Abstract
Bacterial resistance to the glycopeptide antibiotic teicoplanin shows some important differences from the closely related compound vancomycin. They are currently poorly understood but may reflect significant differences in the mode of action of each antibiotic. Streptomyces coelicolor possesses a vanRSJKHAX gene cluster that when expressed confers resistance to both vancomycin and teicoplanin. The resistance to vancomycin is mediated by the enzymes encoded by vanKHAX, but not by vanJ. vanHAX effect a reprogramming of peptidoglycan biosynthesis, which is considered to be generic, conferring resistance to all glycopeptide antibiotics. Here, we show that vanKHAX are not in fact required for teicoplanin resistance in S. coelicolor, which instead is mediated solely by vanJ. vanJ is shown to encode a membrane protein oriented with its C-terminal active site exposed to the extracytoplasmic space. VanJ also confers resistance to the teicoplanin-like antibiotics ristocetin and A47934 and to a broad range of semisynthetic teicoplanin derivatives, but not generally to antibiotics or semisynthetic derivatives with vancomycin-like structures. vanJ homologues are found ubiquitously in streptomycetes and include staP from the Streptomyces toyocaensis A47934 biosynthetic gene cluster. While overexpression of staP also conferred resistance to teicoplanin, similar expression of other vanJ homologues (SCO2255, SCO7017, and SAV5946) did not. The vanJ and staP orthologues, therefore, appear to represent a subset of a larger protein family whose members have acquired specialist roles in antibiotic resistance. Future characterization of the divergent enzymatic activity within this new family will contribute to defining the molecular mechanisms important for teicoplanin activity and resistance.
INTRODUCTION
The development of resistance to existing antibiotics, coupled with a sustained decline in the success rate for the discovery of new ones, is leading to a point in the future where many infections could essentially be untreatable by the compounds available. A detailed understanding of the molecular mechanisms by which antibiotics can fail to be active is vital knowledge for the future design of new, more effective compounds. Such information is often linked intimately to the drug's mode of action and therefore provides unique insights that can be used to help devise novel compounds or new ways of prolonging the therapeutic usefulness of existing ones. The glycopeptide antibiotics vancomycin and teicoplanin are currently especially reserved in the clinic for the last-resort treatment of infections resistant to the antibiotics in mainstream use. Glycopeptide antibiotics inhibit bacterial cell wall biosynthesis, and both vancomycin and teicoplanin are known to bind to the d-alanyl-d-alanine (d-Ala-d-Ala) terminus of peptidoglycan (PG) precursors and so block the formation of a mature PG cell wall (8). However, the two antibiotics show some important differences in their structures and activities (discussed below), and to date, only resistance to vancomycin has been characterized in detail.
Inducible resistance to vancomycin is due to the activity of resistance genes clustered together either on the bacterial chromosome or on transmissible plasmids (3, 15). The number of genes present in the resistance cluster can vary, but the “core” cluster consists of five genes, vanSRHAX. The VanHAX proteins are required for remodeling cell wall precursor biosynthesis to produce molecules with d-alanyl-d-lactate (d-Ala-d-Lac) termini that exhibit significantly lower binding affinity for glycopeptide antibiotics, thus drastically reducing their antimicrobial effects (2, 5, 10). Activation of the transcription of the vanHAX genes is usually regulated by a VanR/VanS two-component response regulator/sensor histidine kinase system. Teicoplanin and vancomycin differ in the structures of their aglycones (the peptide of the molecule), in their glycosylation patterns, and in the presence of a long fatty acid chain attached to teicoplanin that is absent in vancomycin (12, 36, 41) (Fig. 1). In the clinic, the most commonly encountered vancomycin-resistant enterococcal infections have been classified as VanA or VanB type: VanA strains also exhibit inducible resistance to teicoplanin but VanB strains do not (1). It has been proposed that the observed teicoplanin sensitivity of VanB strains is due to the fact that teicoplanin fails to induce the sensor kinase in the resistance cluster, VanSB (6, 7). How teicoplanin can escape recognition by VanSB, resulting in failure to trigger the resistance system and keeping the cell sensitive to teicoplanin, is not clear, although it has been suggested that the lipid moiety can serve to anchor teicoplanin in the bacterial membrane and physically prevent it from interacting productively with the VanS sensor domain (9, 12, 41). In addition, through the chemoenzymatic synthesis of a spectrum of teicoplanin and vancomycin derivatives, Dong and colleagues showed definitively that the key functional difference between teicoplanin and vancomycin is the presence or absence of the lipid moiety: removal of the lipid from teicoplanin prevents it from killing VanB-type enterococci, whereas addition of an appropriate lipid side chain to vancomycin makes it an effective antibiotic against VanB strains (13). Three lines of evidence also suggest that the presence of a lipid side chain contributes to a difference in the mode of action for teicoplanin compared to vancomycin: (i) teicoplanin is more active against a variety of Gram-positive bacteria than vancomycin; (ii) teicoplanin and vancomycin bind to d-Ala-d-Ala and inhibit both transpeptidation and transglycosylation, but vancomycin exerts its major effect on transpeptidation, whereas lipidated glycopeptides, such as teicoplanin, inhibit transglycosylation more strongly; (iii) teicoplanin resistance requires a more complete elimination of d-Ala-d-Ala-ending precursors than vancomycin resistance (5, 11, 16, 28, 43). Arthur et al. demonstrated that a membrane protein gene, vanZ, present in the resistance cluster of clinical isolates of VanA-type enterococci, is responsible for low-level teicoplanin resistance but did not characterize its biological function (4). Three other separate studies of teicoplanin resistance in clinical isolates of staphylococci and enterococci each identified a membrane protein (all with similar sizes between 35 and 41 kDa) that was highly expressed in the resistant strains and specifically responsible for the teicoplanin resistance (20, 34, 42). However, the biological functions of these membrane proteins have also not been studied further, and even the sequence of each protein has not been revealed. Thus, the exact mode of action of teicoplanin and the mechanism of its resistance is currently incompletely understood.
Fig 1.
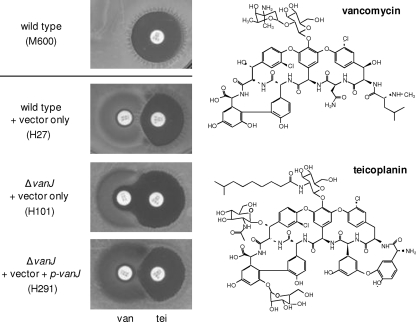
vanJ encodes novel teicoplanin resistance. Paper discs containing vancomycin (30 μg/ml) (van) and teicoplanin (30 μg/ml) (tei) were placed adjacent to each other on freshly spread lawns of S. coelicolor wild-type, ΔvanJ null mutant, or vanJ complemented strains. Bioassay plates were incubated for 2 days at 30°C prior to imaging, and the change in shape of the growth-inhibitory halo around the teicoplanin disc in strain H101 indicates the resistance activity of VanJ (as discussed in the text). The structures of vancomycin and teicoplanin are shown on the right.
In previous work, we studied the molecular mechanisms of resistance to glycopeptide antibiotics using a model actinomycete, Streptomyces coelicolor (23). It was the first case of glycopeptide antibiotic resistance identified and characterized in detail in a nonpathogenic, non-glycopeptide-producing bacterium. It is, however, a close relative of most of the known glycopeptide antibiotic producer strains and shows a pattern of susceptibility to glycopeptide antibiotics similar to that of the VanB-type Enterococcus, i.e., it is sensitive to teicoplanin but highly resistant to vancomycin. S. coelicolor possesses an inducible cluster of seven genes, vanSRJKHAX, in which the VanS sensor is activated by vancomycin but not by teicoplanin (23, 31). The cluster contains the interesting novel features vanJ and vanK, and with the exception of vanJ, all the van genes in S. coelicolor have been studied in detail, and their roles in resistance to vancomycin have been determined (22, 24, 25). vanJ has no counterpart among previously characterized glycopeptide resistance genes (including the teicoplanin resistance activities discussed above) and is predicted to encode a membrane protein of unknown function. Here, we present the first detailed investigation of the role of vanJ in conferring resistance to glycopeptide antibiotics. We report for the first time a family of vanJ homologues that are present ubiquitously in streptomycete genomes and compare the functions of a selected number of these homologues with that of vanJ. We envision that a detailed study of the functions of vanJ and its homologues will significantly extend our understanding of resistance to glycopeptide antibiotics and also of their mode of action.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains and plasmids used in this study are described and listed in Table 1. All the gene complementation and gene overexpression strains created in this study were constructed by conjugal transfer from Escherichia coli strain ET12567(pUZ8002) carrying the appropriate pMS81 or pIJ10257 derivative. Exconjugants were selected with hygromycin (80 μg/ml). For liquid culture, spores of S. coelicolor strains were germinated by heat shock treatment (23) prior to inoculation into the stated medium. Minimal liquid medium (NMMP) (29) was used for the growth curve and microscopic observation of S. coelicolor cells. For protein preparation, S. coelicolor cells were grown for 16 h in tryptic soy broth (TSB) liquid medium. Except where described below, media and culture conditions were as given previously (29).
Table 1.
Plasmids and bacterial strains used in this study
| Plasmids and strains | Description or genotype | Source or reference |
|---|---|---|
| Plasmids | ||
| pMS81 | ϕBT1 attP-int-derived integration vector for the conjugal transfer of DNA from E. coli to Streptomyces spp. (Hygr) | 17 |
| pIJ10257 | 330-bp ermEp (KpnI-PstI) with ribosome binding site and multicloning site from pIJ8723 cloned into pMS81 cut with KpnI-NsiI (Hygr) | 24 |
| pIJ790 | Modified λRED recombination plasmid pKD20 | 18 |
| pGN030 | pMS81 carrying vanJ with its own promoter sequence (p-vanJ) | This study |
| pGN037 | pMS81 carrying p-vanJ with a C-terminal 6-histidine tag (C-6His tag); a stop codon (UGA) was introduced in frame immediately before the annotated start codon | This study |
| pGN038 | pMS81 carrying vanJp with a C-6His tag | This study |
| pGN034 | Short vanJ (annotated vanJ sequence) with a C-6His tag cloned in pIJ10257 under ermEp control | This study |
| pGN035 | Long vanJ (including an extra 117 bp at the 5′ end) with a C-6His tag cloned in pIJ10257 under ermEp control | This study |
| pDU001 | vanS cloned in pIJ10257 under ermEp control | Novotna and Hong (unpublished) |
| pGN091 | vanS with a C-6His tag cloned in pIJ10257 under ermEp control | This study |
| pGN024 | Long vanJ cloned in pIJ10257 under ermEp control | This study |
| pGN026 | Short staP (annotated staP sequence) cloned in pIJ10257 under ermEp control | This study |
| pGN027 | Long staP (including an extra 246 bp at the 5′ end) cloned in pIJ10257 under ermEp control | This study |
| pKV33 | SCO2255 gene cloned in pIJ10257 under ermEp control | This study |
| pKV35 | SCO7017 gene cloned in pIJ10257 under ermEp control | This study |
| pCH32 | SAV5946 gene cloned in pIJ10257 under ermEp control | This study |
| Streptomyces strains | ||
| S. coelicolor M600 | SCP1− SCP2− | 29 |
| S. coelicolor H27 | M600 + pMS81 | This study |
| S. coelicolor H2077 | M600 + pIJ10257 | This study |
| S. coelicolor H2050 | M600 + pGN024 | This study |
| S. coelicolor H357 | M600 + pKV33 | This study |
| S. coelicolor H358 | M600 + pKV35 | This study |
| S. coelicolor H359 | M600 + pCH32 | This study |
| S. coelicolor H2141 | M600 + pGN026 | This study |
| S. coelicolor H2148 | M600 + pGN027 | This study |
| S. coelicolor J3201 | ΔvanRS SCP1− SCP2− | 25 |
| S. coelicolor H102 | J3201 + pIJ10257 | This study |
| S. coelicolor H243 | J3201 + pGN024 | This study |
| S. coelioclor H2135 | J3201 + pGN026 | This study |
| S. coelicolor H2143 | J3201 + pGN027 | This study |
| S. coelicolor J3220 | ΔvanJ::apr SCP1− SCP2− | 23 |
| S. coelicolor H101 | J3220 + pMS81 | This study |
| S. coelicolor H215 | J3220 + pIJ10257 | This study |
| S. coelicolor H291 | J3220 + pGN030 | This study |
| S. coelicolor H2389 | J3220 + pGN038 | This study |
| S. coelicolor H2391 | J3220 + pGN037 | This study |
| S. coelicolor H2087 | J3220 + pGN024 | This study |
| S. coelicolor H2280 | J3220 + pGN034 | This study |
| S. coelicolor H2281 | J3220 + pGN035 | This study |
| S. coelicolor H2137 | J3220 + pGN026 | This study |
| S. coelicolor H2145 | J3220 + pGN027 | This study |
| S. coelicolor J3200 | ΔvanS SCP1− SCP2− | 25 |
| S. coelicolor H2276 | J3200 + pGN091 | This study |
| S. coelicolor J3226 | ΔvanHAX::apr SCP1− SCP2− | 23 |
| S. coelicolor H2270 | J3226 + pIJ10257 | This study |
| S. coelicolor H256 | J3226 + pGN024 | This study |
| S. coelicolor H2139 | J3226 + pGN026 | This study |
| S. coelicolor H2147 | J3226 + pGN027 | This study |
| S. coelicolor H296 | ΔSCO2255::apr SCP1− SCP2− | This study |
| S. coelicolor H347 | ΔSCO7017::apr SCP1− SCP2− | This study |
| S. toyocaensis NRRL15009 | A47934 producer; wild type | 37 |
| S. avermitilis ATCC 31267 | Avermectin producer; wild type | 44 |
| S. avermitilis H334 | ΔSAV5946::apr | This study |
| E. coli strains | ||
| ET12567(pUZ8002) | ET12567 containing helper plasmid pUZ8002 | 35 |
| BW25113(pIJ790) | BW25113 containing helper plasmid pIJ790 | 18 |
Antibiotic susceptibility tests.
All susceptibility tests, including evaluation of MICs, were performed on MMCGT agar medium (23). For MIC determination, confluent lawns of approximately 107 spores were spread onto MMCGT plates containing a range of different antibiotic concentrations, and the results were evaluated after 5 days of incubation at 30°C. For disc diffusion bioassays, commercial antibiotic discs were purchased from Oxoid if available, placed onto freshly spread agar plate spore lawns, and incubated for 2 days at 30°C. Where necessary, homemade antibiotic discs were prepared by applying the desired concentration of antibiotic stock solution onto 6-mm-diameter paper discs purchased from Whatman. Balhimycin was a kind gift from Wolfgang Wohlleben. Chloroeremomycin was a kind gift from Dudley Williams. A47934 was a kind gift from Gerard Wright. The chemoenzymatically synthesized vancomycin and teicoplanin derivatives used for this study were kindly provided by Daniel Kahne, Chris Walsh, and their coworkers. All other antibiotics used for this study were purchased from Sigma-Aldrich.
Construction of mutant strains.
SCO2255, SCO7017, and SAV5946 null mutant strains, in which the entire coding sequence of each gene was replaced with a cassette carrying the apramycin resistance gene (apr) and oriT of RK2, were constructed by PCR targeting of cosmids 1G2 (for SCO2255), 1H10 (for SCO7017), and CL_222_D10 (for SAV5946) using the published method (18). Briefly, each cosmid was introduced into E. coli BW25113 carrying pIJ790, and the target gene was disrupted by electroporation of the cells with the PCR-amplified apr-oriT cassette, generated using primers carrying the appropriate gene-specific extensions (using the oligonucleotides with KO as part of the name in Table 2). The disruption was confirmed by restriction digestion and PCR analysis (using the oligonucleotides with TEST as part of the name in Table 2) of isolated cosmid DNA. The resulting cosmids (1G2/SCO2255::apr, 1H10/SCO7017::apr, and CL_222_D10/SAV5946::apr) were then introduced into E. coli ET12567 carrying pUZ8002 and transferred into S. coelicolor M600 and Streptomyces avermitilis (for the SAV5946 null mutant) by conjugation. Exconjugants of S. coelicolor were selected with 50 μg/ml apramycin on soya flour mannitol (SFM) agar (29), while exconjugants of S. avermitilis were selected with 5 μg/ml apramycin on M4 agar (30). The resulting mutant strains, generated by double-crossover interaction, were identified by their apramycin-resistant (Aprr), kanamycin-sensitive (Kans) phenotype and designated as listed in Table 1.
Table 2.
Oligonucleotides used in this study
| Name of oligonucleotide | Nucleotide sequence (5′–3′)a |
|---|---|
| vanJ-L stop c113g anti | CCACCCGTCACAGCCGTCTCCGAG |
| vanJ-L stop c113 | CTCGGAGACGGCTGTGACGGGTGG |
| pJsc EcoNI IF F | CGCGGATCGCCTCCGCCAGGTAGGGCTCGTCCTCGAC |
| pJsc C-6HIS HindIII IF R | CCTAGGATCCAAGCTTTCA GTGGTGGTGGTGGTGGTGCCAGCTGACCCCCGCCGCCAC |
| vanJsc C-tag Xmnl F | AGGCGGCCGGGAACGGGTTCGGCTTCACCTGGCCGGCGAAG |
| vanJsc C-6His HindIII R | CCTAGGATCCAAGCTTTCAGTGGTGGTGGTGGTGGTGCCAGCTGACCCCCGCCGC |
| vanSsc C-tag StuI F | TCAGCCCCCACCAGGCCTCGACCCTCACCGAACCCTTC |
| vanSsc C-6His HindIII R | CCTAGGATCCAAGCTTTCAGTGGTGGTGGTGGTGGTGCCTGCCGGTGTGCGGAGC |
| LvanJ-I | CATATGCGTCAGACCTCACCACCA |
| vanJ II | CTCGAGTCACCAGCTGACCCCCGCCGC |
| vanJst I | CATATGGAGACCTTCCTGCCGTG |
| vanJst-L I | CATATGATCAATGAGCACCTGCG |
| vanJst II | GATATCTTAATTAATCACCAGCTGATTCGGGCCG |
| 2255 KO I | GGCGACGGTGTGCACAGCAGGCAGTGAGGCAACGGTATGATTCCGGGGATCCGTCGACC |
| 2255 KO II | CAAAGACTCTCAGGGCGGGTATTCCGCAGACGAATTCTATGTAGGCTGGAGCTGCTTC |
| 7017 KO I | CCGGGTCGGGCGAGCTGGACGCGCGGGCGCGGACGGGTGATTCCGGGGATCCGTCGACC |
| 7017 KO II | ACGAAAGGCTAACGCAAACACCGCAAGCGGCTTGCTTCATGTAGGCTGGAGCTGCTTA |
| SCO2255 FLANK F | CATATGGCGCAGCAGGCGTACAT |
| SCO2255 FLANK R | TTAATTAAGACGAATTCTAGGAAGCCG |
| SCO7017L FLANK F | CATATGGTGGAGCGGGTGGGCCGT |
| SCO7017 FLANK R | TTAATTAAGGCTTGCTTCAGTCCCAGG |
| SCO2255 TEST F | CGGCACAGTGCGGAACTCC |
| SCO2255 TEST R | GGTCTCGCCTTTCGAGAGC |
| SCO7017 TEST F | TTCGGTGGTGAAGACGACG |
| SCO7017 TEST R | CCCCGGTTTGCCGTTGAGC |
| SAV_5946 KO F | AGTGACGGTGTGCACGGCAGGTTGTGAGGCGACGGTATGTGTAGGCTG GACCTGCTTC |
| SAV_5946 KO R | ACGGAACAAATAGGCTCGGGCCGAGTATTCCACGAGCTAATTCCGGGGATCCGTCGACC |
| SAV_5946 TEST F | ACCACTAATGTGCGCGTTTC |
| SAV_5946 TEST R | AGAGGCCTTTCACGAGCAAG |
| SAV_5946 pIJ10257 F | AATTCATATGGCGCAGGCGTACGTGAC |
| SAV_5946 pIJ10257 R | AATTAAGCTTCTAGGAAGCAGTTGCGGAG |
Restriction sites used for cloning are underlined.
Construction of vectors for complementation and gene overexpression.
To construct pGN030, a plasmid to test complementation in trans of the vanJ mutant, a 1.2-kb DNA fragment containing vanJ, including its own promoter sequence, was obtained by PstI digestion of cosmid H66 and ligated into the NsiI site of pMS81. To drive constitutive expression of vanJ and other homologues in S. coelicolor, the appropriate gene (long vanJ, short and long staP, SCO2255, SCO7017, and SAV5946) was cloned in pIJ10257 (24) under the control of the ermE* promoter (ermEp), resulting in plasmids pGN024, pGN026, pGN027, pKV33, pKV35, and pCH32, respectively. For this cloning, each gene was first amplified by PCR using the primers in Table 2 to incorporate appropriate upstream and downstream restriction sites. All PCR products were initially cloned into the vector pGEM-T Easy (Promega) and verified by sequencing prior to restriction digestion and ligation into pIJ10257. For Western blot analysis, constructs pGN034, pGN035, pGN037, pGN038, and pGN091, expressing proteins with six histidines (His tag) at the C terminus, were prepared from their nontagged counterparts using the In-Fusion PCR cloning system (Clontech). A restriction fragment carrying the 3′ end of a gene was replaced by a corresponding PCR product in which the sequence for six histidines had been introduced immediately upstream of the stop codon. To create constructs pGN034 and pGN035, Xmnl/HindIII fragments of pGN023 and pGN024 were replaced by a PCR product amplified with primers vanJsc C-tag Xmnl F and vanJsc C-6His HindIII R. In order to create the construct pGN038, an EcoNI/HindIII fragment of pGN030 was replaced by a PCR product amplified with primers pJsc EcoNI IF F and pJsc C-6HIS HindIII IF R. In the pGN091 construct, a StuI/HindIII fragment of pDU001 was replaced by a PCR product amplified with primers vanSsc C-tag StuI F and vanSsc C-6His HindIII R (Table 2). Construct pGN037, carrying p-vanJ with a C-terminal His tag and with a premature stop codon introduced immediately upstream of the annotated start codon, was prepared from pGN030 as described above, except that the EcoNI/HindIII fragment in one step was replaced by the two PCR products, which overlap the site of desired mutation, creating a stop codon. The first PCR product was amplified with primers pJsc EcoNI IF F and vanJ-L stop c113g anti and the second with primers vanJ-L stop c113 and pJsc C-6HIS HindIII IF R. The design of In-Fusion primers (Table 2) and vector-fragment recombination was performed according to the manufacturers' recommendations. The genomic DNA was used as a template for all PCR amplification described here. The resulting plasmids were verified by sequencing and introduced into appropriate strains by conjugal transformation (Table 1).
Cell fractionation and Western blot analysis.
Cells from 15 ml of culture in TSB medium were harvested by centrifugation and washed in a buffer containing 50 mM Tris-Cl (pH 7.6) and 150 mM NaCl. The wet cell pellet was then weighed and resuspended in 3 ml of 50 mM Tris-Cl (pH 7.6) containing 1 mM dithiothreitol and 1× protease inhibitor cocktail (Complete; Roche) for each gram of the pellet. The cells were disrupted by ultrasonication (9 times for 5 s each time at an amplitude of 7 μm) using a Soniprep 150 disintegrator (MSE; United Kingdom) at 30-s intervals to allow the suspension to cool down. Unbroken cells and debris were removed by centrifugation at 16,000 × g for 30 min at 4°C. A portion of the lysate was stored at −20°C for analysis as the whole-cell fraction. Membranes were separated from the cytosol by ultracentrifugation at 60,000 × g for 45 min at 4°C in a Beckman Coulter Optima L-100 ultracentrifuge and resuspended in an equal volume of 50 mM Tris-Cl (pH 7.6) containing 1% SDS and 1× protease inhibitor cocktail. Equal volumes of cell fractions were mixed with 6× sample loading buffer and were boiled prior to being loaded onto a 10% polyacrylamide gel. SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblot transfer onto a polyvinylidene difluoride (PVDF) membrane (Immobilon P; Millipore) were performed using standard methods. For detection of His-tagged proteins, a 1/10,000 dilution of a mouse His-tagged monoclonal antibody (Novagen) was used. The secondary antibody was stabilized peroxidase-conjugated goat anti-mouse (H+L) in 1/2,000 dilution (Thermo Scientific). Signal was detected by incubation of membranes with Immobilon Western chemiluminescent horseradish peroxidase (HRP) substrate (Millipore) and by subsequent exposure (5 to 30 min) to X-ray film (Amersham Hyperfilm ECL; GE Healthcare).
Protoplast shaving.
Protoplasts were prepared as described previously (26). Briefly, cells from 7.5 ml of TSB culture were washed and resuspended in 4 ml of P buffer (29) containing 1× protease inhibitor cocktail (Complete; Roche) and 2 mg/ml lysozyme. After 1 h of incubation at 30°C, the protoplasts were filtered through cotton wool and centrifuged at 1,500 × g for 10 min at 4°C. The protoplast pellet was then resuspended in 200 μl of P buffer, and 50-μl aliquots were incubated with 2 μg of trypsin (Sequencing Grade Modified Trypsin; Promega) for 15 to 90 min at 30°C. For the negative control, identical incubation conditions were used, but without the addition of trypsin. When required, 1% Triton X-100 was added before incubation. Digestion was stopped by addition of 1× protease inhibitor cocktail. Protoplasts were harvested (1,500 × g; 5 min; 4°C), and the supernatant was stored for analysis as an indication of spontaneous lysis during the treatment. The protoplast pellet was resuspended in P buffer containing protease inhibitors and lysed by sonication on ice for two bursts of 2 s each at an amplitude of 5 μm, separated by 20-s rests on ice. For the trypsin digestion of intact cells, the same procedure was followed, but without the lysozyme treatment step. All samples were stored at −80°C until analysis by immunoblotting as described above.
RESULTS
vanJ encodes novel teicoplanin resistance.
Although S. coelicolor carries a complete set of glycopeptide resistance genes, it is sensitive to teicoplanin, because teicoplanin fails to induce expression of the van gene cluster (23, 31). Exposure of a growing lawn of S. coelicolor spores to a paper disc containing teicoplanin, therefore, results in a large circular halo of growth inhibition immediately surrounding the disc (Fig. 1, top). However, when discs containing vancomycin and teicoplanin were analyzed next to each other on the bioassay plate, the presence of vancomycin caused a D-shaped (rather than the expected O-shaped) inhibitory zone to appear around the teicoplanin disc (Fig. 1, second from top). This indicates that the wild-type cells growing in the presence of vancomycin as a result of the induction of van gene expression were also now able to grow in the presence of teicoplanin. In contrast, when the same assay was performed using a lawn of the vanJ null mutant strain, an O-shaped zone around teicoplanin was produced (Fig. 1, third from top). The D-shaped inhibitory-zone phenotype was fully restored by supplying a copy of vanJ in trans (Fig. 1, bottom), indicating the absence of any polar effects on van gene cluster transcription in the mutant strain and suggesting that vanJ is responsible for the observed teicoplanin resistance.
Vancomycin-induced expression of vanHAX does not confer resistance to teicoplanin in S. coelicolor.
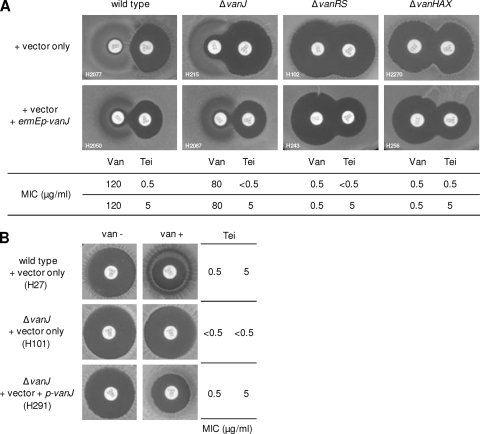
Although the data in Fig. 1 suggest that vanJ is responsible for teicoplanin resistance, it does not exclude the possibility that resistance arises from a combination of both vanJ and vanHAX expression. To distinguish these possibilities, we constructed a vanJ overexpression vector, pGN024, in which a single copy of vanJ is expressed from the strong constitutive ermE* promoter (ermEp). When introduced into both the S. coelicolor M600 wild type and the ΔvanJ null mutant strains, pGN024 resulted in a marked reduction in the size of the halo of growth inhibition surrounding the teicoplanin disc and a corresponding 10-fold increase in the MIC of the drug (Fig. 2A). No significant changes in the response to vancomycin were observed. Interestingly, similar results were obtained following overexpression of vanJ in ΔvanHAX or ΔvanRS mutant strains, indicating that vanKHAX gene expression is not required for the observed increase in resistance to teicoplanin and confirming that vanJ expression is incapable of conferring vancomycin resistance (Fig. 2A). The ΔvanHAX strain lacks the VanHAX enzymes, while the ΔvanRS strain lacks the VanR/VanS two-component response regulator system essential for switching on the expression of all the chromosomal van genes. Neither strain is therefore capable of reprogramming its cell wall biosynthesis to produce d-Ala-d-Lac PG precursors, an event conventionally thought to be crucial for resistance to all glycopeptide family antibiotics (so that the ΔvanHAX and ΔvanRS strains are completely sensitive to vancomycin [Fig. 2A]). This reprogramming is clearly not primarily required for the teicoplanin resistance observed in this experiment, since constitutive expression of vanJ alone was sufficient to increase resistance to teicoplanin in the absence of either vanHAX or vanRS. To confirm this result further, we scored the levels of teicoplanin resistance in wild-type, ΔvanJ mutant, and vanJ complement strains in the presence and absence of vancomycin in the medium. As shown in Fig. 2B, an increased level of teicoplanin resistance (indicated by a reduction in the size of the inhibitory halo around the teicoplanin disc) was observed only when both vanJ (i.e., in strains H27 and H291) and vancomycin were present. When vanJ is absent (strain H101), activation of expression of the vanRSKHAX genes by vancomycin has no effect on the level of teicoplanin resistance. The MIC levels determined for these strains confirm that the presence of a functional VanJ protein increases resistance to teicoplanin by about 10-fold (Fig. 2B). This is in good agreement with the results obtained for ΔvanJ in Fig. 1, where intact copies of the vanRS, vanK, and vanHAX genes are present and can be expressed to wild-type levels following induction by vancomycin. Transcriptional analysis has confirmed that the vanHAX operon can be strongly expressed in the absence of vanJ (data not shown), and the observed high level of resistance to vancomycin in the ΔvanJ mutant is fully consistent with this. All these data indicate that vanJ encodes a novel teicoplanin resistance activity and that this resistance to teicoplanin is completely independent of the reprogramming of cell wall PG precursor biosynthesis by the VanHAX proteins. The mode of action of teicoplanin must therefore be significantly different from that of vancomycin.
Fig 2.
Teicoplanin resistance in S. coelicolor is due to the activity of VanJ and not that of VanHAX. (A) Constitutive expression of vanJ increases resistance to teicoplanin. S. coelicolor wild-type, ΔvanJ, ΔvanRS, and ΔvanHAX strains expressing ermEp-vanJ increased resistance to teicoplanin but had no effect on vancomycin. The MICs for vancomycin and teicoplanin of these strains are shown and correlate with the bioassay results. (B) Teicoplanin resistance in wild-type, ΔvanJ null mutant, and vanJ complemented strains were scored in the presence and absence of vancomycin. Teicoplanin (30 μg/ml) was applied to a paper disc placed on a fresh lawn of spores of each strain spread on MMCGT agar containing (van +) or lacking (van −) 10 μg/ml vancomycin and incubated for 2 days at 30°C. The MIC of teicoplanin under these conditions is shown. As in Fig. 1, H291 contains vanJ under the control of its native promoter and therefore requires vancomycin for its induction.
Overexpression of vanJ increases resistance to a broad range of teicoplanin subfamily glycopeptide antibiotics.
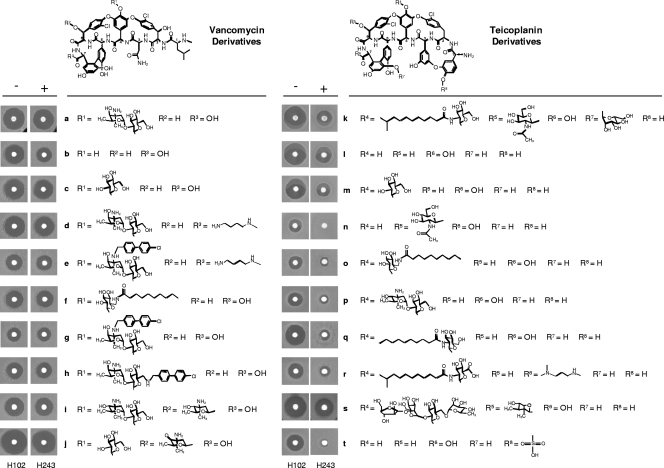
To determine the specificity of VanJ in conferring resistance to the antibiotic activity of glycopeptides with differing chemical structures, bioassay disc experiments were performed using a range of teicoplanin and vancomycin derivative compounds (11, 13, 14, 16, 28). For the indicator strain, overexpression of vanJ in a ΔvanRS mutant background, as opposed to a wild-type background, avoided any confusion with possible effects mediated via induction of expression of the other van genes (vanKHAX) by the compound under test. Constitutive expression of vanJ markedly increased resistance to all the teicoplanin derivatives tested in the bioassay (Fig. 3). In contrast, there was little or no protective effect against exposure to the vancomycin derivatives, with the exception of the vancomycin aglycone (Fig. 3b). All the known glycopeptide antibiotics have a core structure consisting of a linear heptapeptide backbone with a polysaccharide moiety attached to the fourth amino acid residue from the N terminus. For example, vancomycin (Fig. 3a) has a disaccharide at residue 4, while ristocetin A (Fig. 3s) contains a tetrasaccharide. Chloroeremomycin (Fig. 3i) is similar in structure to vancomycin but differs by having an additional monosaccharide moiety attached to the sixth amino acid residue. As such, the majority of glycopeptide antibiotics can be classified into two types based on the nature of the residues at positions 1 and 3. Vancomycin (Fig. 3a); chloroeremomycin (Fig. 3i), and balhimycin (Fig. 3j) are assigned to the same group, since they possess nonaromatic amino acids in these positions. Members of the second group possess aromatic side chains within these amino acid residues that can be cross-linked to each other. Teicoplanin (Fig. 3k), ristocetin A (Fig. 3s), and A47934 (Fig. 3t) all possess cross-linked aromatic groups bridged by a biaryl ether oxygen atom between the two aromatic chains (32). The common feature of all the teicoplanin derivatives against which VanJ was active in the bioassay is the biaryl ether oxygen atom between the two aromatic chains in residues 1 and 3, the structures of which otherwise differ in their patterns of glycosylation and/or lipidation.
Fig 3.
Structure-activity bioassay experiments characterizing the efficacy of VanJ. Paper discs containing approximately 30 μg/ml of each glycopeptide shown were placed on freshly spread lawns of a ΔvanRS indicator strain carrying the ermEp-vanJ overexpression plasmid pGN024 (+, strain H243) or only the empty vector (−, strain H102). Plates are shown after 2 days of incubation at 30°C.
In addition to the glycopeptide antibiotics, a range of other antibiotics known to target cell wall biosynthesis (penicillin G, tunicamycin, bacitracin, flavomycin, fosfomycin, and d-cycloserine), the cell membrane (polymixin B), or intracellular targets (apramycin, rifampin, and thiostrepton) were also tested in the bioassay, but overexpression of vanJ had no effect on the level of resistance to any (data not shown). The protective effect of VanJ function is therefore specific to glycopeptide antibiotics related to teicoplanin.
Characterization of VanJ. (i) VanJ is encoded by a leaderless message.
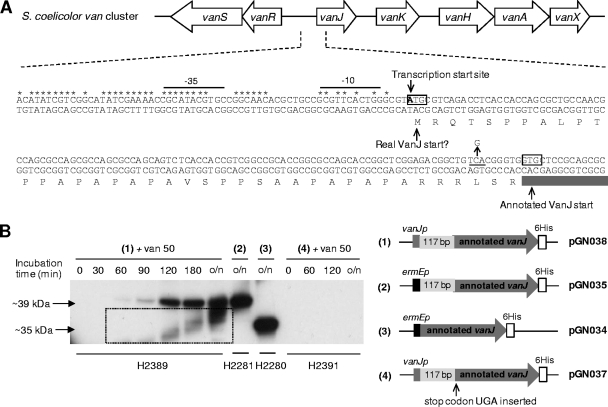
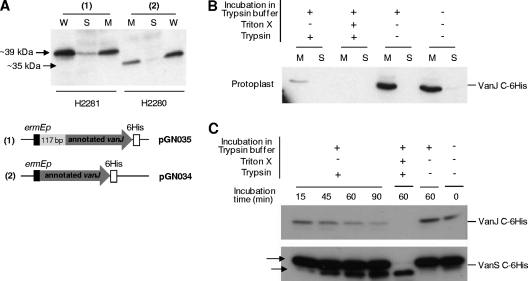
Previous analysis of the transcriptional regulation of vanRSJKHAX expression in S. coelicolor identified four VanR-dependent transcription units, vanRS, vanJ, vanK, and vanHAX (23). With the exception of the vanJ transcript, the start site for each coincided with the A of the annotated AUG translation initiation codon, indicating that of the four, only translation of the vanJ message is mediated via a conventional 5′ mRNA leader and ribosome binding site. However, transcription of vanJ does commence with an AUG codon that is 117 bp upstream and in frame with the annotated translational start site (Fig. 4A), raising the possibility that it could encode two proteins from the alternative translation initiation codons. To investigate this, plasmids pGN034 and pGN035, encoding C-terminal His-tagged versions of the short (the length of the annotated vanJ gene) and long forms of vanJ (respectively) expressed from the constitutive ermEp, were constructed and transformed into the ΔvanJ mutant strain. Both constructs were able to complement the vanJ mutant phenotype, but complementation by the shorter annotated version was only partial (data not shown). Immunoblot analysis using an anti-His antibody detected proteins with apparent molecular masses of 35 kDa and 39 kDa, confirming correct expression of the different protein forms in the two strains (Fig. 4B, lanes in sections 2 and 3). Comparison with a strain expressing a similarly His-tagged version of VanJ, but from its natural promoter sequence, detected only the 39-kDa protein band following induction of transcription by exposure to vancomycin (Fig. 4B, lanes in section 1). Consistent with this, introduction of a UGA translational stop codon between the two putative translational start sites (Fig. 4A) yielded a construct that both failed to complement the ΔvanJ strain and failed to yield any detectable protein following vancomycin treatment (Fig. 4B, lanes in section 4). Translation of the vanJ transcript therefore occurs only from the most upstream AUG start codon, and like the other van gene transcripts, it is therefore also a leaderless message. Kaberdina et al. recently reported that exposure of E. coli to the antibiotic kasugamycin caused the production of abnormally small 61S ribosomes that, strikingly, were proficient in selectively translating leaderless mRNA in preference to transcripts possessing a conventional 5′ mRNA leader sequence. Perhaps the exclusive use of leaderless messages to encode all the proteins in the van gene cluster represents a strategy to ensure their efficient translation, even in the event of any antibiotic-induced damage to ribosomes (27).
Fig 4.
(A) Genetic organization of the vancomycin resistance cluster in S. coelicolor showing details of the nucleotide sequence and annotation of the 5′ end of vanJ and its upstream region. Nucleotides in the conserved van promoter region are marked with asterisks, and the putative −35 and −10 sequences are indicated. The transcription start site (23) is shown, together with the annotated and alternative VanJ translational start codons. Amino acids encoded only from the alternative (most upstream) start codon are shaded in light gray, and the C-to-G mutation introducing a TGA translational stop codon between the two initiator codons is indicated with an arrow. (B) VanJ is expressed only from the upstream alternative start codon and is encoded by a leaderless message. His-tagged VanJ proteins expressed in the ΔvanJ null mutant strain using the plasmids illustrated were detected by immunoblot analysis with an anti-His antibody. For strains carrying pGN037 or pGN038, proteins were extracted from samples taken immediately before (0 min) and at the indicated times after the induction of expression by treatment with 50 μg/ml vancomycin. o/n indicates cultures were incubated overnight (approximately 18 h). Bands appearing in the box are an artifact arising from the exposure of the film.
(ii) VanJ is membrane localized, and its putative active site is oriented to face outside the cell.
Secondary-structure analysis predicts that VanJ contains three transmembrane α-helices in its N terminus (TMpred [http://www.ch.embnet.org]) and a conserved pfam03372 endonuclease/exonuclease/phosphatase domain at its C terminus. To analyze the subcellular localization of VanJ and to determine the orientation of the putative enzyme active site, strain H2281 constitutively expressing a C-terminally His-tagged version of the VanJ protein was subjected to cell fractionation and protoplast-shaving experiments (Fig. 5). Cell disruption, followed by ultracentrifugation of the crude cell extracts to separate the cytosol and membrane fractions, indicated that His-tagged VanJ localized almost exclusively to the membrane fraction (Fig. 5A, first 3 lanes). Analysis of strain H2280, expressing the shortened form of the VanJ protein, produced a similar result (Fig. 5A, second 3 lanes). The short form of VanJ possesses all three predicted transmembrane domains and is capable of partial complementation of the vanJ mutant phenotype, so its correct localization was not altogether surprising. Membrane-localized VanJ could be oriented so that the enzyme domain contained within its C terminus, which presumably mediates the novel teicoplanin resistance activity, is located either intra- or extracellularly. To determine this orientation, protoplasts were isolated from strain H2281, expressing the long form of VanJ bearing a His tag at its C terminus. Trypsin was then used to shave off the portion of VanJ that is exposed extracellularly on the protoplasts' surfaces, similar to previous studies (26). Compared to an untreated control, trypsin digestion dramatically decreased the amount of VanJ protein detected in the membrane fraction isolated from the protoplasts, suggesting that the active C terminus of VanJ is exposed on the outside of the cytoplasmic membrane (Fig. 5B). VanJ was not detectable in the soluble fraction obtained after protoplast incubation in the absence of trypsin, indicating the absence of protoplast lysis under the incubation conditions used. Simultaneous treatment of protoplasts with both trypsin and the detergent Triton X, however, resulted in the complete disappearance of VanJ from both the membrane and soluble fractions due to protoplast lysis. Similar experiments with intact cells (nonprotoplasted) always resulted in comparable amounts of VanJ in the isolated membrane fraction, indicating that trypsin cannot access the VanJ protein in the cell membrane in the presence of an intact cell wall (data not shown). To confirm these observations further, a time course trypsin digestion experiment was performed, and the results were compared to those of a control experiment similarly following the change in abundance of a C-terminally His-tagged VanS protein (Fig. 5C). The VanS sensor kinase is also known to be membrane localized (31) and possesses two domains, a sensor loop that is exposed on the outside of the membrane and a C-terminal cytoplasmic kinase. As expected, the His-tagged VanS protein was readily detectable during the entire 90 min of trypsin exposure due to protection of the C-terminal tag from digestion by localization within the cell. In contrast, His-tagged VanJ protein decreased in abundance during incubation and was almost undetectable after 90 min. Interestingly, two different bands corresponding to His-tagged VanS were observed during the experiment, one corresponding to the size of the full-length VanS protein and a smaller band that increased in abundance during the trypsin incubation. The externally located sensory loop of VanS contains a trypsin cleavage site, and the appearance and size of the low-molecular-weight form in the experiment is consistent with digestion at this site removing the N terminus of the protein. These control experiments fully support the conclusion that the C terminus of VanJ is oriented to the external face of the membrane. This orientation, importantly, limits the identity of potential substrates that the enzyme can come into contact with and informs the rational use of an in vitro screening approach to determine the nature of the substrate.
Fig 5.
(A) VanJ is predominantly localized in the membrane. Western immunoblot analysis indicates that the vast majority of His-tagged VanJ protein detectable in whole cells (W) subfractionates to the membrane fraction (M) and not the soluble cytoplasmic fraction (S). Both the short (annotated) and long (experimentally determined) forms of VanJ are membrane localized. (B) Protoplast-shaving experiments demonstrate that VanJ is exposed on the external face of the membrane. Protoplasts prepared from the H2281 strain expressing His-tagged VanJ were incubated with or without trypsin (as indicated) before being lysed and separated into membrane (M) and soluble cytoplasmic (S) fractions. Immunoblotting against the C-terminally located His tag indicates it is removed on exposure of the protoplasts to trypsin, consistent with external localization. (C) Time course protoplast-shaving experiments showing the gradual disappearance of His-tagged VanJ following treatment with trypsin. Experiments were performed as described for panel B and are compared to a similar analysis of His-tagged VanS (using strain H2276) as both a positive and a negative control. The arrows indicate both the appearance of a smaller N-terminally truncated form of VanS (positive control) and the net persistence of the two detectable forms of His-tagged VanS due to intracellular localization of the C-terminal tag (negative control).
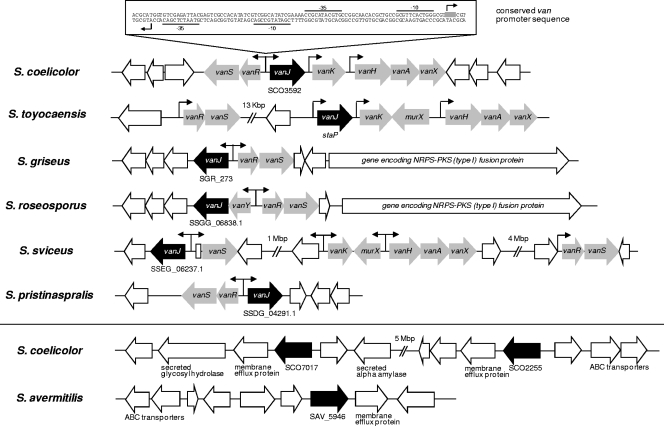
VanJ homologues are ubiquitous in actinomycetes.
Although VanJ has no counterpart in previously characterized antibiotic resistance proteins, two homologues, SCO7017 and SCO2255, are present in the S. coelicolor chromosome. Both are predicted to be membrane proteins with unknown functions and to possess the Pfam endonuclease/exonuclease/phosphatase domain at their C termini. Through the recent explosion in microbial genome sequencing, many more homologues (40 to 80% amino acid identity) to VanJ have been found, all residing in actinomycete species. Interestingly, the closest homologues (70 to 80% amino acid identity) are predominantly located adjacent to predicted vanRS two-component systems (often also with associated vanHAX genes) and possess a van promoter motif sequence, implying a glycopeptide resistance function analogous to that of VanJ in S. coelicolor (Fig. 6). In addition, in Streptomyces griseus and Streptomyces roseosporus, the vanRSJ genes are located immediately adjacent to putative secondary-metabolite-biosynthetic clusters that could conceivably encode the production of unknown antibiotic compounds. The homologues with lower identity (40 to 60%), including SCO7017 and SCO2255, all lack the van promoter sequence motif, but both their predicted transmembrane domains and their genomic contexts are indicative of roles in cell envelope physiology. Consistent with this, the results of transcriptome analyses indicate that expression of both of these homologues is significantly induced following exposure to three antibiotics that target distinct stages of cell wall biosynthesis: vancomycin, moenomycin A, and bacitracin (21). In the same experiment, transcription of vanJ was induced only by vancomycin, suggesting that all three homologues have roles to play in combating cell wall damage caused by the antibiotics but that they are regulated differently. In contrast to S. coelicolor, S. avermitilis possesses only a single vanJ homologue (SAV5946) and no close orthologues of vanRS. It is not resistant to vancomycin. Bioinformatics analysis suggests that SAV5946 is orthologous to SCO2255; the nucleotide sequences of both their open reading frames (ORFs) and putative promoter regions are almost identical.
Fig 6.
Homologues (black arrows) of VanJ are widespread in Streptomyces strains. Above the line, the genetic context of putative orthologues that show high sequence homology (70 to 80% amino acid identity) to VanJ and are located adjacent to close homologues of other van resistance genes (gray arrows) is shown. The locations and orientations of conserved van promoter sequences are marked with arrows. Below the line, the context of the other VanJ homologues present in S. coelicolor and S. avermitilis that exhibit a lower sequence identity (50 to 60%), as discussed in the text, is shown.
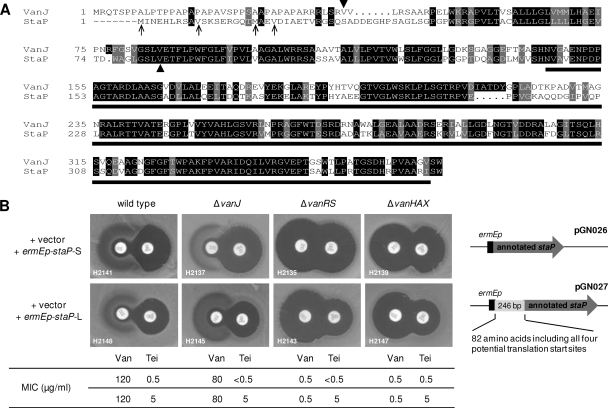
Functional characterization of selected VanJ homologues. (i) S. toyocaensis StaP is orthologous to VanJ.
The gene cluster responsible for the biosynthesis of the glycopeptide antibiotic A47934 in Streptomyces toyocaensis possesses a homologue of S. coelicolor vanJ named staP (37). VanJ and StaP share 75% amino acid sequence identity but are least homologous toward their N termini, where the identity of the translation start site for StaP is uncertain from sequence data alone (Fig. 7A). Constitutive ermEp-driven expression of the annotated (short) staP sequence in the S. ceolicolor ΔvanJ, ΔvanHAX, and ΔvanRS mutant strains had no effect in a teicoplanin resistance bioassay (Fig. 7B). In contrast, however, similar expression of a longer sequence that included the most upstream of four possible alternative translational start codons increased teicoplanin resistance to levels comparable to those of constitutive vanJ expression. StaP and VanJ are therefore functional orthologues, although the published translational start site for StaP has previously been misassigned.
Fig 7.
(A) Amino acid sequence alignment of VanJ and StaP. The annotated translational start sites of both VanJ and StaP are marked with arrowheads, and the alternative translational start sites in the StaP sequence are indicated by arrows. The black bar beneath the amino acid sequence alignment represents the conserved pfam03372 endonuclease/exonuclease/phosphatase domain. (B) StaP is an orthologue of VanJ. Constitutive expression of the annotated staP gene (pGN026) in S. coelicolor has no effect on teicoplanin resistance, but constitutive expression of staP encoding all four potential translational start codons (pGN027) increased resistance to teicoplanin, analogous to the results for VanJ shown in Fig. 2A. MIC values are indicated beneath the bioassay plates, and the staP gene inserts present in pGN026 and pGN027 are illustrated on the right.
(ii) SCO2255 and SCO7017 in S. coelicolor and SAV5946 in S. avermitilis do not confer teicoplanin resistance.
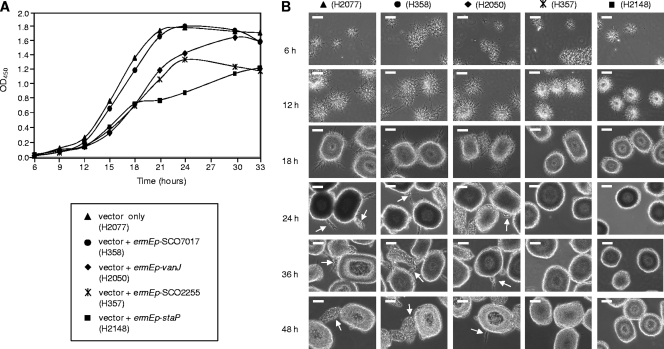
Constitutive ermEp-driven expression of the vanJ homologue SCO2255, SCO7017, or SAV5946 in the teicoplanin resistance bioassay using the S. coelicolor wild-type strain did not result in any measurable change in resistance (data not shown). The overexpression strains were similarly unaltered in their resistance to a range of other glycopeptide antibiotics tested, and the constructed deletion mutant strains also exhibited no change in their susceptibilities to antibiotic treatments (data not shown). None of these homologues, therefore, encode the same teicoplanin resistance activity as VanJ and StaP. Interestingly, however, overexpression of either SCO2255 or staP in S. coelicolor resulted in a marked inhibition of growth in liquid culture (Fig. 8A). Microscopy indicated that this coincided with noticeable changes in the appearance of the mycelia in the cultures, with the strains expressing SCO2255 and staP exhibiting mycelial pellets that were both noticeably smoother and smaller than the strains that grew more normally (Fig. 8B). In liquid culture, spores germinate to produce mycelial hyphae that grow in a radial pattern to form a pellet that eventually becomes almost smooth on its surface. As the diameter of the pellet increases, new mycelial hyphae emerge through the smooth periphery (as indicated by arrows in Fig. 8B), giving rise to a second phase of active growth (33). Strains overexpressing SCO2255 or staP are defective in the latter process, and the mycelial pellets remain smooth in appearance (Fig. 8B). In addition, we were not able to obtain any transformants when attempting to introduce the staP overexpression vector pGN027 into S. toyocaensis, but the empty pIJ10257 vector transformed readily, and also, overexpression of SAV5946 (an orthologue of SCO2255) in S. coelicolor severely reduced colony size, indicative of growth inhibition (data not shown). Although the molecular basis for these observations is as yet unclear, these data likely reflect interesting differences in roles or potencies between the different homologues.
Fig 8.
Overexpression of certain VanJ homologues results in the inhibition of growth of S. coelicolor during liquid culture. (A) Growth curves in NMMP liquid medium of the S. coelicolor M600 wild-type strain overexpressing vanJ (diamond), staP (square), SCO2255 (asterisk), or SCO7017 (circle) or carrying only the empty vector (triangle). Germinated spores of each strain were inoculated into 50 ml of NMMP, and the absorbance at 450 nm was measured every 3 h. The results shown are the average of three cultures. (B) Microscopic observation of the appearance and size of mycelial clumps formed during the growth of each strain. Samples were taken at the times indicated and analyzed using phase-contrast microscopy with ×40 magnification. Experiments were performed at least three times, and representative fields of view are shown. Mycelial clumps in strains overexpressing SCO2255 (H357; asterisk) or staP (H2148; square) were significantly smoother and smaller than the other strains, suggesting abnormal growth. The arrows indicate new mycelial hyphae emerging through the smooth peripheries of old mycelial pellets in some 24-h, 36-h, and 48-h samples. The scale bars represent 100 μm.
DISCUSSION
VanJ increases resistance to teicoplanin in S. coelicolor but is not required for resistance to vancomycin. It is therefore unlikely to be involved in the remodeling of cell wall precursor biosynthesis to produce molecules with pentapeptide chains terminating in d-Ala-d-Lac, the process vital for conferring resistance to vancomycin and most other glycopeptide antibiotics. Indeed, liquid chromatography-mass spectrometry (LC-MS) analysis of PG precursor composition failed to identify any differences between wild-type and ΔvanJ mutant strains treated with vancomycin or differences resulting from strong constitutive overexpression of vanJ (data not shown). We also exclude the possibility that VanJ may directly inactivate teicoplanin, since incubation of the antibiotic in the presence of a vanJ overexpression strain failed to reduce its activity (data not shown).
VanJ and its homologues all contain a predicted endonuclease/exonuclease/phosphatase (pfam03372) domain. This large family of proteins includes Mg2+-dependent endonucleases and a large number of phosphatase enzymes involved in intracellular signaling, e.g., AP endonuclease, DNase I, synaptojanin (an inositol-1,4,5-trisphosphate phosphatase), sphingomyelinase (SMase), and nocturnin. SMase (also known as sphingomyelin [SM] phosphodiesterase) is a hydrolase enzyme that is involved in sphingolipid metabolism in eukaryotes. SMase is a member of the DNase I superfamily of enzymes and is responsible for breaking SM down into phosphocholine and ceramide by cleavage of a phosphate bond (19). The homology to VanJ suggests that VanJ, too, may possess lipid phosphatase activity and may therefore function by cleaving adjacent to a phosphate group present in a membrane lipid. Given the evident role of VanJ in conferring resistance to the glycopeptide antibiotic teicoplanin and the observed extracellular orientation of the enzyme active site, one potential substrate is undecaprenol pyrophosphate (C55-PP). C55-PP is a precursor to lipid II and therefore a key lipid intermediate in the synthesis of bacterial cell wall PG precursors. During cell wall biosynthesis, C55-PP is freed from lipid II and dephosphorylated by the C55-PP phosphatase to generate undecaprenol phosphate (C55-P), which is flipped back into the cytoplasm across the cell membrane and recycled for further rounds of PG precursor synthesis. We hypothesize that VanJ (and StaP) may be required for this recycling, catalyzing extracellular cleavage of the phosphodiester bond joining C55-P to the phospho-MurNAc(-pentapeptide)-GlcNAc in lipid II. This would increase recycling of C55-P, and thereby C55-PP, which could in turn increase resistance to teicoplanin. If true, this would indicate that a slowing (or blocking) of C55-P recycling is a significant part of the inhibitory activity of teicoplanin, but not vancomycin. The membrane-anchoring role of the lipid side chain present in the teicoplanin structure (but not vancomycin) may contribute to these differences in activity. The structure-activity studies, however, indicate that the presence of a biaryl ether oxygen bridge has more influence than lipidation, suggesting that this could be the basis for the distinct mode of action. Alternatively, given the previous observation that comprehensive elimination of PG precursors ending in d-Ala-d-Ala is important for teicoplanin resistance in Enterococcus faecium (5), it is possible that VanJ may function by specifically degrading d-Ala-d-Ala-containing lipid II molecules via its phosphatase activity. Other plausible explanations for the function of VanJ include the possibility that it acts by modifying membrane phospholipids (e.g., phosphatidylethanolamine, phosphatidylserine, phosphatidylglycerol, phosphatidylcholine, phosphatidylinositol, or C55-PP) or lipoteichoic acid (LTA). LTAs are more commonly found in low-G+C-content bacteria, where they are believed to play an important role in regulating cell wall autolysins, but prototypical polyglycerophosphate (PGP) LTAs have recently been identified in the high-G+C S. coelicolor (38, 39, 40).
Future detailed characterization of the function of VanJ at the molecular level, in addition to defining a novel mechanism of resistance, could also be central to obtaining a more complete understanding of the mode of action of teicoplanin. Comparing and contrasting the functions of VanJ and StaP with those more universally conserved homologues whose activity does not result in increased teicoplanin resistance may also throw new light on the relationship between antibiotic resistance mechanisms and the processes important for cell envelope integrity.
ACKNOWLEDGMENTS
We thank Wolfgang Wohlleben, Dudley Williams, Gerard Wright, Christopher Walsh, and Daniel Kahne for kindly providing glycopeptide antibiotics to test; Haruo Ikeda for providing CL_222_D10 cosmid DNA; and Lionel Hill for technical assistance with LC-MS analysis of peptidoglycan precursors. We also thank Andy Hesketh and Mark Buttner for helpful discussions, and the former also for comments on the manuscript.
This work was funded by grants from the Medical Research Council (G0700141) and the Royal Society (516002.K5877/ROG).
Footnotes
Published ahead of print 9 January 2012
REFERENCES
- 1. Arthur M, Quintiliani R. 2001. Regulation of VanA- and VanB-type glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 45:375–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arthur M, Molinas C, Courvalin P. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 174:2582–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arthur M, Molinas C, Reynolds P, Courvalin P. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arthur M, Depardieu F, Molinas C, Reynolds P, Courvalin P. 1995. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene 154:87–92 [DOI] [PubMed] [Google Scholar]
- 5. Arthur M, Depardieu F, Reynolds P, Courvalin P. 1996. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol. Microbiol. 21:33–44 [DOI] [PubMed] [Google Scholar]
- 6. Baptista M, Depardieu F, Courvalin P, Arthur M. 1996. Specificity of induction of glycopeptide resistance genes in Enterococcus faecalis. Antimicrob. Agents Chemother. 40:2291–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baptista M, Depardieu F, Reynolds P, Courvalin P, Arthur M. 1997. Mutations leading to increased levels of resistance to glycopeptide antibiotics in VanB-type enterococci. Mol. Microbiol. 25:93–105 [DOI] [PubMed] [Google Scholar]
- 8. Barna JCJ, Williams DH. 1984. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu. Rev. Microbiol. 38:339–357 [DOI] [PubMed] [Google Scholar]
- 9. Beauregard DA, Williams DH, Gwynn MN, Knowles DJC. 1995. Dimerization and membrane anchors in extracellular targeting of vancomycin group antibiotics. Antimicrob. Agents Chemother. 39:781–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bugg TDH, et al. 1991. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 30:10408–10415 [DOI] [PubMed] [Google Scholar]
- 11. Chen L, et al. 2003. Vancomycin analogues active against vanA-resistant strains inhibit bacterial transglycosylase without binding substrate. Proc. Natl. Acad. Sci. U. S. A. 100:5658–5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cooper MA, Williams DH. 1999. Binding of glycopeptide antibiotics to a model of a vancomycin-resistant bacterium. Chem. Biol. 6:891–899 [DOI] [PubMed] [Google Scholar]
- 13. Dong SD, et al. 2002. The structural basis for induction of VanB resistance. J. Am. Chem. Soc. 124:9064–9065 [DOI] [PubMed] [Google Scholar]
- 14. Eggert US, et al. 2001. Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science 294:361–364 [DOI] [PubMed] [Google Scholar]
- 15. Evers S, Courvalin P. 1996. Regulation of VanB-type vancomycin resistance gene expression by the VanSB-VanRB two-component regulatory system in Enterococcus faecalis V583. J. Bacteriol. 178:1302–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ge M, et al. 1999. Vancomycin derivatives that inhibit peptidoglycan biosynthesis without binding D-Ala-D-Ala. Science 284:507–511 [DOI] [PubMed] [Google Scholar]
- 17. Gregory MA, Till R, Smith MCM. 2003. Integration site for Streptomyces phage ϕBT1 and development of site-specific integration vectors. J. Bacteriol. 185:5320–5323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hannun YA, Obeid LM. 2002. The ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J. Biol. Chem. 277:25847–25850 [DOI] [PubMed] [Google Scholar]
- 20. Hayden MK, Trenholme GM, Schultz JE, Sahm DF. 1993. In vivo development of teicoplanin resistance in a VanB Enterococcus faecium isolate. J. Infect. Dis. 167:1224–1227 [DOI] [PubMed] [Google Scholar]
- 21. Hesketh A, et al. 2011. Genome-wide dynamics of a bacterial response to antibiotics that target the cell envelope. BMC Genomics 12:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hong H-J, Hutchings MI, Buttner MJ. 2008. Vancomycin resistance VanRS two-component systems. Adv. Exp. Med. Biol. 631:200–213 [DOI] [PubMed] [Google Scholar]
- 23. Hong H-J, et al. 2004. Characterization of an inducible vancomycin resistance system in Streptomyces coelicolor reveals a novel gene (vanK) required for drug resistance. Mol. Microbiol. 52:1107–1121 [DOI] [PubMed] [Google Scholar]
- 24. Hong H-J, Hutchings MI, Hill L, Buttner MJ. 2005. The role of the novel Fem protein VanK in vancomycin resistance in Streptomyces coelicolor. J. Biol. Chem. 280:13055–13061 [DOI] [PubMed] [Google Scholar]
- 25. Hutchings MI, Hong H-J, Buttner MJ. 2006. The vancomycin resistance VanRS signal transduction system of Streptomyces coelicolor. Mol. Microbiol. 59:923–935 [DOI] [PubMed] [Google Scholar]
- 26. Hutchings MI, Hong H-J, Leibovitz E, Sutcliffe IC, Buttner MJ. 2006. The SigE cell envelope stress signal of Streptomyces coelicolor is influenced by a novel lipoprotein, CseA. J. Bacteriol. 188:7222–7229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaberdina AC, Szaflarski W, Nierhaus KH, Moll I. 2009. An unexpected type of ribosomes induced by kasugamycin: a look into ancestral times of protein synthesis? Mol. Cell 33:227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kerns R, et al. 2000. The role of hydrophobic substituents in the biological activity of glycopeptides antibiotics. J. Am. Chem. Soc. 122:12608–12609 [Google Scholar]
- 29. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 30. Komatsu M, Tsuda M, Omura S, Oikawa H, Ikeda H. 2008. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc. Natl. Acad. Sci. U. S. A. 105:7422–7427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koteva K, et al. 2010. A vancomycin photoprobe identifies the histidine kinase VanSsc as a vancomycin receptor. Nat. Chem. Biol. 6:327–329 [DOI] [PubMed] [Google Scholar]
- 32. Loll PJ, Axelsen PH. 2000. The structural biology of molecular recognition by vancomycin. Annu. Rev. Biophys. Biomol. Struct. 29:265–289 [DOI] [PubMed] [Google Scholar]
- 33. Manteca A, Alvarez R, Salazar N, Yagüe P, Sanchez J. 2008. Mycelium differentiation and antibiotic production in submerged cultures of Streptomyces coelicolor. Appl. Environ. Microbiol. 74:3877–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Hare MD, Reynolds PE. 1992. Novel membrane proteins present in teicoplanin-resistant, vancomycin-sensitive, coagulase-negative Staphylococcus spp. J. Antimicrob. Chemother. 30:753–768 [DOI] [PubMed] [Google Scholar]
- 35. Paget MSB, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. 1999. Evidence that the extracytoplasmic function sigma factor, σE, is required for normal cell wall structure in Streptomcyes coelicolor A3(2). J. Bacteriol. 181:204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pootoolal J, Neu J, Wright GD. 2002. Glycopeptide antibiotic resistance. Annu. Rev. Pharmacol. Toxicol. 42:381–408 [DOI] [PubMed] [Google Scholar]
- 37. Pootoolal J, et al. 2002. Assembling the glycopeptide antibiotic scaffold: the biosynthesis of A47934 from Streptomyces toyocaensis. Proc. Natl. Acad. Sci. U. S. A. 99:8962–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Potekhina NV, Streshinskaya GM, Novitskaya GV, Naumova IB. 1983. Isolation of lipoteichoic acid from Streptomyces levoris. Mikrobiologiia 52:340–343 [PubMed] [Google Scholar]
- 39. Rahman O, Dover LG, Sutcliffe IC. 2009. Lipoteichoic acid biosynthesis: two steps forwards, one step sideways? Trends Microbiol. 17:219–225 [DOI] [PubMed] [Google Scholar]
- 40. Rahman O, Cummings SP, Sutcliffe IC. 2009. Phenotypic variation in Streptomyces sp. DSM 40537, a lipoteichoic acid producing actinomycete. Lett. Appl. Microbiol. 48:226–229 [DOI] [PubMed] [Google Scholar]
- 41. Sharman GJ, et al. 1997. The roles of dimerization and membrane anchoring in activity of glycopeptide antibiotics against vancomycin-resistant bacteria. J. Am. Chem. Soc. 119:12041–12047 [Google Scholar]
- 42. Shlaes DM, et al. 1993. Teicoplanin-resistant Staphylococcus aureus expresses a novel membrane protein and increases expression of penicillin-binding protein 2 complex. Antimicrob. Agents Chemother. 37:2432–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sinha Roy R, et al. 2001. Direct interaction of a vancomycin derivative with bacterial enzymes involved in cell wall biosynthesis. Chem. Biol. 8:1095–1106 [DOI] [PubMed] [Google Scholar]
- 44. Waksman SA, Henrici AT. 1943. The nomenclature and classification of the actinomycetes. J. Bacteriol. 46:337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]