAC1071.11 | NA | NADH-dependent flavin oxidoreductase | ++ | ++ | 0.03 | 0.015 |
| SPAC1556.02c | sdh1 | Succinate dehydrogenase Sdh1 | +++ | + | 0.25 | 0.12 |
| SPAC4G8.08 | mrs3b | Mitochondrial iron ion transporter | − | + | 0.5 | 0.12 |
| Amino acid metabolism | ||||||
| SPAC3A12.17c | cys12 | Cysteine synthase Cys12 | +++ | + | 0.12 | 0.06 |
| SPBC3B8.03 | lys9b | Saccharopine dehydrogenase | +++ | + | 0.12 | 0.12 |
| SPAC3H1.07 | aru1 | Arginase Aru1 | + | − | 0.5 | 0.25 |
| SPAC24H6.10c | aro3b | Phospho-2-dehydro-3-deoxyheptonate aldolase | + | − | 0.25 | 0.25 |
| Nucleic acid metabolism | ||||||
| SPAC644.14c | rhp51 | Recombinase Rhp51 | +++ | + | 0.12 | 0.03 |
| SPBC29A3.14c | trt1 | Telomerase reverse transcriptase 1 protein | − | + | 0.5 | 0.12 |
| SPAPB1E7.02c | mcl1 | DNA polymerase alpha accessory factor | + | + | 0.25 | 0.06 |
| SPCC736.09c | NA | TRAX | + | + | 1.0 | 0.25 |
| SPAC30.02c | kti12b | Elongator complex-associated protein | +++ | − | 0.03 | 0.12 |
| SPBC36.07 | iki3 | Elongator subunit Iki3 | +++ | + | 0.25 | 0.06 |
| SPCC31H12.08c | ccr4 | CCR4-Not complex subunit Ccr4 | ++ | + | 0.25 | 0.12 |
| SPAC9G1.12 | cpd1 | tRNA (m1A) methyltransferase complex subunit Cpd1 | +++ | ++ | 0.25 | 0.03 |
| SPBC25D12.06c | NA | RNA helicase | +++ | + | 0.25 | 0.06 |
| Lipid metabolism | ||||||
| SPBC31F10.02 | NA | Acyl-CoA thioesterase | +++ | + | 0.12 | 0.06 |
| SPCC1235.15 | dga1 | Diacylglycerol O-acyltransferase | +++ | + | 0.12 | 0.06 |
| SPAC1786.01c | tgl4b | Triacylglycerol lipase | +++ | + | 0.5 | 0.06 |
| Meiosis | ||||||
| SPBC359.06 | mug14 | Adducin | ++ | + | 0.25 | 0.12 |
| SPAC3A11.02 | cps3c | Zinc finger protein Cps3 | +++ | + | 0.12 | 0.03 |
| SPBC577.12 | mug71 | Endoribonuclease | + | + | 0.12 | 0.12 |
| SPCC338.08 | ctp1 | CtIP-related endonuclease | + | + | 0.25 | 0.06 |
| SPAC110.02 | pds5 | Cohesin-associated protein Pds5 | + | + | 0.25 | 0.12 |
| SPAC15A10.03c | rhp54 | Rad54 homolog Rhp54 | ++ | + | 0.25 | 0.06 |
| Other functions | ||||||
| SPAC1D4.05c | erd1b | Erd1 homolog | ++ | + | 0.12 | 0.03 |
| SPBC1709.14 | png1b | Peptide N-glycanase | +++ | + | 0.12 | 0.03 |
| SPBC365.14c | uge1 | UDP-glucose 4-epimerase | + | + | 0.5 | 0.06 |
| SPAC19B12.08 | atg4 | Atg8 deconjugator Atg4 | + | + | 0.5 | 0.12 |
| SPAC823.16c | mug179 | WD repeat protein involved in autophagy Atg18b | ++ | ++ | 0.5 | 0.06 |
| SPCC794.10 | ugp1b | UTP-glucose-1-phosphate uridylyltransferase | + | + | 0.25 | 0.06 |
| SPBP35G2.14 | puf1b,c | RNA binding protein | + | + | 0.25 | 0.12 |
| SPAC12G12.03 | cip2 | RNA binding protein Cip2 | + | ++ | 0.015 | 0.015 |
| Unknown functions | ||||||
| SPAC19A8.11c | irc6b | Recombination protein Irc6 | +++ | − | 0.06 | 0.25 |
| SPCC1020.07 | NA | Haloacid dehalogenase-like hydrolase | +++ | + | 0.25 | 0.06 |
| SPBC2A9.11c | thp3b | Nuclear export factor | +++ | + | 0.12 | 0.06 |
| SPAC56E4.07c | NA | N-Acetyltransferase | +++ | + | 0.12 | 0.06 |
| SPAC57A10.08cc | NA | Carboxylic ester hydrolase activity | +++ | ++ | 0.06 | 0.03 |
| SPBC660.05 | wwm1b,c | Conserved fungal protein | +++ | + | 0.12 | 0.06 |
| SPAC22F8.03cd | NA | Sequence orphan | +++ | ++ | 0.12 | 0.06 |
| SPCC622.14 | gcs1b | GTPase-activating protein | +++ | ++ | 0.12 | 0.03 |
| SPCC1739.01 | lee1b,c | zf-CCCH-type zinc finger protein | +++ | + | 0.12 | 0.06 |
| SPAC1805.14d | NA | Sequence orphan | + | + | 0.12 | 0.12 |
Gene description as indicated in the Sanger Center S. pombe database. WD, tryptophan-aspartic acid; MBF, MluI cell cycle box binding complex; CtIP, CtBP (carboxy-terminal binding protein) interacting protein; TRAX, translin-associated protein X homolog.
The common name is taken from the orthology of S. cerevisiae.
The gene is conserved in fungi only.
The gene is identified in S. pombe only.
Values obtained by using the MIC-2 (50% growth inhibition) endpoint.
+++, severely sensitive; ++, moderately sensitive; +, mildly sensitive; −, not sensitive; Clot, clotrimazole; Terb, terbinafine; NA, the common gene name is not applicable.
Notably, among the 109 genes, 13 genes are conserved only in fungi (Sanger Center S. pombe database) (Table 1). These genes include sft1+, ada3+, spt8+, mub1+ (SPBC31F10.10c), git3+, cgr1+, fta5+, pal1+, cps3+, puf1+ (SPBP35G2.14), lee1+ (SPCC1739.01), wwm1+ (SPBC660.05), SPBC25D12.06, SPCC297.05, SPAC56E4.07, and SPAC57A10.08c. It is attractive to suggest that these genes that are unique to fungi may provide novel targets for the development of powerful, effective combination therapeutic strategies against life-threatening fungal diseases.
Functional categories of proteins by which absence renders cells sensitive to ergosterol biosynthesis inhibitors.
Our results showed that the genes by which gene deletion confers sensitivity to the ergosterol biosynthesis inhibitors are classified into 15 functional categories, including those relating to ergosterol biosynthesis (4 genes), membrane trafficking (19 genes), histone acetylation and deacetylation (6 genes), ubiquitination (9 genes), signal transduction (13 genes), ribosome biosynthesis and assembly (6 genes), regulation of transcription and translation (4 genes), cell wall organization and biogenesis (5 genes), mitochondrion function (3 genes), amino acid metabolism (4 genes), nucleic acid metabolism (9 genes), lipid metabolism (3 genes), meiosis (6 genes), other known functions (8 genes), and other unknown functions (10 genes) (Table 1). Below, we discuss each of the main categories:
Ergosterol biosynthesis pathway.
The first group of the genes that are involved in ergosterol biosynthesis was identified. As shown in Table 1, the deletions of the sts1+, spo9+, and kes1+ genes exhibited hypersensitivity to both inhibitors of ergosterol biosynthesis, namely, clotrimazole and terbinafine, and the deletion of the dap1+ gene exhibited hypersensitivity to clotrimazole but not to terbinafine. The sts1+ gene encodes the C-24(28) sterol reductase Sts1, which is located downstream of Erg1 and Erg11 (21), and the spo9+ gene encodes farnesyl pyrophosphate synthetase, which is located upstream of Erg1 and Erg11 (43). Both the sts1+ and spo9+ genes are responsible for catalyzing an enzymatic reaction in the ergosterol biosynthesis pathway; therefore, the deletion of either the sts1+ or spo9+ gene may cause a synthetic effect on the biosynthesis and result in hypersensitivity to ergosterol biosynthesis inhibitors. Next, the kes1+ gene, encoding a homolog of the oxysterol binding protein, may play an important role in sterol metabolism similar to that of its orthologs in budding yeast (22) and may cause a similar synthetic effect. Another gene is the dap1+ gene, encoding an ortholog of the budding yeast Dap1p, a heme binding protein related to cytochrome b5 that activates Erg11p. Our result showing that dap1 deletion mutant cells are hypersensitive to clotrimazole is consistent with a previous report on budding yeast Dap1p by Craven et al. (11).
Membrane trafficking.
The largest group of genes by which gene deletion confers sensitivity to the ergosterol biosynthesis inhibitors is involved in membrane trafficking (Table 1). This group includes 19 genes, namely, apl5+, apl6+, aps3+, apm3+, apm1+, vps1+, vps45+, vps32+, sip1+, sft1+, sft2+, gos1+, cat1+, gms1+, zhf1+, gga1+ (SPBC25H2.16c), uga4+ (SPCC794.03), ent3+ (SPCC794.11c), and SPBC725.10. Adapter protein complex 3 (AP-3) is a heterotetramer composed of 2 large adaptins (Apl5 and Apl6), a medium adaptin (Apm3), and a small adaptin (Aps3) that is involved in membrane trafficking to the vacuole (14). Interestingly, the deletion of any of the AP-3 subunits leads to hypersensitivity to ergosterol biosynthesis inhibitors (Table 1). The apm1+ gene, encoding a μ1 subunit of AP-1 that plays a key role in Golgi/endosome trafficking and vacuole fusion, was also identified in the clotrimazole- and terbinafine-sensitive screen. Next is the vacuolar protein-sorting pathway (VPS), which mediates the localization of proteins from the trans-Golgi network to the vacuole via a prevacuolar endosome compartment in yeast cells (4, 30). In our screen, 3 VPS genes, vps1+, vps32+, and vps45+, by which gene deletion leads to hypersensitivity to ergosterol biosynthesis inhibitors were identified. In addition, Sip1, a large HEAT (huntingtin, elongation factor 3, PR65/A subunit of protein phosphatase 2A, and lipid kinase Tor)-repeat-containing protein that forms a complex with Pof6, playing an essential role in endocytosis, cytokinesis, and cell division (23), was also identified. Moreover, the Golgi transport-related proteins Sft1, Sft2, and Gos1; the UDP-galactose transporter Gms1; the cationic amino acid transporter Cat1; and the zinc ion transporter Zhf1 were also identified in this screen. Another gene identified was the gga1+ gene, which encodes a homolog of adaptin that is involved in transport from the Golgi apparatus to the vacuole. The next gene identified is SPBC725.10, which is predicted to be involved in the cytoplasm/mitochondrial transport of heme (Sanger Center S. pombe gene database). Still other genes identified are the ent3+ gene, encoding a homolog of the ENTH/VHS domain protein Ent3p, which is predicted to be involved in transport from the Golgi apparatus to the endosome, and uga4+, encoding a homolog of an amino acid permease that is involved in the transmembrane transport of amino acid. All these data suggest that both the Golgi-to-endosome and endosome-to-vacuole stages of transport play major roles in defining sensitivity to ergosterol biosynthesis inhibitors. In the ergosterol biosynthesis pathway, gene products such as farnesylpyrophosphate and geranylgeranylpyrophosphate are essential isoprenoids for the function of small GTPases such as Rab and Ras. In our previous report, we found that two Rab GTPases, namely, Ypt3 and Ryh1, are involved in fission yeast membrane trafficking (8, 19), and the overexpression of rep1+, which encodes the Rab escort protein, suppressed the phenotype of an hmg1-1 mutant, an allele of the hmg1+ gene that encodes the sterol biosynthetic enzyme HMGR (17). Taken together, it is possible that the inhibition or mutation of the ergosterol biosynthesis pathway may reduce the production of isoprenoids, thereby affecting the function of the small GTPase Rab.
Histone acetylation and deacetylation.
Another major group of genes identified comprises pathways involved in histone acetylation and deacetylation. This group includes ada2+, ada3+, spt8+, lsd1+, cph1+, and SPCC126.13c. The ada2+, ada3+, and spt8+ genes encode the Spt-Ada-Gcn5 acetyltransferase (SAGA) complex subunits that are involved in histone acetylation. Next is the lsd1+ gene, which encodes the histone demethylase SWIRM1, which is involved in targeting histone tails for continuous acetylation and deacetylation. In addition, the cph1+ gene and SPCC126.13c, encoding Clr6 histone deacetylase-associated PHD (plant homeo domain) protein 1 (Cph1) and the histone deacetylase complex subunit, respectively, are involved in histone deacetylation. These data suggest that in fission yeast, both histone acetylation and histone deacetylation are important for clotrimazole and terbinafine sensitivity. In support of this, Smith et al. reported previously that in budding yeast, strains with a deletion of SAGA transcriptional adaptor/HAT (histone acetyltransferase) complex genes (e.g., ADA3 and SPT7) markedly enhanced azole sensitivity (36).
Ubiquitination.
Several of the genes identified encode ubiquitin ligase proteins, including pub1+, ubp8+, pli1+, brl2+, rkr1+ (SPBC21D10.09c), SPBC14F5.10c, and SPCC1223.01. In addition, cdt2+, encoding an adaptor protein, and ddb1+, encoding a damaged DNA binding protein, which form a Ddb1-Cul4-Cdt2 ubiquitin ligase complex with the COP9 signalosome complex (CSN), a Cullin-4 ubiquitin ligase (Pcu4) (27), were also identified in this screen. All these results suggest that various ubiquitination processes play important roles in defining sensitivity to the inhibitors of ergosterol biosynthesis. It was reported previously that an inhibitor of squalene synthetase encoded by ERG9 promotes the ubiquitin-ligase-mediated vacuolar degradation of the tryptophan permease Tat2p in budding yeast, suggesting the involvement of ubiquitin ligase in membrane trafficking (12). We suggest that the deletion of ubiquitin ligase genes may affect membrane trafficking, thereby causing hypersensitivity to antifungal drugs, as described above.
Signal transduction.
Several of the genes identified are involved in signal transduction, with the largest group being involved in the glucose/cyclic AMP (cAMP) signaling pathway, including git1+, git3+, git5+, and gpa2+/git8+. The git (glucose-insensitive transcription) genes encode components of a cAMP signaling pathway required for adenylate cyclase activation (20, 39). In addition, the deletion of 3 protein kinase genes, namely, ppk33+, pka1+, and rrd1+ (SPAC4F10.04), conferred hypersensitivity to the ergosterol biosynthesis inhibitors. Also identified were those genes relating to the mitogen-activated protein kinase (MAPK) signal pathway, including the rnc1+ gene, encoding a K homology (KH)-type RNA binding protein that negatively regulates the MAPK kinase kinase (MAPKKK) cascade (37); the rho2+ gene, encoding the Rho family GTPase Rho2 that acts upstream of the MAPK signal pathway (28); and the cch1+ gene, encoding a putative subunit of the Ca2+ channel that is involved in the calcium-dependent calcineurin signal pathway (15). Both the glucose/cAMP signal pathway and the MAPK pathway are the two major intracellular signaling pathways that play a crucial role in various biological functions and control many cellular processes. Although the mechanisms are still unclear, we speculate that these pathways may influence ergosterol biosynthesis, and vice versa.
Other biological processes and unknown functions.
Genes modulating other biological processes, such as the regulation of transcription and translation, ribosome biogenesis and assembly, cell wall organization and biogenesis, mitochondrion function, amino acid metabolism, nucleic acid metabolism, lipid metabolism, and meiosis, also contribute to hypersensitivity to the inhibitors of ergosterol biosynthesis, namely, clotrimazole and terbinafine, upon gene deletion (Table 1). In addition, not all genes identified in this screen were grouped according to known functions in relation to ergosterol biosynthesis, because a number of the mutants showed a clear hypersensitivity to these inhibitors, yet the functions as indicated are apparently unrelated to ergosterol biosynthesis. Also identified were several open reading frames (ORFs) that encode proteins of unknown function, including irc6+, thp3+ (SPBC2A9.11c), wwm1+ (SPBC660.05), gcs1+ (SPCC622.14), lee1+ (SPCC1739.01), SPAC56E4.07, SPAC57A10.08c, SPAC22F8.03c, SPCC1020.07, and SPAC1805.14, and all of these need to be further characterized.
Nearly three-quarters of clotrimazole- and terbinafine-sensitive mutants showed defects in cell wall integrity.
In addition, we examined the growth of the clotrimazole- or terbinafine-sensitive mutants on plates containing 2 μg/ml micafungin, another antifungal drug that inhibits (1,3)-β-d-glucan synthase, which is essential for cell wall synthesis. As shown in Table 2, among the 109 ergosterol biosynthesis inhibitor-sensitive mutants, 57 mutants (or 52.3%) showed hypersensitivity to micafungin, suggesting that these mutants are defective in cell wall integrity. The other 52 mutants (or 47.7%) grew equally well compared to wild-type cells on plates containing micafungin. This finding prompted us to examine whether the ergosterol biosynthesis inhibitor-sensitive mutants showed altered resistance to cell wall-damaging agents such as β-glucanase. β-Glucanase treatments of wild-type cells and Δpmk1 cells, which lack a MAPK regulating the cell wall integrity of fission yeast (38), were also performed as a negative control and a positive control, respectively. As shown in Fig. 3C, the optical density (OD) of wild-type cells at 120 min was 78%, and that of Δpmk1 cells was 47% (the value before the addition of the enzyme was taken as 100%). Our results showed that 24 mutants (or 22.0%), with an OD of <60% at 120 min, were lysed significantly faster than wild-type cells (Table 2). Specifically, 6 mutants, namely, the Δkap1, Δpub1, Δend4, Δugp1, Δpal1, and Δbrl2 mutants, were lysed even faster than the Δpmk1 cells (Fig. 3C). Furthermore, it was observed that 41 gene deletion mutants (or 37.6%) showed osmoremedial ergosterol biosynthesis inhibitor sensitivity; that is, the addition of 1.2 M sorbitol suppressed the clotrimazole- and terbinafine-sensitive growth defect of the deletion mutant cells (Table 2). The Δgpa2 deletion mutant, a representative example of the ergosterol biosynthesis inhibitor-sensitive mutants, is shown in Fig. 3A and B.
Table 2.
Summary of genes responding to cell wall integrity
| Phenotype | Genes |
|---|---|
| Micafungin sensitivity | apm1,eaps3,evps45,epub1,epal1,evps1,bsft1,bada3,bcph1,bpka1,bgpa2,bgit1,brpl2001,b SPBC25D12.06,brkr1,a,brrd1,a,bkti12,a,birc6,a,bccr4,cgos1,cbrl2,cddb1,cnut2,cend4,crpl3202,ccip2,cugp1,a,cgga1,a,cent3,aerd1,apuf1,asts1, spo9, kes1, apl6, apm3, sip1, lsd1, SPBC725.10, SPCC126.13c, cdt2, SPBC14F5.10c, rho2, rnc1, yox1, rep2, iws1, aah3, SPAC1071.11, rhp51, trt1, iki3, mug14, pds5, rph54, uge1, SPAC57A10.08c |
| Osmoremedial ergosterol biosynthesis inhibitor sensitivity | apm1,eaps3,evps45,epub1,epal1,evps1,bsft1,bada3,bcph1,bpka1,bgpa2,bgit1,brpl2001,b SPBC25D12.06,brkr1,a,brrd1,a,bkti12,a,birc6,a,b SPCC297.05,dada2,dapl5,dnog2,apng1,athp3,awwm1,agcs1,avps32, uga4, gms1, zhf1, cat1, ppk33, git5, cgr1, sdh1, cys12, dga1, atg4, SPCC1020.07, SPAC56E4.07, SPAC22F8.03c |
| β-Glucanase sensitivity | apm1,eaps3,evps45,epub1,epal1,egos1,cbrl2,cddb1,cnut2,cend4,crpl3202,ccip2,cugp1,a,cgga1,a,cccr4,cada2,d SPCC297.05,dapl5,dlee1,afta5, spt8, kap1, cch1, sft2 |
The common name is taken from the orthology of S. cerevisiae.
The gene disruption exhibited two of the three phenotypes, that is, micafungin sensitivity and osmoremedial ergosterol biosynthesis inhibitor sensitivity.
The gene disruption exhibited two of the three phenotypes, that is, micafungin sensitivity and β-glucanase sensitivity.
The gene disruption exhibited two of the three phenotypes, that is, osmoremedial ergosterol biosynthesis inhibitor sensitivity and β-glucanase sensitivity.
Genes by which gene disruption exhibited all three phenotypes, that is, micafungin sensitivity, osmoremedial ergosterol biosynthesis inhibitor sensitivity, and β-glucanase sensitivity.
Fig 3.
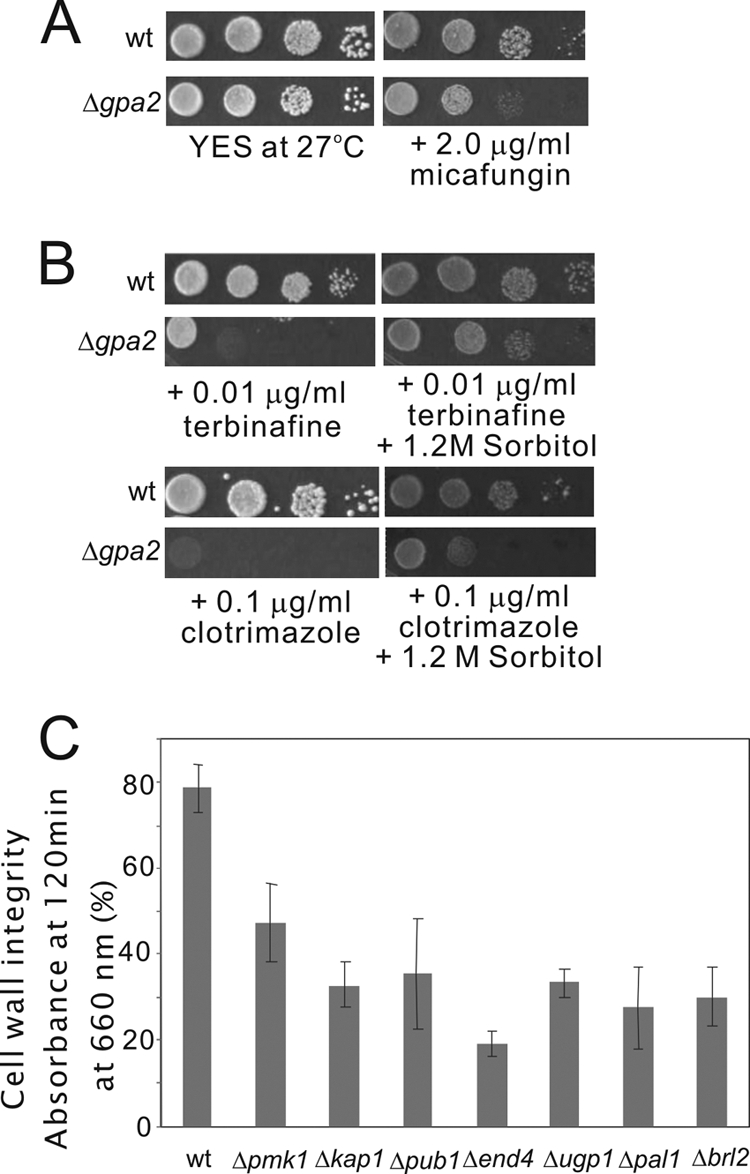
Defects in cell wall integrity in some of the ergosterol biosynthesis inhibitor-sensitive mutants. (A) The Δgpa2 mutants, a representative example of the ergosterol biosynthesis inhibitor-sensitive mutants, were hypersensitive to micafungin, a (1,3)-β-d-glucan synthase inhibitor. Cells were spotted onto plates containing YES or YES plus 2.0 μg/ml micafungin and incubated at 27°C for 4 days. (B) High osmolarity suppressed the ergosterol biosynthesis inhibitor-sensitive phenotype of the Δgpa2 mutants. Wild-type cells and Δgpa2 mutant cells were spotted onto each plate as indicated. (C) Cell wall digestion of the deletion mutants and wild-type cells by β-glucanase. Cells exponentially growing in YES medium were harvested, incubated with β-glucanase at 27°C, and subjected to vigorous shaking. Cell lysis was monitored by measurements of the optical density at 660 nm. The data shown are representative of multiple experiments.
Taken together, 82 mutants (or nearly three-quarters) of all genes by which gene deletions confer hypersensitivity to ergosterol biosynthesis inhibitors were found to express three additional phenotypes, including micafungin sensitivity, β-glucanase sensitivity, and osmoremedial ergosterol biosynthesis inhibitor sensitivity, respectively. Among them, five mutants, namely, the Δpub1, Δvps45, Δapm1, Δpal1, and Δaps3 mutants, expressed all three phenotypes, and 26 mutants expressed two of the three phenotypes correlated with cell wall integrity (Table 2). These findings suggest that cell wall integrity is one of the resultant effects of the sensitivity of these cells to the ergosterol biosynthesis inhibitors and that another mechanism is also involved in the hypersensitivity to ergosterol biosynthesis inhibitors.
Identification of clotrimazole- and terbinafine-resistant S. pombe deletion mutants.
We also screened for genes by which gene deletion renders cells resistant to the antifungal agents that are inhibitors of ergosterol biosynthesis, namely, clotrimazole and terbinafine. As shown in Table 3, we identified 11 genes by which gene deletion confers resistance to the ergosterol biosynthesis inhibitors. These genes include php3+, ubr11+, amo1+, apl3+, tom7+, ssr4+, tom13+, nat2+ (SPBC106.07c), mss116+ (SPBC691.04), SPBC1861.05, and SPCC1442.05c. It is interesting that tom13+ deletion mutants exhibited resistance only to terbinafine and not to clotrimazole. The genes mentioned are categorized into the following groups: mitochondrion function (5 genes), ubiquitination (1 gene), membrane trafficking (1 gene), cell polarity (1 gene), chromatin remodeling (1 gene), and other unknown functions (2 genes). Of the 11 resistant mutants, 5 mutants correspond to genes that are involved in mitochondrion function, and this result is consistent with a previous report that the inhibition of mitochondrial function resulted in fluconazole resistance in a wild-type Saccharomyces cerevisiae strain (26). A relationship between mitochondrial DNA deficiency and resistance to azoles was demonstrated recently for azole-resistant isolates of Candida glabrata (6, 13). More recently, Ferrari et al. reported that the loss of mitochondrial functions associated with azole resistance in Candida glabrata also resulted in enhanced virulence in mice (18). Although the mechanisms involved in these resistant mutants are still unknown, our current findings with fission yeast may provide important clues to a better understanding of the mechanisms of the emergence of antifungal drug resistance. As described above, the deletion of mitochondrial genes confers both sensitivity and resistance to these drugs (Table 1). These results suggest that mitochondria are involved in the regulation of the ergosterol biosynthesis pathway and that the sensitive and resistant genes may play opposite roles in their regulation.
Table 3.
S. pombe genes identified in a screen for clotrimazole- and terbinafine-resistant genese
| Category and systematic gene name | Common gene name | Gene descriptiona | Spot assay result with: |
MIC (μg/ml)d |
||
|---|---|---|---|---|---|---|
| Clot | Terb | Clot | Terb | |||
| Wild type | 2.0 | 0.25 | ||||
| Mitochondrial function | ||||||
| SPAC23C11.08 | php3 | CCAAT binding factor complex subunit Php3 | +++ | +++ | 4.0 | 0.5 |
| SPCC1442.05cc | NA | Conserved fungal protein | ++ | + | 2.0 | 0.25 |
| SPBC27B12.10c | tom7 | Mitochondrial TOM complex subunit Tom7 | ++ | ++ | 4.0 | 0.5 |
| SPBC691.04 | mss116b | Mitochondrial ATP-dependent RNA helicase Mss116 | ++ | ++ | 2.0 | 0.5 |
| SPBC713.08 | tom13c | Mitochondrial TOM complex subunit Tom13 | − | + | 2.0 | 0.5 |
| Ubiquitination | ||||||
| SPAC15A10.11 | ubr11 | N-end-recognizing protein | ++ | + | 4.0 | 0.25 |
| Membrane trafficking | ||||||
| SPBC691.03c | apl3 | AP-2 adaptor complex subunit Alp3 | ++ | +++ | 4.0 | 0.5 |
| Cell polarity | ||||||
| SPBC15D4.10c | amo1 | Nuclear rim protein Amo1 | + | + | 2.0 | 0.5 |
| Chromatin remodeling | ||||||
| SPBP23A10.05 | ssr4c | SWI/SNF and RSC complex subunit | + | + | 2.0 | 0.5 |
| Unknown functions | ||||||
| SPBC1861.05 | NA | Carbohydrate kinase | ++ | ++ | 2.0 | 0.5 |
| SPBC106.07c | nat2b | N-alpha-acetylation-related protein | + | ++ | 2.0 | 0.5 |
Gene description as indicated in the Sanger Center S. pombe database. TOM, translocase of outer membrane; RSC, remodel the structure of chromatin.
The common name is taken from the orthology of S. cerevisiae.
The gene is conserved in fungi only.
Values obtained by using the MIC-2 (50% growth inhibition) endpoint.
+++, strongly resistant; ++, moderately resistant; +, mildly resistant; −, not resistant; NA, the common gene name is not applicable.
Complementation of the growth phenotypes of deletion strains by the wild-type genes.
To confirm that the sensitivity was due to a specific gene deletion and not a consequence of secondary mutations during the screening process, we proceeded to revert the phenotype by the complementation of the strains with their specific genes. We selected strains with mutations that showed the highest level of sensitivity to the drugs. These strains included apm1, rho2, pka1, and vps45 deletion mutants. As shown in Fig. 4, all the strains tested could be efficiently complemented by plasmids containing wild-type copies of the genes. These results suggest that the sensitive phenotypes of these strains were indeed caused by the deletion of specific genes.
Fig 4.
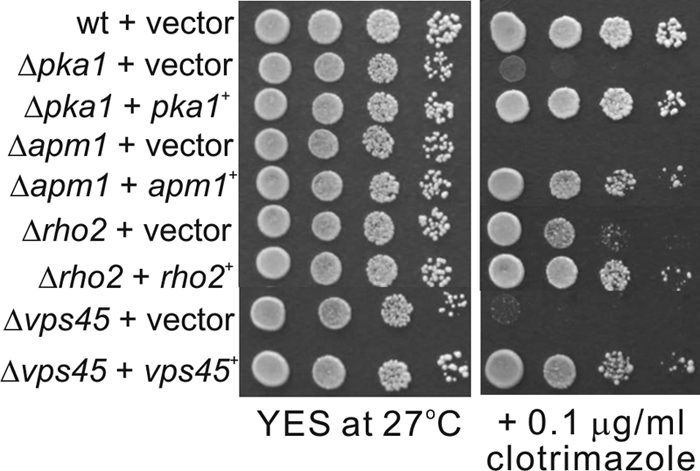
Complementation of the growth phenotypes of deletion strains by the wild-type genes. Deletion strains were transformed with expression plasmids containing their specific wild-type genes or the empty vector and tested for complementation. Serial dilutions of stationary cultures were spotted onto the plates as indicated and then incubated at 27°C for 4 days.
Responses of clotrimazole- and terbinafine-sensitive or -resistant mutants to other antifungal drugs.
We also tested the growths of the strains isolated in this screen on plates containing other antifungals, including fluconazole, another azole, and amphotericin B (AmB), which binds to ergosterol and destabilizes the cell membrane. To test for drug hypersensitivity, different concentrations of the drugs, ranging from 10 to 30 μg/ml of fluconazole and 15 to 20 μg/ml of AmB, were used. To test for drug resistance, different concentrations of the drugs, ranging from 100 to 300 μg/ml of fluconazole and 30 to 32 μg/ml of AmB, were used. As expected, all of the mutants showed the same response to fluconazole compared to the response to clotrimazole, while the potency of fluconazole was significantly weaker than that of clotrimazole. The results obtained with AmB are summarized in Table 4. Among the 109 mutants that were sensitive and 11 mutants that were resistant to ergosterol biosynthesis inhibitors, including clotrimazole and terbinafine, 34 of them showed sensitivity to AmB, whereas 4 of them showed resistance to AmB (Table 4). Specifically, two mutants, namely, the Δapl3 and Δamo1 mutants, exhibited resistance to ergosterol biosynthesis inhibitors but exhibited sensitivity to AmB, whereas 4 mutants, namely, the Δsts1, Δkes1, Δada2, and Δada3 mutants, exhibited resistance to AmB but exhibited sensitivity to ergosterol biosynthesis inhibitors (Table 4). On the other hand, 32 mutants showed sensitivity to both the ergosterol biosynthesis inhibitors and AmB. These results may be helpful for the selection and combination of various antifungal drugs to enhance their therapeutic efficacy.
Table 4.
S. pombe genes by which gene deletion confers sensitivity or resistance to amphotericin B
| Phenotype | Genes |
|---|---|
| Amphotericin B sensitivity | apl3,damo1,dapm1,cvps45,clsd1,ccph1,cpub1,crep2,cpal1,cend4,cmrs3,a,crhp54,ccip2,c SPAC1805.14,csft2,cgga1,a,cent3,a,ccdt2,cddb1,crho2,crrd1,a,circ6,a,cmug179,cctp1,cpds5,ciki3,crpl2001,crpl1101,ciws1,caah3,crhp51,c SPAC1071.11,cccr4,catg4,c |
| Amphotericin B resistance | sts1,bkes1,bada2,bada3b |
The common name is taken from the orthology of S. cerevisiae.
The gene disruption exhibited sensitivity to ergosterol biosynthesis inhibitors but exhibited resistance to AmB.
The gene disruption exhibited sensitivity to both ergosterol biosynthesis inhibitors and AmB.
The gene disruption exhibited resistance to ergosterol biosynthesis inhibitors but exhibited sensitivity to AmB.
Determination of ergosterol levels of several deletion mutants that exhibited altered sensitivity to antifungal drugs.
To examine whether the ergosterol content of the strains correlates with the altered sensitivity to ergosterol biosynthesis inhibitors, we determined the ergosterol levels of several strains that showed the highest levels of sensitivity or resistance to the drugs. The results are shown in Fig. 5. Regarding the strains that exhibit resistance to clotrimazole and terbinafine, the ergosterol content of php3 deletion mutants was dramatically increased compared with that of wild-type cells, whereas that of apl3 mutants was only slightly increased. Regarding the strains that exhibited sensitivity to clotrimazole and terbinafine, the ergosterol contents of the SPBC947.04, gpa2, rpl2001, and apm1 mutants were dramatically decreased, whereas those of the ddb1, apl5, cat1, and rho2 mutants were similar to that of the wild-type cells. These findings indicate that the sensitivity to ergosterol biosynthesis inhibitors correlates with the ergosterol level in some but not all the deletion strains.
Fig 5.
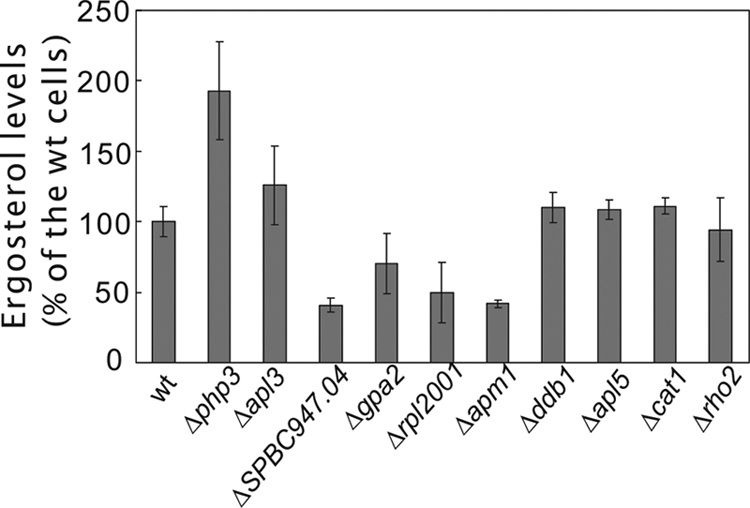
Ergosterol levels of deletion mutants that exhibit altered sensitivity to antifungal drugs. The cells indicated were grown to saturation in YES at 27°C, and ergosterol from the strains was extracted and levels of ergosterol were determined as described in Materials and Methods. The ergosterol content of wild-type cells was taken as 100%, and the ergosterol contents of the other deletion mutants were calculated as a percentage of that of the wild-type cells. Each value represents the average from at least three independent experiments.
In conclusion, the first genomewide screen of haploid deletion mutants in fission yeast was performed to determine altered sensitivities to antifungal drugs, including clotrimazole and terbinafine, in this study. Here, we have identified 109 mutants that were sensitive and 11 mutants that were resistant to these antifungals which are inhibitors of ergosterol biosynthesis. To our knowledge, this is the first report of a global screening for altered sensitivity to these antifungal drugs in a fungal model organism, and the genes or pathways identified in our study may provide valuable insights into the development of novel antifungal drugs that enhance the therapeutic efficacy of existing drugs and help minimize the occurrence of an increasing frequency of drug resistance.
ACKNOWLEDGMENTS
We thank Susie O. Sio for critical reading of the manuscript.
This work was supported by the National Natural Science Foundation of China (grant no. 30900795 and 31071094) and research grants from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print 17 January 2012
REFERENCES
- 1. Alamgir M, Erukova V, Jessulat M, Azizi A, Golshani A. 2010. Chemical-genetic profile analysis of five inhibitory compounds in yeast. BMC Chem. Biol. 10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson JB. 2005. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat. Rev. Microbiol. 3:547–556 [DOI] [PubMed] [Google Scholar]
- 3. Arndt GM, Atkins D. 1996. pH sensitivity of Schizosaccharomyces pombe: effect on the cellular phenotype associated with lacZ gene expression. Curr. Genet. 29:457–461 [PubMed] [Google Scholar]
- 4. Bankaitis VA, Johnson LM, Emr SD. 1986. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. U. S. A. 83:9075–9079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartolommei G, Devaux N, Tadini-Buoninsegni F, Moncelli M, Apell HJ. 2008. Effect of clotrimazole on the pump cycle of the Na,K-ATPase. Biophys. J. 95:1813–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brun S, et al. 2003. Relationships between respiration and susceptibility to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 47:847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen SC, Sorrell TC. 2007. Antifungal agents. Med. J. Aust. 187:404–409 [DOI] [PubMed] [Google Scholar]
- 8. Cheng H, et al. 2002. Role of the Rab GTP-binding protein Ypt3 in the fission yeast exocytic pathway and its connection to calcineurin function. Mol. Biol. Cell 13:2963–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clinical and Laboratory Standards Institute 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard CLSI document M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 10. Cowen LE. 2008. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat. Rev. Microbiol. 6:187–198 [DOI] [PubMed] [Google Scholar]
- 11. Craven RJ, Mallory JC, Hand RA. 2007. Regulation of iron homeostasis mediated by the heme-binding protein Dap1 (damage resistance protein 1) via the P450 protein Erg11/Cyp51. J. Biol. Chem. 282:36543–36551 [DOI] [PubMed] [Google Scholar]
- 12. Daicho K, et al. 2007. The ergosterol biosynthesis inhibitor zaragozic acid promotes vacuolar degradation of the tryptophan permease Tat2p in yeast. Biochim. Biophys. Acta 1768:1681–1690 [DOI] [PubMed] [Google Scholar]
- 13. Defontaine A, et al. 1999. In-vitro resistance to azoles associated with mitochondrial DNA deficiency in Candida glabrata. J. Med. Microbiol. 48:663–670 [DOI] [PubMed] [Google Scholar]
- 14. Dell'Angelica EC. 2009. AP-3-dependent trafficking and disease: the first decade. Curr. Opin. Cell Biol. 21:552–559 [DOI] [PubMed] [Google Scholar]
- 15. Deng L, et al. 2006. Real-time monitoring of calcineurin activity in living cells: evidence for two distinct Ca2+-dependent pathways in fission yeast. Mol. Biol. Cell 17:4790–4800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Enoch DA, Ludlam HA, Brown NM. 2006. Invasive fungal infections: a review of epidemiology and management options. J. Med. Microbiol. 55:809–818 [DOI] [PubMed] [Google Scholar]
- 17. Fang Y, et al. 2009. Pleiotropic phenotypes caused by an opal nonsense mutation in an essential gene encoding HMG-CoA reductase in fission yeast. Genes Cells 14:759–771 [DOI] [PubMed] [Google Scholar]
- 18. Ferrari S, et al. 2011. Loss of mitochondrial functions associated with azole resistance in Candida glabrata results in enhanced virulence in mice. Antimicrob. Agents Chemother. 55:1852–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He Y, et al. 2006. Genetic and functional interaction between Ryh1 and Ypt3: two Rab GTPases that function in S. pombe secretory pathway. Genes Cells 11:207–221 [DOI] [PubMed] [Google Scholar]
- 20. Ivey FD, et al. 2010. Activated alleles of the Schizosaccharomyces pombe gpa2+ Galpha gene identify residues involved in GDP-GTP exchange. Eukaryot. Cell 9:626–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iwaki T, et al. 2008. Multiple functions of ergosterol in the fission yeast Schizosaccharomyces pombe. Microbiology 154:830–841 [DOI] [PubMed] [Google Scholar]
- 22. Jiang B, Brown JL, Sheraton J, Fortin N, Bussey H. 1994. A new family of yeast genes implicated in ergosterol synthesis is related to the human oxysterol binding protein. Yeast 10:341–353 [DOI] [PubMed] [Google Scholar]
- 23. Jourdain I, et al. 2009. Identification of a conserved F-box protein 6 interactor essential for endocytosis and cytokinesis in fission yeast. Biochem. J. 420:169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kennedy PJ, et al. 2008. A genome-wide screen of genes involved in cadmium tolerance in Schizosaccharomyces pombe. Toxicol. Sci. 106:124–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim DU, et al. 2010. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 28:617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kontoyiannis DP. 2000. Modulation of fluconazole sensitivity by the interaction of mitochondria and erg3p in Saccharomyces cerevisiae. J. Antimicrob. Chemother. 46:191–197 [DOI] [PubMed] [Google Scholar]
- 27. Liu C, et al. 2005. Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4-Ddb1-CSN ubiquitin ligase. EMBO J. 24:3940–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma Y, Kuno T, Kita A, Asayama Y, Sugiura R. 2006. Rho2 is a target of the farnesyltransferase Cpp1 and acts upstream of Pmk1 mitogen-activated protein kinase signaling in fission yeast. Mol. Biol. Cell 17:5028–5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martinez-Rossi NM, Peres NT, Rossi A. 2008. Antifungal resistance mechanisms in dermatophytes. Mycopathologia 166:369–383 [DOI] [PubMed] [Google Scholar]
- 30. Miyatake M, et al. 2007. Valproic acid affects membrane trafficking and cell-wall integrity in fission yeast. Genetics 175:1695–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 [DOI] [PubMed] [Google Scholar]
- 32. Moreton RS. 1989. Yeast lipid estimation by enzymatic and nuclear magnetic resonance methods. Appl. Environ. Microbiol. 55:3009–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Munn AL, Heese-Peck A, Stevenson BJ, Pichler H, Riezman H. 1999. Specific sterols required for the internalization step of endocytosis in yeast. Mol. Biol. Cell 10:3943–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Onyewu C, Blankenship JR, Del Poeta M, Heitman J. 2003. Ergosterol biosynthesis inhibitors become fungicidal when combined with calcineurin inhibitors against Candida albicans, Candida glabrata, and Candida krusei. Antimicrob. Agents Chemother. 47:956–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schuman P, et al. 1998. Mucosal candidal colonization and candidiasis in women with or at risk for human immunodeficiency virus infection. HIV Epidemiology Research Study (HERS) Group. Clin. Infect. Dis. 27:1161–1167 [DOI] [PubMed] [Google Scholar]
- 36. Smith WL, Edlind TD. 2002. Histone deacetylase inhibitors enhance Candida albicans sensitivity to azoles and related antifungals: correlation with reduction in CDR and ERG upregulation. Antimicrob. Agents Chemother. 46:3532–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sugiura R, et al. 2003. Feedback regulation of MAPK signalling by an RNA-binding protein. Nature 424:961–965 [DOI] [PubMed] [Google Scholar]
- 38. Toda T, et al. 1996. The fission yeast pmk1+ gene encodes a novel mitogen-activated protein kinase homolog which regulates cell integrity and functions coordinately with the protein kinase C pathway. Mol. Cell. Biol. 16:6752–6764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang L, Griffiths K, Jr, Zhang YH, Ivey FD, Hoffman CS. 2005. Schizosaccharomyces pombe adenylate cyclase suppressor mutations suggest a role for cAMP phosphodiesterase regulation in feedback control of glucose/cAMP signaling. Genetics 171:1523–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wilson LS, et al. 2002. The direct cost and incidence of systemic fungal infections. Value Health 5:26–34 [DOI] [PubMed] [Google Scholar]
- 41. Witzke A, Lindner K, Munson K, Apell HJ. 2010. Inhibition of the gastric H,K-ATPase by clotrimazole. Biochemistry 49:4524–4532 [DOI] [PubMed] [Google Scholar]
- 42. Xia L, Jaafar L, Cashikar A, Flores-Rozas H. 2007. Identification of genes required for protection from doxorubicin by a genome-wide screen in Saccharomyces cerevisiae. Cancer Res. 67:11411–11418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ye Y, et al. 2007. Geranylgeranyl diphosphate synthase in fission yeast is a heteromer of farnesyl diphosphate synthase (FPS), Fps1, and an FPS-like protein, Spo9, essential for sporulation. Mol. Biol. Cell 18:3568–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]