Abstract
The marine snail Aplysia californica produces escapin, an l-amino acid oxidase, in its defensive ink. Escapin uses l-lysine to produce diverse products called escapin intermediate products of l-lysine (EIP-K), including α-amino-ε-caproic acid, Δ1-piperidine-2-carboxylic acid, and Δ2-piperidine-2-carboxylic acid. EIP-K and H2O2 together, but neither alone, is a powerful bactericide. Here, we report bactericidal mechanisms of escapin products on Escherichia coli. We show that EIP-K and H2O2 together cause rapid and long-lasting DNA condensation: 2-min treatment causes significant DNA condensation and killing, and 10-min treatment causes maximal effect, lasting at least 70 h. We isolated two mutants resistant to EIP-K plus H2O2, both having a single missense mutation in the oxidation regulatory gene, oxyR. A complementation assay showed that the mutated gene, oxyR(A233V), renders resistance to EIP-K plus H2O2, and a gene dosage effect leads to reduction of resistance for strains carrying wild-type oxyR. Temperature stress with EIP-K does not produce the bactericidal effect, suggesting the effect is due to a specific response to oxidative stress. The null mutant for any single DNA-binding protein—Dps, H-NS, Hup, Him, or MukB—was not resistant to EIP-K plus H2O2, suggesting that no single DNA-binding protein is necessary to mediate this bactericidal effect, but allowing for the possibility that EIP-K plus H2O2 could function through a combination of DNA-binding proteins. The bactericidal effect of EIP-K plus H2O2 was eliminated by the ferrous ion chelator 1,10-phenanthroline, and it was reduced by the hydroxyl radical scavenger thiourea, suggesting hydroxyl radicals mediate the effects of EIP-K plus H2O2.
INTRODUCTION
Many natural and synthesized chemicals have been identified as bactericidal agents that target and inactivate essential metabolic pathways. One class of bactericidal agents is the reactive oxygen species (ROSs). ROSs are highly active free radicals containing oxygen, and they include peroxide and superoxide. Hydrogen peroxide (H2O2) is a relatively weak peroxide of ROSs, but it generates a powerful ROS, hydroxyl radical (HO·), through the Fenton reaction (14, 15, 23).
H2O2 and other ROSs can have diverse deleterious effects on cells. They can interact with membrane lipids and proteins, resulting in loss of membrane fluidity and structural integrity, which in turn can affect cell integrity and function (14, 15, 23). They can also damage DNA and cause single-strand breaks that accumulate in cells (3, 15, 17, 23). Such DNA damage can block DNA replication and transcription (14, 15, 23).
Bacteria have evolved mechanisms to protect themselves from ROSs. They decompose ROSs using enzymes, such as catalase and glutathione peroxidase, or using antioxidants, such as ascorbic acid. This protective mechanism can be induced by stress. For example, Escherichia coli treated with a nonlethal dose (10 μM) of H2O2 induced the production of 30 proteins and a tolerance for higher doses of H2O2 (7). Twelve of these proteins were “early,” defined as being synthesized at a maximal rate during the first 10 min following addition of H2O2, and their synthesis returned to normal rates by 30 min after addition. The other 18 proteins were “late,” with their maximum rate of synthesis 10 to 30 min after the addition of H2O2. Notably, 9 of these 12 early proteins belong to the oxyR regulon, including catalase. Bacteria are highly sensitive to H2O2 in the oxyR deletion mutant (7).
ROSs are produced in a variety of contexts. They are generated during cellular respiration associated with normal metabolism. Stressors, such as starvation and induced oxidative stress, can cause bacteria to produce and accumulate high levels of ROSs, which they can use in competitive interactions (25, 30, 32). ROSs are also produced by other organisms as natural antimicrobial agents. For example, a marine snail, the sea hare Aplysia californica, produces an l-amino acid oxidase, aplysianin A (9), which is likely used to prevent bacterial fouling of the sea hare's egg capsules (12).
A paralog of aplysianin A, called escapin, has been purified from the ink gland of the sea hare (34). Escapin normally functions as an antipredatory chemical defense (12), but it also has broad antimicrobial activity, both bacteriostatic and bactericidal (34), indicative of its evolutionary roots (12, 13). Escapin uses l-lysine as its primary substrate to produce diverse products (21, 34). Escapin deaminates l-lysine to generate α-amino-ε-caproic acid, H2O2, and ammonia (NH3). α-Amino-ε-caproic acid forms an equilibrium mixture of several compounds, which we collectively call escapin intermediate products of l-lysine (EIP-K). EIP-K reacts with H2O2 to produce a mixture of decarboxylation products called escapin end products of lysine (EEP-K). Interestingly, the mixture of EIP-K plus H2O2, but not EIP-K, EEP-K, H2O2, or EEP-K plus H2O2, shows rapid, powerful, and long-lasting bactericidal activity, much more than H2O2 alone (22). The composition of EIP-K equilibrium mixture changes with the pH level (21). A cyclic deaminated product, Δ1-piperidine-2-carboxylic acid, dominates at any pH level, but the linear form, α-amino-ε-caproic acid, becomes more prevalent at lower pH values (21), which is biologically significant because sea hare ink is acidic (27). This increase in the relative amount of α-amino-ε-caproic acid in EIP-K at low pH values is correlated with an increase in bactericidal activity (22). Furthermore, the synergistic bactericidal effect of EIP-K and H2O2 requires their simultaneous, not sequential, presence (22). Together, these results suggest that the powerful bactericidal effect of EIP-K plus H2O2 is due to synergy involving α-amino-ε-caproic acid and H2O2 (22).
The goal of this study is to understand mechanisms underlying the bactericidal effect of EIP-K plus H2O2. We pursued two lines of experiments. In the first set, we followed up on our observation that EIP-K plus H2O2 causes DNA condensation. DNA condensation is commonly associated with antimicrobial agents that inhibit protein synthesis, most notably chloramphenicol (35). If DNA condensation is long lasting, it can be bactericidal (2). Consequently, we hypothesized that DNA condensation is a major cause of the bactericidal effect of EIP-K plus H2O2. To test this idea, we examined the time course of DNA condensation in relation to treatment with EIP-K plus H2O2 and examined whether DNA condensation and bactericidal activity occurred under the same treatment conditions. Our second line of investigation into the bactericidal mechanism of EIP-K plus H2O2 was to isolate mutant bacterial strains resistant to EIP-K plus H2O2, followed by comparison of the mutants' genotype relative to the wild-type strain. Given that our two mutants had a single missense mutation in the oxidation regulatory gene, oxyR, we performed complementation assays, and we used ferrous ion chelators, hydroxyl radical scavengers, and temperature stress to evaluate if the bactericidal and DNA condensation effects of EIP-K plus H2O2 are due to oxidative stress created by free radicals.
MATERIALS AND METHODS
Preparation of oxidative products of l-amino acids by escapin.
To produce EIP-K, 55 mM l-lysine monohydrochloride (Sigma-Aldrich, St. Louis, MO), 1 mg/ml escapin, and 0.13 mg/ml catalase (Sigma-Aldrich, MO) in double-deionized water (ddH2O) were incubated at 30°C with shaking for up to 24 h. This solution was filtered using an Amicon Ultra-4 centrifugal filter device (Millipore Corp., Billerica, MA) to remove escapin and catalase and then stored at −80°C until it was used further. The purity of EIP-K was checked by nuclear magnetic resonance (21). The concentration of EIP-K is expressed as the starting concentration of l-lysine monohydrochloride (21, 22).
Bacterial strains.
The bacterial strains used in our experiments include the following: (i) E. coli strain MC4100 (from John Beckwith, Harvard Medical School); (ii) E. coli resistant strains 1 and 2 (RS1 and RS2) (isolated from E. coli strain MC4100, as described below); (iii) E. coli strain NT3 (MC4100 lac+) and individual DNA-associated protein (derived from NT3) mutant E. coli strains, including ΔHupA, ΔHns, ΔHimA, and ΔMukB (from Nancy Trun, Duquesne University); and (iv) E. coli strain ZK126 (W3110 tna-2 ΔlacU169) and its stationary-phase DNA-binding protein mutant, ΔDps, E. coli strain (from Roberto Kolter, Harvard Medical School).
Bacterial-culture preparation.
E. coli MC4100 was used as a test strain and also as a parental strain for the generation of two strains resistant to EIP-K plus H2O2. The cells were stored as a −80°C stock. For culturing the cells, the stocks were incubated at 37°C overnight in Luria-Bertani (LB) medium, and the overnight culture was diluted 100 times to regrow until it reached log phase (density, ∼3 × 108 cells/ml; optical density at 600 nm [OD600], ∼0.5). After washing with phosphate-buffered saline (PBS) (containing 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, and 0.24 g KH2PO4 in 1 liter solution, pH 7.3), the bacteria were resuspended in PBS to a density of 6 ×108 cells/ml. Experiments on the ΔHupA, ΔHns, ΔHimA, and ΔMukB mutant strains and their parental strain (NT3) were performed at 30°C.
RS1.
E. coli MC4100 cultured cells were treated with 13.75 mM EIP-K plus 3 mM H2O2, which are the most effective conditions for the bactericidal assay (22), and spread onto solid LB plates. Surviving colonies were taken from the plates and reinoculated until they reached a density of ∼3 × 108 cells/ml (log phase; OD600, ∼0.5) in LB medium. The cells were washed with PBS and then treated with EIP-K plus H2O2 and spread onto solid LB plates as before. This process was repeated four times until it yielded a colony that was insensitive to treatment with EIP-K plus H2O2, as measured by less than 1 log unit reduction in the number of bacterial CFU. This ensured resistance rather than persistence.
RS2.
Bacteria from the culture preparation were treated for 40 min with a mutagen, 2% ethyl methanesulfonate (Sigma-Aldrich). This was followed by repeated treatment with EIP-K plus H2O2 as described above for isolation of RS1.
Nucleoid staining to evaluate DNA condensation.
To stain DNA, bacteria were washed with PBS and incubated for 10 min in 10 μg/ml DNA staining agent, Hoechst 33342 (Molecular Probes, Eugene, OR). The bacteria were then placed between a microscope slide (Superfrost; Fisher Scientific, Waltham, MA) and a cover glass with mounting solution, glycerol, and anti-fading agent, triethylenediamine (DABCO; Sigma-Aldrich). Images were captured using a Nikon Eclipse 80i microscope under ×1,000 magnification. Images of stained cells were captured in transmitted light to observe the shape and location of cells and/or in UV light to observe the distribution of DNA in the cells. When images were taken from samples during 1.5 to 70 h of treatment, the samples were kept at room temperature.
The size and shape of nucleoid staining, and thus DNA condensation, were analyzed using CellProfiler cell image analysis software (Broad Institute; http://www.cellprofiler.org). The length of the major axis and the form factor (which represents shape, with 0.0 indicating a line and 1.0 indicating a perfect circle) of the nucleoid of each cell were quantified. Data from >50 cells were used for each treatment, and the treatments were statistically analyzed using analysis of variance (ANOVA), followed by Scheffé post hoc tests.
Bactericidal assay.
Bacteria (with PBS, pH 7.3) prepared as described above were treated with 13.75 mM EIP-K plus 3 mM H2O2 for different time periods (0, 2, 4, 6, 8, and 10 min) in the time course experiments and for 10 min in all other experiments. Experiments were performed at 37°C in a Thermomixer (Eppendorf, Hamburg, Germany) for all strains except ΔMukB and its parental strain (NT3), for which they were performed at 30°C. Samples were serially diluted in log-unit steps with PBS, and serial dilutions were plated on petri dishes with solid LB agar. The plates were incubated overnight at 37°C, and counts of viable cells were determined by enumeration of CFU with appropriate dilutions.
Genomic DNA preparation.
Genomic DNA of RS1, RS2, and their parental strain, MC4100, were purified with a Qiagen Genomic-tip 100/G and a Genomic DNA Buffer Set (Qiagen Inc., Valencia, CA), using the manufacturer's protocols. To remove high-affinity DNA-binding proteins and to ensure better DNA quality for the whole-genome sequence, the DNA solution was purified in two sequential steps: (i) DNA was first treated with a mixture of equal volumes of phenol and chloroform-isopropanol (24:1) to separate it into phenol and aqueous phases; (ii) the aqueous phase was mixed with an equal volume of chloroform-100% isopropanol (24:1 mixture). After purification, the DNA was precipitated using 66% ethanol with 0.1 M sodium acetate (NaOAc, pH 5.2), and the pellet was washed with 70% ethanol. DNA was reconstituted in 10 mM Tris buffer (pH 8.0) for sequencing.
Localization of mutations.
Whole-genome sequencing was conducted by the Interdisciplinary Center for Biotechnology Research of the University of Florida (http://www.biotech.ufl.edu), using a 454 Genome Sequencer (GS-FLX; Roche, Branford, CT). The results for RS1 and RS2 were mapped against the genome sequence of the parental strain, MC4100. Fifteen mutations were detected in RS1 and 22 mutations in RS2. Eleven mutations were common to both RS1 and RS2 but different from the parental strain, and these were further tested by high-fidelity PCR (Roche) and DNA sequencing using an ABI sequencer. To avoid bias from a single replication mistake during the PCR process, the entire PCR product, not just a single cloning product, was sequenced. This process led to the identification of a single gene mutation in both RS1 and RS2: oxyR.
Cloning of the oxyR gene.
Primers were designed to amplify oxyR of MC4100 (wild-type oxyR) and oxyR of RS1 [oxyR(A233V)] for complementation assays. The 5′ primer included an NdeI restriction site to provide a start codon and no redundant nucleotide in the N terminus (5′-GGATGGATACATATGAATATTCGT-3′). The 3′ end included an XhoI restriction site (5′-TTAAACGGTCTCGAGTTAAACCGC-3′). PCR was applied using a high-fidelity PCR system (Roche). Wild-type oxyR and oxyR(A233V) fragments of the expected size were individually cloned into a pET29a vector carrying a T7 promoter (Novagen, Madison, WI). The mutated fragments were confirmed by DNA sequencing using an ABI sequencer.
Complementation assay.
Plasmids pET29a, pET29a-oxyRwt (wild-type oxyR), and pET29a-oxyR(A233V) [oxyR(A233V)] were transformed into E. coli MC4100 or RS1. A single colony was taken from each transformed plate and cultured to stationary phase (OD600 > 2.0). Cultures were diluted to an OD600 of ∼0.1 and cultured anew. The same treatments were applied as in the bactericidal assay.
RESULTS
EIP-K plus H2O2 causes rapid and long-lasting DNA condensation in E. coli MC4100.
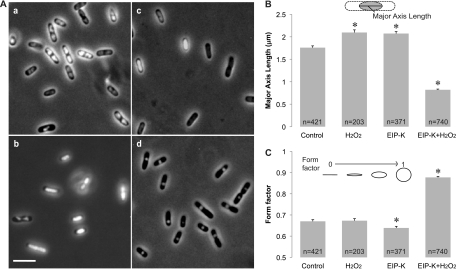
The structure of the DNA of E. coli MC4100 was visualized by examining cells stained with Hoechst under simultaneous UV and transmitted illumination. Cells that were treated for 10 min with either ddH2O (control), 13.75 mM EIP-K, or 3 mM H2O2 showed a variety of nucleoid morphologies, including cells with relaxed DNA in which the nucleoid filled most of the cell volume and cells with condensed DNA in which the nucleoid had various irregular shapes (Fig. 1). On the other hand, the nucleoid of cells treated with 13.75 mM EIP-K plus 3 mM H2O2 for 10 min had a significantly different morphology (Fig. 1A, d) than the control or cells undergoing the other treatments: they were nearly uniform spheres and significantly shorter (Fig. 1B) and rounder (Fig. 1C) and thus more condensed. DNA condensation caused by EIP-K plus H2O2 is further indicated by the fact that ∼80% of cells under this treatment had a form factor of >0.8 and ∼50% had a form factor of >0.9, whereas only ∼20% of the control cells had a form factor of >0.8 and ∼5% had a form factor of >0.9 (data not shown).
Fig 1.
DNA condensation in E. coli MC4100. (A) Light micrographs of DNA condensation under different treatments. (a) Control (ddH2O, used as a solvent in the other treatments); (b) 3 mM H2O2; (c) 13.75 mM EIP-K; (d) 13.75 mM EIP-K plus 3 mM H2O2. Cells were stained with Hoechst to label DNA, and images were taken by simultaneously presenting UV and transmitted light. Scale bar, 5 μm. (B and C) Quantification of DNA condensation using the length of the major axis of the nucleoid (B) and the form factor, where 1.0 is a perfect circle and 0.0 is a straight line (C). The treatments in panels B and C were the same as in panel A. The values are means and standard errors of the mean (SEM), and the number of cells (n) used for each treatment is shown in the bar. The asterisks indicate that treatments are significantly different from the control. In panel B, there is a significant treatment effect (one-way ANOVA; F[3,1731] = 389.17; P < 0.001), and post hoc Scheffé tests (α [level of significance] = 0.05) show that (H2O2) = (EIP-K) > (control) > (EIP-K plus H2O2). In panel C, there is a significant treatment effect (one-way ANOVA; F[3,1731] = 355.01; P < 0.001), and post hoc Scheffé tests (α = 0.05) show that all treatments differ from each other, except the control and H2O2.
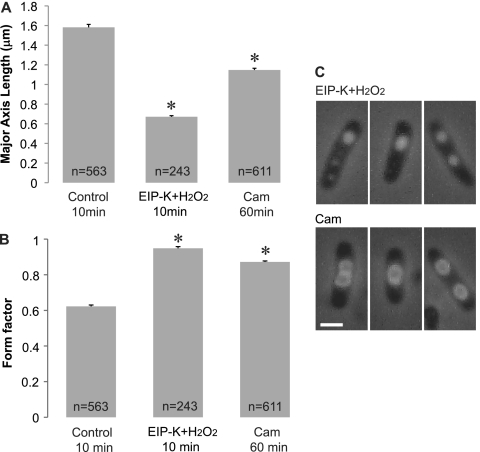
DNA condensation has been reported for treatment with antibiotics, such as chloramphenicol (CAM), that inhibit protein synthesis (35). In our experiments, either CAM or EIP-K plus H2O2 induced DNA condensation, but to different degrees (Fig. 2A) and with different forms (Fig. 2B). EIP-K plus H2O2 compacted DNA into a tighter area, with nucleoid diameters around half that induced by CAM. EIP-K plus H2O2 induced solid and homogeneous condensation, whereas CAM induced heterogeneous condensation with a dense ring and a hollow center (Fig. 2C).
Fig 2.
Effect of chloramphenicol, an inhibitor of protein synthesis, on DNA condensation in comparison to that of EIP-K plus H2O2. The treatments included control (ddH2O, used as a solvent in the other treatments), 13.75 mM EIP-K plus 3 mM H2O2, and 100 μg/ml CAM. The values are means and SEM, and the number of cells (n) for each treatment is shown in the bar. There is a significant treatment effect for major axis length (one-way ANOVA; F[2,1413] = 526.68; P < 0.001) and form factor (F[2,1413] = 394.99; P < 0.001). Post hoc Scheffé tests (α = 0.05) show that all treatments differ from each other in major axis length, as well as in form factor. The asterisks indicate that the treatments are significantly different from the control.
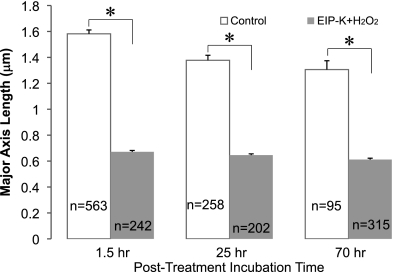
The effect of treatment of cells with EIP-K plus H2O2 for 10 min on nucleoid structure was long lasting. The cells retained similar high degrees of DNA condensation at 1.5, 25, or 70 h after a 10-min treatment with EIP-K plus H2O2 (Fig. 3). The DNA of control cells showed a low degree of condensation at 1.5, 25, or 70 h, and the nucleoid of control cells was significantly longer than that of cells treated with EIP-K plus H2O2 (Fig. 3). Thus, DNA condensation lasts much longer than the time of exposure to EIP-K plus H2O2, with a 10-min treatment causing condensation lasting at least 70 h.
Fig 3.
Brief treatment with EIP-K plus H2O2 causes long-lasting DNA condensation in E. coli MC4100. The treatment was a 10-min incubation in either control (ddH2O; open bars) or 13.75 mM EIP-K plus 3 mM H2O2 (shaded bars), followed by incubation in PBS buffer for either 1.5, 25, or 70 h. The values are means and SEM, and the number of cells (n) for each treatment is shown in the bar. For each treatment time, the control values are significantly greater than the EIP-K plus H2O2 values, as indicated by the asterisks (Students t test; P < 0.001).
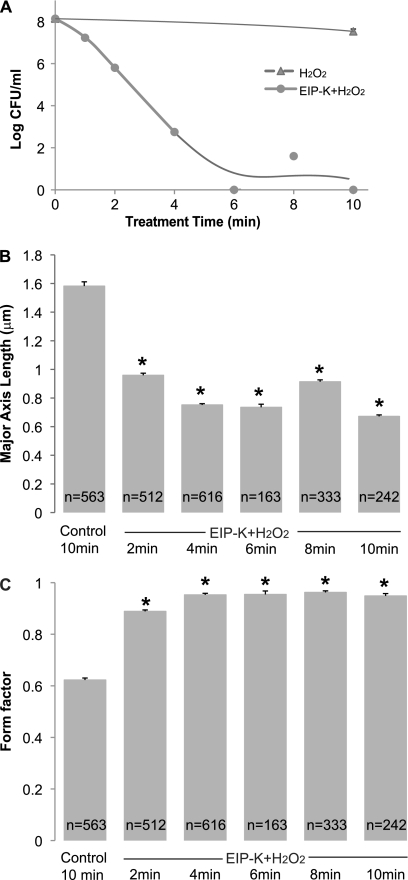
Incubation of E. coli MC4100 in EIP-K plus H2O2 caused changes in both killing and DNA condensation. The shortest treatment time, 2 min, produced a 2-log-unit bactericidal effect, and incubation for 6 to 10 min caused a maximal (8-log-unit) effect (Fig. 4A). Similarly, a 2-min treatment with EIP-K plus H2O2 caused robust DNA condensation (Fig. 4B).
Fig 4.
Effect of time of treatment with 13.75 mM EIP-K plus 3 mM H2O2 on bactericidal activity (A) and DNA condensation (B an C) in E. coli MC4100. The values are means and standard errors of the mean for three experiments, each run in duplicate or triplicate. (A) There is a significant effect of treatment time with EIP-K plus H2O2 on the number of CFU (one-way ANOVA, F[7,8] = 83.86; P < 0.0001), and post hoc Scheffé tests (α = 0.05) show that the untreated group (time = 0) is significantly different than all treated groups. (B and C) The negative control, “control 10 min,” is 10-min exposure to ddH2O, and the number of cells (n) for each treatment is shown in the bars. One-way ANOVA for nucleoid length (B) shows a significant effect of treatment time (F[5,2433] = 320.35; P < 0.001), and post hoc Scheffé tests (α = 0.05) show that the control group is significantly different than the other groups, as indicated by asterisks. One-way ANOVA for form factor (C) shows a significant effect of treatment time (F[5,2433] = 394.99; P < 0.001), and post hoc Scheffé tests (α = 0.05) show that the control group has a nucleoid that is significantly rounder than the other groups, as indicated by asterisks.
E. coli mutants resistant to EIP-K plus H2O2 treatment are mapped at oxyR.
Two E. coli colonies resistant to EIP-K plus H2O2, designated RS1 and RS2, were isolated and identified as described in Materials and Methods. They showed similar phenotypes, so only data for RS1 are described here. RS1 displayed only a very weak bactericidal effect (<1-log-unit reduction in the number of CFU) at various concentrations of either EIP-K or H2O2 (data not shown). Treatment of RS1 cells with 13.75 mM EIP-K plus 3 mM H2O2 also did not cause DNA condensation, as the form factor and length of cells were within 10% of those of wild-type cells (data not shown). The effect of pH on the bactericidal action of EIP-K plus H2O2 was examined for RS1, since pH is an important factor in bactericidal activity for the parental strain (22). For both RS1 and the parental strain, the bactericidal effect of EIP-K plus H2O2 was greater at pH 6 than at pH 8, although the magnitude of the effect was much greater for the parental strain (22) than for RS1 (data not shown).
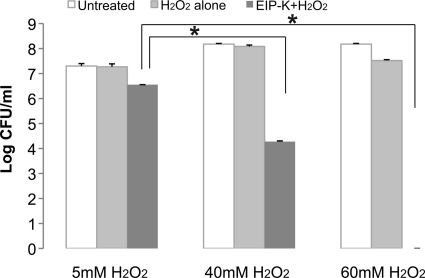
A high concentration of H2O2 is lethal for the parental strain, and 15 mM H2O2 produced a 4-log-unit bactericidal effect (data not shown). Higher concentrations of H2O2 were tested on RS1. For RS1 cells, H2O2 concentrations up to 40 mM were not bactericidal, and 60 mM H2O2 produced only a slight bactericidal effect (<1-log-unit reduction in the number of CFU) (Fig. 5); however, a 4-log-unit bactericidal effect was produced by treatment with 40 mM H2O2 plus 13.75 mM EIP-K, and a maximal effect was produced by 60 mM H2O2 plus 13.75 mM EIP-K (Fig. 5).
Fig 5.
Bactericidal effect of high concentrations of H2O2 with EIP-K on a resistant E. coli strain. White bars, control (untreated; time zero); light-gray bars, treated for 10 min with H2O2 at the indicated concentration; dark-gray bars, treated for 10 min with H2O2 at the indicated concentration plus 13.75 mM EIP-K. The values are means and standard errors of the mean for three experiments, each run in triplicate. Two-way ANOVA shows a significant treatment effect (F[2,13] = 96.02; P = 0.0000001), a significant concentration effect (F[2,13] = 57.15; P = 0.0000001), and a significant treatment-concentration interaction effect (F[4,13] = 26.87; P = 0.000004). The asterisks indicate that the effect of EIP-K plus H2O2 on the resistant strain is significantly affected by the concentration of H2O2.
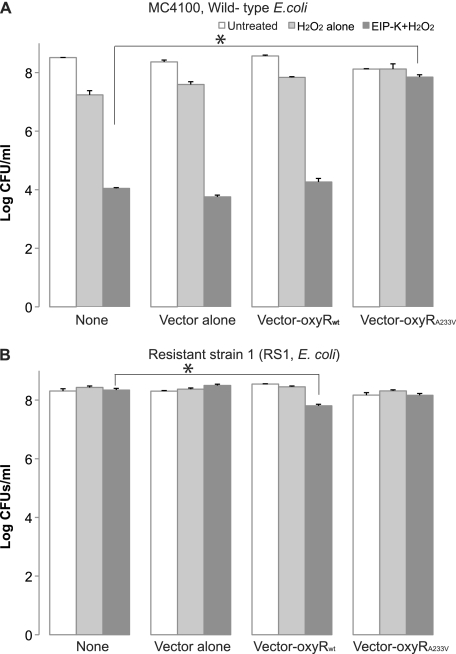
Of the 11 mutations that we identified by genomic sequencing as common to RS1 and RS2, only one was confirmed by high-fidelity PCR. It was a single missense mutation in oxyR, with a C-to-T transition that changes the amino acid in position 233 from alanine to valine [i.e., oxyR(A233V)]. A complementation assay was conducted to determine whether oxyR(A233V) is the sole mutated gene responsible for resistance to EIP-K plus H2O2 in resistant strains. Parental E. coli MC4100 without any plasmid, E. coli MC4100 carrying the empty-vector plasmid, and E. coli MC4100 carrying the wild-type oxyR plasmid were not resistant to EIP-K plus H2O2, but E. coli MC4100 carrying the oxyR(A233V) plasmid showed resistance to treatment (Fig. 6A). Resistance to treatment also occurred when RS1 was used as a host for various plasmids. A small but statistically significant decrease (1 log unit) in the number of CFU occurred for RS1 carrying the wild-type oxyR plasmid (Fig. 6B), probably due to a dose effect from the plasmid. Since an unknown leakage of expression occurred, we were able to observe the plasmid effect on E. coli MC4100, a strain lacking the factor (T7 RNA polymerase) essential to overproduce genes carried by the T7 promoter. This result indicates that the mutation in oxyR(A233V) renders resistance to EIP-K plus H2O2, and a gene dosage effect leads to reduction of resistance for resistant strains carrying wild-type oxyR.
Fig 6.
Complementation assay of E. coli MC4100 (A) and E. coli resistant strain 1 with plasmids containing the wild-type or mutant oxyR gene (B), using pET29a as the vector. The conditions were as follows: no vector (None), vector alone, vector plus parental oxyR (oxyRwt), and vector plus the mutant oxyR gene [oxyR(A233V)]. Each bacterial type was treated with control (ddH2O), 3 mM H2O2, and 13.75 mM EIP-K plus 3 mM H2O2. The values are means and SEM for four experiments, each run in duplicate. The asterisks indicate significant differences between cells without any plasmid (None) and any of the vector conditions under EIP-K-plus-H2O2 treatment. (A) Two-way ANOVA for the wild-type strains shows that the plasmid type effect is not significant (F[3,19] = 2.57; P = 0.085), the treatment effect is significant (F[3,19] = 91.01; P < 0.00000001), and the interaction effect is significant (F[6,19] = 9.02; P < 0.0001). Given the significant interaction effect, we performed a one-way ANOVA for the EIP-K-plus-H2O2 treatment and found a significant effect (F[3,7] = 46.64; P = 0.00005), and Scheffé post hoc tests show that the bactericidal effect of vector plus oxyR(A233V) is significantly less than that of vector plus wild-type oxyR or vector alone. (B) Two-way ANOVA for resistant E. coli strain RS1 shows a significant plasmid effect (F[3,21] = 6.33; P = 0.0031), a significant treatment effect (F[2,21] = 7.38; P = 0.0037), and a significant treatment-plasmid interaction effect (F[6,21] = 13.85; P = 0.000002). A subsequent one-way ANOVA for EIP-K plus H2O2 shows a significant effect (F[3,9] = 22.11; P = 0.00017), and Scheffé post hoc tests show that the bactericidal effect of vector–wild-type oxyR, but not vector-oxyR(A233V) or vector alone, is greater than that of the control.
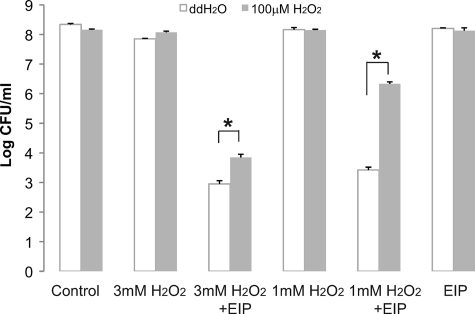
The oxyR(A233V) strain overexpresses antistress proteins, especially oxidative-stress proteins (8), and these antistress proteins can be induced in E. coli by incubation in low concentrations of H2O2 (7). Thus, we wanted to test if our E. coli MC4100 strain can be induced to tolerate higher concentrations of EIP-K plus H2O2 when incubated in a low concentration of H2O2. Indeed, such incubation resulted in E. coli with lower sensitivity to EIP-K plus H2O2 (Fig. 7).
Fig 7.
Effect of adaptation to H2O2 on the bactericidal activity of H2O2 plus EIP-K in E. coli MC4100. Bacteria were pretreated with 100 μM H2O2 or the same amount of ddH2O for 1 h, followed by a bactericidal assay using control (ddH2O), 1 or 3 mM H2O2 alone, or 1 or 3 mM H2O2 plus 13.75 mM EIP-K. The values are means and SEM for one experiment run in duplicate. Two-way ANOVA shows a significant adaptation effect (F[1,19] = 16.48; P = 0.003), a significant treatment effect (F[2,9] = 16.25; P = 0.001), and a significant adaptation-treatment interaction effect (F[2,9] = 16.21; P = 0.001). The asterisks indicate that H2O2 pretreatment reduces the bactericidal effect of H2O2 plus EIP-K.
Evaluation of the roles of DNA-associated proteins in mediating the bactericidal and DNA condensation effects of EIP-K plus H2O2.
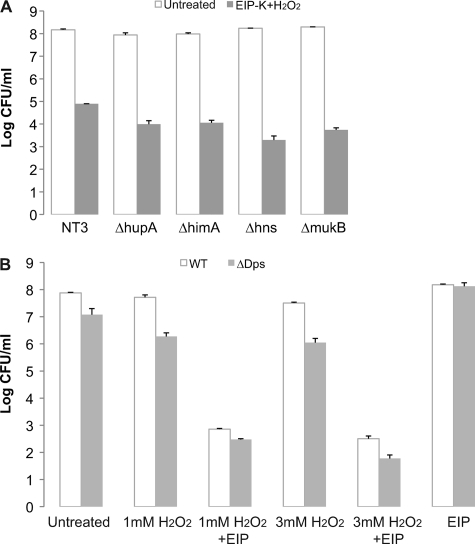
We hypothesized that EIP-K plus H2O2 causes DNA condensation by targeting DNA-binding proteins. Two candidate targets are Dps (DNA-binding protein from starved cells) and H-NS (histone-like nucleoid-structuring protein). According to our hypothesis, H2O2 induces oxidative stress, which triggers OxyR to overexpress Dps, which self-aggregates and binds to DnaA, thus causing DNA condensation (5). Also according to our hypothesis, EIP-K stabilizes the DNA-Dps complex by inhibiting the activity of the Dps-dissociating enzyme, ClpX, or by stabilizing the Dps-DnaA complex and thus preventing DNA replication. EIP-K plus H2O2 might also cause DNA condensation and induce the bactericidal effect through H-NS, since these effects can result from overexpression of H-NS (29). To examine the involvement of Dps, we used a Δdps E. coli mutant strain and overproduced Dps in wild-type E. coli. Our results showed that treatment of the Δdps E. coli strain with EIP-K plus H2O2 caused DNA condensation (data not shown) and bactericidal activity (Fig. 8B) and that EIP-K treatment of cells overproducing Dps did not cause DNA condensation (data not shown) or bactericidal activity (see Fig. S1A in the supplemental material). To test the potential role of H-NS in DNA condensation mediated by EIP-K plus H2O2, we performed experiments with a Δhns E. coli mutant strain. Our results showed that treatment of this strain with EIP-K plus H2O2 caused DNA condensation and bactericidal activity (Fig. 8A).
Fig 8.
Effects of null mutations of DNA- associated proteins (A) and a Dps null mutant strain (B) on bactericidal activity. (A) Treatment with 13.75 mM EIP-K plus 3 mM H2O2 and untreated (number of cells at time zero). The values are means and SEM for two experiments, each run in triplicate. NT3 is the parental strain for the Δhup, ΔhimA, Δhns, and ΔmukB mutants, and ZK126 is the parental strain for the Δdps mutant. Two-way ANOVA shows a significant treatment effect (F[1,21] = 68.96; P = 0.0000001), a nonsignificant mutant effect (F[5,21] = 1.27; P = 0.314), and a nonsignificant treatment-mutant interaction effect (F[6,21] = 1.269; P = 0.313. The lack of asterisks indicates that the mutant strains showed effects similar to those of the wild type under EIP-K-plus-H2O2 treatment. (B) Various concentrations of H2O2, alone or with 13.75 mM EIP-K, and 13.75 mM EIP-K alone were tested on a Dps null mutant strain and compared with the wild-type (WT) strain or the Dps null mutant. The values are means and SEM for one experiment run in duplicate.
We also examined the roles of other DNA-associated proteins by testing other mutant strains defective for a single protein, mutants for one subunit of Hu (a histone-like protein; ΔhupA), one subunit of IHF (integration host factor; ΔhimA), and MukB (a chromosome-partitioning protein; ΔmukB). None of these mutants alone was resistant to EIP-K plus H2O2 (Fig. 8A). Thus, our results support the conclusion that no single DNA-binding protein is necessary to mediate this bactericidal effect of EIP-K plus H2O2, but the possibility still remains that DNA-binding proteins mediate this effect but that any can substitute for the effects of the others.
Temperature stress does not substitute for oxidative stress.
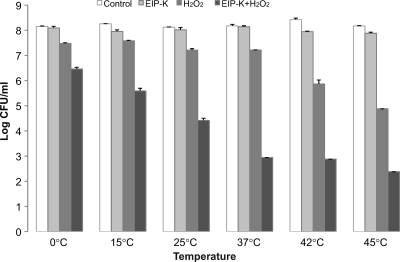
H2O2 generates hydroxyl radicals, which are known to damage cellular components (18, 19). We hypothesized that the bactericidal effect of EIP-K plus H2O2 results in part from the oxidative stress associated with hydroxyl radicals. To examine whether the bactericidal effect is specific to oxidative stress or whether it generalizes to other stressors, we used E. coli MC4100 to examine whether heat or cold shock has the same bactericidal effect as EIP-K plus H2O2. Our results showed that EIP-K alone at any temperature between 0 and 45°C has no significant bactericidal effect (less than 1-log-unit change in CFU) and that H2O2 is required in combination with EIP-K to evoke the bactericidal effect (Fig. 9). Thus, heat or cold shock response does not substitute for oxidative stress from H2O2.
Fig 9.
Effect of temperature on the bactericidal effect of EIP-K plus H2O2. Bacteria were grown at 37°C to exponential phase and then treated with ddH2O (control), 13.75 mM EIP-K, 3 mM H2O2, or 13.75 mM EIP-K plus 3 mM H2O2, each at six temperatures between 0 and 45°C. The values are means and SEM for one experiment run in duplicate. Two-way ANOVA shows a significant treatment effect (F[3,26] = 253.90; P = 0.0000001), a significant temperature effect (F[5,26] = 3.25; P = 0.026), and a significant treatment-temperature interaction effect (F[15,26] = 6.53; P = 0.00002).
Hydrogen peroxide functions as a hydroxyl radical form.
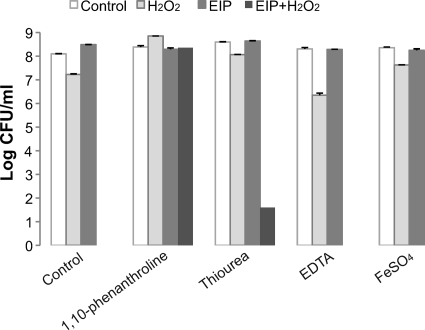
Hydrogen peroxide is reactive with ferrous ions, leading to the generation of hydroxyl radicals (14, 23). To test whether H2O2 or hydroxyl radicals are involved in the bactericidal effect of EIP-K plus H2O2, we used FeSO4 (10 μM) to generate hydroxyl radicals, a ferrous ion chelator (100 μM 1,10-phenanthroline) to prevent the formation of hydroxyl radicals, or a hydroxyl radical scavenger (10 mM thiourea) to remove hydroxyl radicals that were generated (Fig. 10). The bactericidal effect of EIP-K plus H2O2 was eliminated by the presentation of 1,10-phenanthroline and significantly reduced by thiourea. EDTA at the same concentration as 1,10-phenanthroline (100 μM) did not affect bactericidal activity, demonstrating a specific effect for ferrous ions.
Fig 10.
Effects of ferrous ion chelators, hydroxyl radical scavengers, or ferrous ions on the bactericidal activity of EIP-K plus H2O2. Bacteria were pretreated with a ferrous chelator, 1,10-phenanthroline (100 μM), and hydroxyl radical scavengers, mannitol (100 μM) and thiourea (10 mM), for 30 min, followed by treatment with ddH2O (control), 13.75 mM EIP-K, 3 mM H2O2, or 13.75 mM EIP-K plus 3 mM H2O2. EDTA (100 μM) and FeSO4 (10 μM) were presented at the same time as the treatment with escapin products. The values are means and SEM for two experiments, each run in duplicate. Two-way ANOVA shows a significant treatment effect (F[3,23] = 145.34; P = 0.0000001), a significant chelator effect (F[4,23] = 128.50; P = 0.0000001), and a significant treatment-chelator interaction effect (F[12,23] = 58.57; P = 0.0000001).
DISCUSSION
Broad bactericidal activity has been found for escapin, an l-amino acid oxidase, and l-lysine, its substrate, both present in the defensive ink secretion of the sea hare A. californica (34). The products of the escapin-lysine reaction have been identified (10), and among them, H2O2 and EIP-K together account for the powerful bactericidal effect (34). In this study, we examined the mechanism underlying this bactericidal activity in E. coli.
Escapin products cause long-lasting DNA condensation with a time course similar to that of the bactericidal effect.
During the cell cycle, DNA dynamically changes its morphology, unwinding to allow DNA transcription and replication, followed by DNA condensation to allow separation and movement of replicated DNA to form the two daughter cells, again followed by DNA unwinding. Since the growth phases of the cells in our culture system were not synchronized, we observed cells in various stages of DNA condensation. Nonetheless, our results clearly show that brief treatment with the ink secretion of sea hares, and more specifically with EIP-K plus H2O2, causes long-lasting DNA condensation, while treatment with either EIP-K or H2O2 alone does not (Fig. 1 and 2). The condensation results in most (>50%) cells having a spherical and condensed nucleoid (i.e., a form factor of >0.9), which is a form present in only 5% of the control cells. The condensation lasts for at least 70 h after treatment and thus is long lasting. The effect is rapid, as a 2-min exposure causes maximal DNA condensation and also kills 90% of the bacterial cells (Fig. 4).
This long-lasting DNA condensation indicates inhibition of DNA transcription and replication, resulting in an arrested cell cycle and cessation of cell division, even long after removal of the chemical treatment. This bactericidal effect is reminiscent of the E. coli toxin-antitoxin system, where arresting the cell cycle with toxin for more than 6 h in the absence of antitoxin causes cell death (2). This long-lasting DNA condensation associated with cell death also occurs with exposure to other chemicals, such as Cyt1Aa (an insecticidal crystal protein purified from Bacillus thuringiensis) (24) and H-NS (a bacterial DNA-binding protein) (16, 29). It is interesting that the shape of the condensed nucleoid induced by EIP-K plus H2O2 is similar to that induced by these other chemical agents (4). On the other hand, the antibacterial agent chloramphenicol, which inhibits protein synthesis and is bacteriostatic, produces a nucleoid that is condensed, but less so, and with a different shape than that produced by EIP-K plus H2O2: with chloramphenicol, most of the DNA is in the perimeter of the nucleoid and its center is without DNA (11). Collectively, these results suggest that the bactericidal effect of EIP-K plus H2O2 may be similarly mediated by DNA condensation, resulting in inhibition of metabolic reactions, leading to cell death.
OxyR(A233V) confers high resistance to EIP-K plus H2O2.
A change in the amino acid composition or sequence of a protein can alter its structure and consequently lower its affinity for antibiotics, thus giving bacteria containing such proteins more resistance to antibiotics (26, 28). We applied this concept to identify potential targets of EIP-K plus H2O2. In our isolation of strains resistant to EIP-K plus H2O2, bacteria were repeatedly treated with EIP-K plus H2O2 to ensure that they lost sensitivity to the treatment and thus to differentiate between persistent cells and resistant cells. We identified two resistant strains, RS1 and RS2, and showed that they are resistant to H2O2 under conditions lethal to the parental wild-type strain. The parental strain is killed by >10 mM H2O2, but RS1 can tolerate up to 40 mM H2O2 with very little cell death (Fig. 5). Comparison of whole-genome sequences of the parental and mutant strains, coupled with a complementation assay (Fig. 6), showed that both RS1 and RS2 have a single amino acid mutation (A233V) in the oxyR gene and that this gene provides resistance to EIP-K plus H2O2. OxyR is a dual transcriptional regulator protein that senses oxidative stress and regulates antioxidant genes, including the oxidative-stress regulator (oxyR) itself, catalase (katG), alkyl hydroperoxide reductase (ahpC and ahpF), and glutathione reductase (gorA) (31). oxyR is required to sense H2O2 and to induce its regulation, and cells with oxyR deleted are hypersensitive to H2O2 (7). An identical single point mutation of oxyR(A233V) is reported in E. coli strain K-12 and named oxyR2 (8). The oxyR2 mutant strain (also called the constitutive strain) overexpresses OxyR-regulated proteins, such as catalases and peroxidases, in the absence of oxidative stress. Christman et al. showed that oxyR2 has the same level of production of oxyR mRNA as the wild type (8). Therefore, they hypothesized that the alanine-to-valine mutation causes a significant conformational change that affects its regulatory activity. oxyR(A233V) is a constitutive mutant that overproduces anti-oxidative-stress genes, including Dps, catalase (an H2O2 scavenger), and peroxidase genes, under nonstress conditions (7). This may explain the resistance of RS1 and RS2 to EIP-K plus H2O2, since H2O2, an essential component of the bactericidal effect, is removed by overexpression of catalase. This explanation is supported by a lack of bactericidal activity upon treatment of the resistant strains with 40 mM H2O2, but killing occurs at higher concentrations.
The relationship between bactericidal and DNA condensation effects of escapin products.
The close correlation between the bactericidal and DNA condensation effects of EIP-K plus H2O2 suggests that DNA condensation is a potential mechanism behind the bactericidal effect. Is there a link between the oxyR gene and DNA condensation? Does the mutant strain simply eliminate H2O2, or does it also permanently avoid intensive DNA condensation? Knowing this is important in identifying the targets of EIP-K plus H2O2.
Both the bactericidal activity and DNA condensation are absent in the resistant strains, and the complementary assay shows that the single mutated gene oxyR(A233V) is responsible for conferring resistance to EIP-K plus H2O2. This mutation has been widely shown to overexpress anti-stress proteins, especially oxidative-stress proteins (7). Previous studies showed that those anti-stress proteins can be induced in E. coli by incubation in low concentrations of H2O2 (7). Indeed, the bactericidal effect of EIP-K plus H2O2 decreases when bacteria are preincubated in 100 μM H2O2 (Fig. 7). This is similar to the oxyR(A233V) strain, which overproduces catalase, leading to removal of H2O2, which is essential for the bactericidal effect of EIP-K plus H2O2. It is still not clear whether the resistance of this mutant is due to removal of H2O2 or whether the mutant also overexpresses other stress proteins that prevent DNA condensation. However, mutations of oxyR2 [oxyR(A233V), mistakenly called oxyR(A234V) in reference 8] could not be generated by H2O2 treatment alone but could be generated only through random mutagenesis (7, 8). Under our experimental conditions, H2O2 is not lethal, and the damage caused by stress associated with H2O2 is repaired through the bacterial oxidative response. These observations suggest that EIP-K plus H2O2 produce a strong environmental stress response that causes the bactericidal effect or is followed by triggering mutation in genes mediating the global stress regulator oxyR. We hypothesize that the strong environmental stress is oxidative stress. Our results show that temperature stress does not induce the same response (Fig. 9), and thus, the response seems to be specific for oxidative stress.
Mechanisms underlying the effects of escapin products on E. coli.
What are the molecular causes of EIP-K-plus-H2O2-mediated DNA condensation? Although the concentration of H2O2 that we used under our conditions is not lethal to bacteria, the cells may have experienced some degree of oxidative stress. DNA is one target for oxidative stress, and cells have protective mechanisms to ensure DNA fidelity. The most direct protective factors are DNA-binding proteins. Under environmental stress, DNA-binding proteins play important roles in modifying supercoilicity to protect DNA. One DNA-binding protein, Dps, is under OxyR regulation in exponential-phase cells that experience oxidative stress, and Dps is also overexpressed in the oxyR constitutive mutant, oxyR(A233V) (1). Dps self-aggregates, sequesters DNA, and forms a large DNA-Dps condensate to protect cells from oxidative stress. Dps is thought to be a cell cycle checkpoint. Binding of Dps and DnaA, the DNA replication initiator, inhibits the unwinding step and thus interferes with initiation of DNA replication (6). Another DNA-binding protein, H-NS, when overproduced increases the bactericidal activity and causes perfectly spherical DNA condensation (29), as did treatment with EIP-K plus H2O2 in our experiments. H-NS is a repressor of many genes and traps RNA polymerase (10, 16), so when H-NS is overexpressed, it represses metabolic pathways. The H-NS null mutant affects chromosome partitioning and delays initiation of DNA replication (20). Thus, we hypothesized that Dps or H-NS mediates the effect of EIP-K plus H2O2 on DNA condensation. Our tests of this hypothesis, either by using mutant E. coli strains (Δdps and Δhns) or by overexpressing Dps in the wild-type strain, showed neither Dps nor H-NS alone is necessary for EIP-K-plus-H2O2-mediated DNA condensation (Fig. 8; see Fig. S1 in the supplemental material). Our expectation was that the strain overexpressing Dps would be sensitive to EIP-K alone, since the abundance of Dps would mimic the effect of H2O2 and the Δdps mutant that lacked Dps would be resistant to the bactericidal and DNA condensation effects of EIP-K plus H2O2. Our negative results may be due to the ability of Dps, N-HS, and other DNA-binding proteins, such as histone-like proteins or condensin-like proteins, to functionally substitute for each other (33). Consequently, the function of either a Dps or an H-NS null mutant might be substituted for by other DNA-binding proteins.
Hydroxyl radicals may play an important role in the bactericidal activity of EIP-K plus H2O2. Ferrous ions are an essential factor in the production of hydroxyl radicals from hydrogen peroxide (3, 14, 15, 17, 23). We showed that 1,10-phenanthroline, a membrane-permeable ferrous ion chelator, prevents the bactericidal effect while EDTA at the same concentration does not (Fig. 10), demonstrating the specificity of the membrane-permeable chelator for ferrous ion, and not just any ferrous ion. Treatment with thiourea, a remover of hydroxyl radicals, inhibits the bactericidal effect significantly though incompletely (Fig. 10), demonstrating that the effect may be through hydroxyl radicals.
In conclusion, we propose the following model for the bactericidal effect of EIP-K plus H2O2. EIP-K plus H2O2 causes bacterial DNA to condense into a tight spherical nucleoid. A well-organized compacting mechanism, such as that mediated by DNA-binding proteins, is responsible for this effect. This DNA condensation is long lasting, which suggests it is due to damage to DNA-unwinding mechanisms. Arresting the initiation of DNA replication, preventing DNA segregation properties, and negatively affecting enzymes that resolve DNA-binding protein from DNA are possible reasons to keep DNA condensed. Oxidative stress is well known to induce protection by causing bacteria to overproduce anti-oxidative-stress proteins, such as catalase and Dps (1, 7). DNA is a favorite target of ROSs, and DNA condensation is a way to avoid attack from ROSs. EIP-K may play a role in stabilizing the oxidative response from H2O2 and in inducing this long-lasting DNA condensation. In our model, we speculate that EIP-K may affect ClpXP protease, which regulates Dps degradation (30), and then maintain the self-aggregated form of Dps with DNA or maintain Dps-DnaA binding and thus inhibit DNA replication (6). However, we know very little of the effects of EIP-K, and heat shock stress does not induce the same effects as H2O2 when combined with EIP-K. Also, since EIP-K and H2O2 must be present simultaneously to have their effect (22), we hypothesize that the unstable reaction products of H2O2 and EIP-K are the active compounds in this bactericidal effect, and understanding the chemistry of this mixture is important.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jonathan Beckwith (Harvard Medical School), Nancy Trun (Duquesne University), and Roberto Kolter (Harvard Medical School) for providing E. coli MC4100, DNA-associated protein mutants, and the Dps null mutant, respectively. Chung-Dar Lu provided helpful comments throughout the course of the work and on the manuscript.
This work was supported by NSF grants IOS-0324435, -0614685, and -1036742; NIH grant GM-34766; and a grant from the GSU Brains and Behavior Program and by a fellowship from the GSU Molecular Basis of Disease Program.
Footnotes
Published ahead of print 9 January 2012
Supplemental material for this article may be found at http://aac.asm.org.
REFERENCES
- 1. Altuvia S, Almirón M, Huisman G, Kolter R, Storz G. 1994. The dps promoter is activated by OxyR during growth and by IHF and σS in stationary phase. Mol. Microbiol. 13:265–272 [DOI] [PubMed] [Google Scholar]
- 2. Amitai S, Yassin Y, Engelberg-Kulka H. 2004. MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 186:8295–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ananthaswamy HN, Eisenstark A. 1977. Repair of hydrogen peroxide-induced single-strand breaks in Escherichia coli deoxyribonucleic acid. J. Bacteriol. 130:187–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bloomfield VA. 1996. DNA condensation. Curr. Opin. Struct. Biol. 6:334–341 [DOI] [PubMed] [Google Scholar]
- 5. Ceci P, et al. 2004. DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus. Nucleic Acids Res. 32:5935–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chodavarapu S, Gomez R, Vicente M, Kaguni JM. 2008. Escherichia coli Dps interacts with DnaA protein to impede initiation: a model of adaptive mutation. Mol. Microbiol. 67:1331–1346 [DOI] [PubMed] [Google Scholar]
- 7. Christman MF, Morgan RW, Jacobson FS, Ames BN. 1985. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41:753–762 [DOI] [PubMed] [Google Scholar]
- 8. Christman MF, Storz G, Ames BN. 1989. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc. Natl. Acad. Sci. U. S. A. 86:3484–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cummins SF, et al. 2004. Characterization of Aplysia enticin and temptin, two novel water-borne protein pheromones that act in concert with attractin to stimulate mate attraction. J. Biol. Chem. 279:25614–25622 [DOI] [PubMed] [Google Scholar]
- 10. Dame RT, Wyman C, Wurm R, Wagner R, Goosen N. 2002. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 277:2146–2150 [DOI] [PubMed] [Google Scholar]
- 11. Daneo-Moore L, Higgins ML. 1972. Morphokinetic reaction of Streptococcus faecalis (ATCC 9790) cells to the specific inhibition of macromolecular synthesis: nucleoid condensation on the inhibition of protein synthesis. J. Bacteriol. 109:1210–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Derby CD. 2007. Escape by inking and secreting: marine molluscs avoid predators through a rich array of chemicals and mechanisms. Biol. Bull. 213:274–289 [DOI] [PubMed] [Google Scholar]
- 13. Derby CD, Kicklighter CE, Johnson PM, Zhang X. 2007. Chemical composition of inks of diverse marine molluscs suggests convergent chemical defenses. J. Chem. Ecol. 33:1105–1113 [DOI] [PubMed] [Google Scholar]
- 14. Dwyer DJ, Kohanski MA, Collins JJ. 2009. Role of reactive oxygen species in antibiotic action and resistance. Curr. Opin. Microbiol. 12:482–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Farr SB, Kogoma T. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55:561–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grainger DC, Goldberg MD, Lee DJ, Busby SJ. 2008. Selective repression by Fis and H-NS at the Escherichia coli dps promoter. Mol. Microbiol. 68:1366–1377 [DOI] [PubMed] [Google Scholar]
- 17. Imlay JA, Linn S. 1986. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J. Bacteriol. 166:519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Imlay JA, Linn S. 1988. DNA damage and oxygen radical toxicity. Science 240:1302–1309 [DOI] [PubMed] [Google Scholar]
- 19. Imlay JA, Chin SM, Linn S. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640–642 [DOI] [PubMed] [Google Scholar]
- 20. Kaidow A, Wachi M, Nakamura J, Magae J, Nagai K. 1995. Anucleate cell production by Escherichia coli Δhns mutant lacking a histone-like protein, H-NS. J. Bacteriol. 177:3589–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kamio M, et al. 2009. The chemistry of escapin: identification and quantification of the components in the complex mixture generated by an l-amino acid oxidase in the defensive secretion of the sea snail Aplysia californica. Chem. Eur. J. 15:1597–1603 [DOI] [PubMed] [Google Scholar]
- 22. Ko K-C, Wang B, Tai PC, Derby CD. 2008. Identification of potent bactericidal compounds produced by escapin, an l-amino acid oxidase in the ink of the sea hare Aplysia californica. Antimicrob. Agents Chemother. 52:4455–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8:423–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manasherob R, et al. 2003. Compaction of the Escherichia coli nucleoid caused by Cyt1Aa. Microbiology 149:3553–3564 [DOI] [PubMed] [Google Scholar]
- 25. Molina-Quintero LR, Lucas-Elío P, Sanchez-Amat A. 2010. Regulation of the Marinomonas mediterranea antimicrobial protein lysine oxidase by l-lysine and the sensor histidine kinase PpoS. Appl. Environ. Microbiol. 76:6141–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ng EY, Trucksis M, Hooper DC. 1996. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shabani S, Yaldiz S, Vu L, Derby CD. 2007. Acidity enhances the effectiveness of active chemical defensive secretions of sea hares, Aplysia californica, against spiny lobsters, Panulirus interruptus. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 193:1195–1204 [DOI] [PubMed] [Google Scholar]
- 28. Spratt BG. 1994. Resistance to antibiotics mediated by target alterations. Science 264:388–393 [DOI] [PubMed] [Google Scholar]
- 29. Spurio R, et al. 1992. Lethal overproduction of the Escherichia coli nucleoid protein H-NS: ultramicroscopic and molecular autopsy. Mol. Gen. Genet. 231:201–211 [DOI] [PubMed] [Google Scholar]
- 30. Stephani K, Weichart D, Hengge R. 2003. Dynamic control of Dps protein levels by ClpXP and ClpAP proteases in Escherichia coli. Mol. Microbiol. 49:1605–1614 [DOI] [PubMed] [Google Scholar]
- 31. Storz G, Tartaglia LA, Ames BN. 1990. The OxyR regulon. Antonie Van Leeuwenhoek 58:157–161 [DOI] [PubMed] [Google Scholar]
- 32. Tong H, Chen W, Shi W, Qi F, Dong X. 2008. SO-LAAO, a novel l-amino acid oxidase that enables Streptococcus oligofermentans to outcompete Streptococcus mutans by generating H2O2 from peptone. J. Bacteriol. 190:4716–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weinstein-Fischer D, Elgrably-Weiss M, Altuvia SS. 2000. Escherichia coli response to hydrogen peroxide: a role for DNA supercoiling, topoisomerase I and Fis. Mol. Microbiol. 35:1413–1420 [DOI] [PubMed] [Google Scholar]
- 34. Yang H, et al. 2005. Cloning, characterization and expression of escapin, a broadly antimicrobial FAD-containing l-amino acid oxidase from ink of the sea hare Aplysia californica. J. Exp. Biol. 208:3609–3622 [DOI] [PubMed] [Google Scholar]
- 35. Zusman DR, Carbonell A, Haga JY. 1973. Nucleoid condensation and cell division in Escherichia coli MX74T2 ts52 after inhibition of protein synthesis. J. Bacteriol. 115:1167–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.