LETTER
Acquired metallo-β-lactamases (MBLs) are emerging resistance determinants of great clinical concern. They are found in Enterobacteriaceae, Pseudomonas aeruginosa, and other Gram-negative nonfermenters, in which they can confer a very broad spectrum of β-lactam resistance, including the expanded-spectrum cephalosporins and carbapenems, that cannot be reverted by β-lactamase inhibitors currently available for clinical use (2).
The VIM-type enzymes were the second acquired MBLs to be detected, in the late 1990s in Europe (5, 10), and are currently the most widespread in terms of geographical distribution and range of host bacterial species (2). VIM-1 and VIM-2 were the first reported VIM-type allelic variants, in P. aeruginosa isolated in 1997 in Verona, Italy (index strain VR-143/97) (5) and in 1996 in Marseilles, France, respectively (10).
We report here on the characterization of a Pseudomonas clinical isolate producing the VIM-1 MBL, isolated in Genoa, Italy, in 1994, which to our best knowledge represents the earliest known VIM-producing strain.
Pseudomonas sp. AM/94 was isolated in November 1994 from the lower respiratory tract of an inpatient admitted in the general intensive care unit of Genoa University Hospital. The isolate was at the time identified as Pseudomonas fluorescens by the API 20E system (bioMérieux, Marcy l'Etoile, France), with a low grade of discrimination (62%). Reidentification of the isolate by Vitek-2 (bioMérieux) yielded a result of low discrimination between P. aeruginosa (34%), P. fluorescens (33%), and Pseudomonas putida (33%). Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF) analysis, using a Microflex LT benchtop MALDI-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany), identified the isolate as Pseudomonas monteilii (score, 2.03). Molecular identification by amplification and sequencing of 16S rRNA, rpoD, rpoB, and gyrB genes unambiguously identified AM/94 as Pseudomonas mosselii, a recently recognized species of the P. putida group (3, 7).
Susceptibility testing, carried out with Etest (bioMérieux) or with broth microdilution (1) and interpreted according to the EUCAST breakpoints for Pseudomonas spp. (document version 1.3, 2011-01-05, http://www.eucast.org/clinical_breakpoints, accessed October 2011), revealed that AM/94 was resistant to all tested antibiotics except amikacin and colistin (Table 1).
Table 1.
Antimicrobial susceptibility profile of the P. mosselii strain AM/94
| Antibiotica | MIC (mg/liter) |
|---|---|
| Piperacillin-tazobactam* | >128 |
| Trimethoprim-sulfamethoxazole | >128 |
| Aztreonam | 32 |
| Ceftazidime | >128 |
| Cefepime | >128 |
| Imipenem | 64 |
| Meropenem | 64 |
| Amikacin* | 2 |
| Gentamicin* | 16 |
| Ciprofloxacin | 2 |
| Levofloxacin | 4 |
| Colistin | 0.5 |
MICs were determined by broth microdilution or by Etest (*).
MBL activity, assayed spectrophotometrically (5), was detected in a crude extract of AM/94 (specific imipenemase activity was 120 ± 1 nmol/min/mg protein, inhibited by >80% after exposure to 2 mM EDTA).
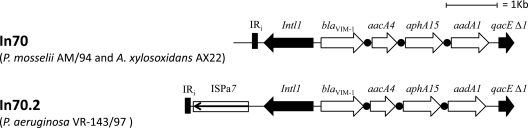
Analysis of acquired MBL genes, including blaVIM, blaIMP, and blaNDM, using PCR and sequencing (4) revealed the presence of the blaVIM-1 allele. PCR mapping and sequencing of the genetic context of the blaVIM-1 gene (12) revealed that the gene was carried on a gene cassette inserted into a class 1 integron whose variable region contained four cassettes (blaVIM-1, aacA4, aphA15, and aadA1) and was identical to that of In70-type integrons previously described in other VIM-1-producing strains from Italy (12). Mapping the region of the cognate Tn402-like transposon flanking the integrase gene, however, revealed that the structure was identical to that flanking In70 detected in plasmid pAX22 from Achromobacter xylosoxidans AX22 (11): it was lacking the ISPa7 insertion sequence present downstream of the intl1 gene in In70.2, the blaVIM-1-containing integron from the VR-143/97 P. aeruginosa index strain (12) (Fig. 1).
Fig 1.
Map of the integron In70 of P. mosselii AM/94, identical to In70 of A. xylosoxidans AX22 (11), compared with the In70.2 variant carried by P. aeruginosa VR-143/97 (12). Open reading frames are indicated by arrows; the attC recombination sites of gene cassettes are indicated by circles.
All together, present results indicate that influx of the blaVIM-1 MBL gene in the clinical setting started at least since the early 1990s and that In70 might have been the original genetic element involved in this influx.
P. mosselii is an environmental species detected in rhizospheric soil (8, 9) and is an overall unusual human opportunistic pathogen. In literature, only one report describes P. mosselii as a cause of infection (a prosthetic valve endocarditis) in human (6). Similar environmental species occasionally acting as opportunistic pathogens most probably play a role as shuttles for acquired MBL genes from their as-yet-unknown natural reservoirs to the clinical setting.
ACKNOWLEDGMENTS
This work was partially supported by funding from the European Commission under the 7th Framework Programme (TROCAR contract HEALTH-F3-2008-223031 and TEMPOtest-QC contract HEALTH-2009-241742).
Footnotes
Published ahead of print 30 January 2012
Contributor Information
Tommaso Giani, Department of Biotechnologies Section of Microbiology University of Siena Siena, Italy.
Erika Coppo, Section of Microbiology Department of Surgical and Diagnostic Integrated Science (DISC) University of Genoa Genoa, Italy.
Vesselina Kroumova, Laboratory of Microbiology and Virology University Hospital Maggiore della Carità Novara, Italy.
Gian Maria Rossolini, Department of Biotechnologies Section of Microbiology University of Siena Siena, Italy.
REFERENCES
- 1. Clinical and Laboratory Standards Institute 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 2. Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect. Dis. 11:381–393 [DOI] [PubMed] [Google Scholar]
- 3. Dabboussi F, et al. 2002. Pseudomonas mosselii sp. nov., a novel species isolated from clinical specimens. Int. J. Syst. Evol. Microbiol. 52:363–376 [DOI] [PubMed] [Google Scholar]
- 4. D'Andrea M, et al. 2011. Persistent carriage and infection by multidrug-resistant Escherichia coli ST405 producing NDM-1 carbapenemase: report on the first Italian cases. J. Clin. Microbiol. 49:2755–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lauretti L, et al. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mclellan E, Partridge D. 2009. Prosthetic valve endocarditis caused by Pseudomonas mosselii. J. Med. Microbiol. 58:144–145 [DOI] [PubMed] [Google Scholar]
- 7. Mulet M, Lalucat JJ, García-Valdés E. 2010. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 12:1512–1530 [DOI] [PubMed] [Google Scholar]
- 8. Naik P, Raman G, Narayanan K, Sakthivel N. 2008. Assessment of genetic and functional diversity of phosphate solubilizing fluorescent pseudomonads isolated from rhizospheric soil. BMC Microbiol. 8:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Naik P, Sahoo N, Goswami D, Ayyadurai N, Sakthivel N. 2008. Genetic and functional diversity among fluorescent pseudomonads isolated from the rhizosphere of banana. Microb. Ecol. 56:492–504 [DOI] [PubMed] [Google Scholar]
- 10. Poirel L, et al. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Riccio M, Pallecchi L, Fontana R, Rossolini GM. 2001. In70 of plasmid pAX22, a blaVIM-1-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob. Agents Chemother. 45:1249–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Riccio M, et al. 2005. Clonal relatedness and conserved integron structures in epidemiologically unrelated Pseudomonas aeruginosa strains producing the VIM-1 metallo-β-lactamase from different Italian hospitals. Antimicrob. Agents Chemother. 49:104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]