Abstract
Drug-resistant viral variants are a major issue in the use of direct-acting antiviral agents in chronic hepatitis C. Ketoamides are potent inhibitors of the NS3 protease, with V55A identified as mutation associated with resistance to boceprevir. Underlying molecular mechanisms are only partially understood. We applied a comprehensive sequence analysis to characterize the natural variability at Val55 within dominant worldwide patient strains. A residue-interaction network and molecular dynamics simulation were applied to identify mechanisms for ketoamide resistance and viral fitness in Val55 variants. An infectious H77S.3 cell culture system was used for variant phenotype characterization. We measured antiviral 50% effective concentration (EC50) and fold changes, as well as RNA replication and infectious virus yields from viral RNAs containing variants. Val55 was found highly conserved throughout all hepatitis C virus (HCV) genotypes. The conservative V55A and V55I variants were identified from HCV genotype 1a strains with no variants in genotype 1b. Topology measures from a residue-interaction network of the protease structure suggest a potential Val55 key role for modulation of molecular changes in the protease ligand-binding site. Molecular dynamics showed variants with constricted binding pockets and a loss of H-bonded interactions upon boceprevir binding to the variant proteases. These effects might explain low-level boceprevir resistance in the V55A variant, as well as the Val55 variant, reduced RNA replication capacity. Higher structural flexibility was found in the wild-type protease, whereas variants showed lower flexibility. Reduced structural flexibility could impact the Val55 variant's ability to adapt for NS3 domain-domain interaction and might explain the virus yield drop observed in variant strains.
INTRODUCTION
Infection with hepatitis C virus (HCV) is a frequent cause of chronic hepatitis with liver cirrhosis and hepatocellular carcinoma as sequelae (1). Until recently, the standard of care (SOC) for patients with chronic hepatitis C virus infection (CHC) has consisted of a combination of pegylated interferon alpha plus ribavirin (Peg-IFN/RBV) administered for 24 to 48 weeks, depending on the HCV genotype. This is only partially effective, with about a 50% sustained viral response (SVR) in patients infected with HCV genotype 1, the most common genotype in Europe and North America (11, 21, 33, 44). The addition of a direct-acting antiviral agent (DAA) targeting the NS3/4A serine protease of HCV significantly improves the SVR rate, and two such drugs have recently been approved for clinical use by the European Medicines Agency (EMA) and the U.S. Food and Drug Administration (FDA). The ketoamide compounds boceprevir (Victrelis) and telaprevir (Incivek) were both designed to mimic the natural substrate of the NS3 protease (16, 19, 22, 23, 30). Clinical trials in treatment-naïve genotype 1-infected patients have revealed significant improvements in the kinetics of the virologic response with the addition of a DAA to the prior SOC, leading to improved SVR rates of up to 74% (16, 19, 22). Despite the progress, however, protease inhibitor (PI) resistance is a major challenge for future treatment. Resistant viral variants exist at low frequencies in untreated patients as part of the viral quasispecies population (29), reflecting the highly replicative nature of HCV infections and the error-prone character of its RNA-dependent RNA polymerase, NS5B.
The NS3 protease plays an essential role in the HCV life cycle by processing nonstructural (NS) proteins from the viral polyprotein downstream of the NS2-3 junction (24). The protease domain of NS3 comprises the amino-terminal third of the protein containing a catalytic triad, H57, D81, and S139, and an “oxyanion hole” at G137. It acts in concert with its cofactor, NS4A, which intercalates into its structure and is required for full enzymatic activity and proper subcellular localization. The carboxy-terminal two-thirds of NS3 consists of a DExD-box RNA helicase domain that is essential for productive viral infection (28). NS3 thus appears to be a critical component of the macromolecular viral RNA replicase that directs the synthesis of new viral RNAs. Genetic evidence indicates that NS3 has an additional distinct function in assembly of infectious virus particles (20, 34). Since viral RNA replication capacity and virus assembly are crucial determinants of viral fitness, mutations in NS3 that contribute to PI resistance can also profoundly influence virus fitness (42). The probability of a resistant variant emerging from the quasispecies population during treatment with a DAA is determined not only by its degree of drug resistance, but also by its fitness. The NS3 domain interdependency might provide for novel molecular mechanisms in treatment escape from ketoamide compounds. Many mutations associated with PI resistance negatively impact the replication of genotype 1a HCV RNA in cell culture, while some have additional effects on the production of infectious virus (34). Compensatory second-site mutations may enhance the fitness of these resistant viruses (42), but current understanding about the underlying molecular mechanisms is rudimentary.
Previous studies identified the V55A variant as resistance-associated amino acid variant for the ketoamide compound boceprevir (27, 37). Furthermore, the V55A variant was found in the long-term follow up of patients up to 5.5 years upon boceprevir treatment completion. Moreover, the variant was dominant already at baseline in one of the patients before any PI exposure (36). Interestingly, Val55 is related to PI resistance, although it is buried in the NS3 protease domain structure, without direct ligand interaction (37). The V55A variant showed medium-level resistance against the ketoamide compounds boceprevir and telaprevir in enzymatic assays and had a negative impact on RNA replication in the Con1 replicon system (37). The viral fitness of the V55A variant has not been determined so far, since no data are available on its infectious virus yield.
In this article, we present a combined in silico and in vitro study on ketoamide resistance and viral fitness in Val55 variants of the NS3 protease. A comprehensive sequence analysis was performed using a public database with worldwide patient isolates to identify Val55 variants from dominant strains. Subsequently, we applied an HCV cell culture system of infection and molecular dynamics (MD) simulations to assess ketoamide resistance, viral variant fitness, and underlying molecular mechanisms in Val55 variants. The potential role of Val55 in the NS3 protease as structural and functional regulatory site is analyzed using a residue-interaction network approach.
MATERIALS AND METHODS
In silico sequence analysis.
We analyzed sequences of the hepatitis C virus NS3 protease deposited in the European HCV Database (euHCVdb) (7), which contains sequences of major variants from around the world. HCV genotypes and subtypes were differentiated according to a consensus proposal for a unified system of HCV genotype nomenclature (35). The sequences in this study were confirmed as HCV genotypes 1a, 1b, 2, 3, 4, 5, 6, and 7. Sequence alignments (not shown) were computed using ClustalW (6) and MUSCLE (10), with minor manual modifications, in the SEAVIEW alignment editor (12). Comprehensive sequence analysis on natural Val55 variants and correlated variants was performed in 676 HCV NS3 isolates from seven genotypes and their corresponding subtypes. We confirmed 202 strains as HCV genotype 1a and 335 strains as HCV genotype 1b. Subsequently, we use the genotype 1a H77 strain (UniProtKB reference no. P27958) for in vitro analysis and a genotype 1a protease from Protein Data Bank (PDB) structure 2OC8 for structure analysis. Both sequences are 98.9% identical, with only two variants between UniProtKB P27958 and PDB 2OC8, i.e., M76T and A149T, respectively.
Cells and reagents.
Huh7 and Huh7.5 cells were provided from Apath and grown in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% fetal calf serum, penicillin, streptomycin, l-glutamine, and nonessential amino acids. Boceprevir (SCH503034) and telaprevir (VX-950) stock solutions were prepared in dimethyl sulfoxide (DMSO). All final dilutions contained 0.5% DMSO.
Plasmids.
pH77S.3 and pH77S.3/GLuc2A are molecular clones of the genotype 1a H77 strain of HCV. Synthetic RNA transcribed from these plasmids replicates in transfected Huh7 cells and produces infectious virus. pH77S.3/GLuc2A also produces secreted Gaussia luciferase (GLuc) reporter protein. The V55A and V55I single-amino-acid variants and the T54S V55I double variant were created in these plasmids by site-directed mutagenesis, using AfeI and BsrGI restriction sites. Sequences of manipulated DNA segments were confirmed by DNA sequencing. pH77S/GLuc2A/AAG is a replication-defective pH77S/GLuc2A mutant. It was generated by inserting an AfeI/AscI restriction fragment containing the GLuc2A sequence between the corresponding sites of pH77S/AAG, in which the GDD motif of the polymerase, NS5B, is replaced with AAG (43).
RNA transcription and transfection.
RNA was synthesized with T7 MEGAScript reagents (Ambion), after linearizing plasmids with XbaI. Following treatment with RNase-free DNase to remove template DNA, RNA was purified using the RNeasy minikit (Qiagen). RNA transfection was carried out with the Trans-IT mRNA transfection kit (Mirus) according to the manufacturer's suggested protocol. Transfection protocols were optimized for determination of antiviral susceptibility, RNA replication capacity, and infectious virus yield. Briefly, for cell-based antiviral activity assays, 150 ng RNA was transfected into 4 × 104 cells/well seeded in 48-well plates. To test the RNA replication capacity of GLuc-containing constructs, 250 ng RNA was transfected into 8 × 104 cells/well in 24-well plates. To test the capacity of RNA to produce infectious virus, 1.25 μg RNA was transfected into 6 × 105 cells/well in 6-well plates.
Luciferase activity assay.
Replication of HCV genotype 1a RNA for the V55A and V55I variants was determined by measuring GLuc activity using the H77S.3/GLuc2A construct (34). Following RNA transfection, cell culture supernatant fluids were collected, and fresh medium was added at 24-h intervals. Secreted GLuc activity was measured in 25-μl aliquots of the supernatant fluids using the GLuc assay kit (New England BioLabs) according to the manufacturer's suggested protocol. The luminescent signal was measured on a Synergy 2 (Bio-Tek) Multi-Mode microplate reader.
Virus yield determination.
Cells were split at a 1:1 ratio 24 h after RNA transfection, and the medium was replaced with fresh medium containing 10% HEPES every 24 h thereafter. Cell supernatant culture fluids collected at 72 h after transfection were assayed for infectious virus in a fluorescent focus virus titration assay described previously (43). Briefly, cells were seeded in 48-well plates at a density of 1 × 105 cells/well 24 h prior to inoculation with 50 to 100 μl of culture medium. Cells were maintained at 37°C in a 5% CO2 environment and fed with 300 μl of medium at 24 h later. Following 48 h of additional incubation, cells were fixed in methanol-acetone (1:1) at room temperature for 9 min and stained with monoclonal antibody C7-50 (Affinity BioReagents, Golden, CO) to the HCV core protein (1:300). After extensive washing, cells were stained with fluorescein isothiocyanate-conjugated goat anti-mouse IgG antibody. Clusters of infected cells staining for core antigen were considered to constitute a single infectious focus-forming unit (FFU). Virus titers were reported as FFU/ml.
Antiviral activity assay.
Wild-type, V55A and V55I variants, and the T54S V55I double variant viral RNAs were transfected as described above with serial dilutions of the ketoamide compounds boceprevir and telaprevir added separately to the growth medium. The medium was replaced with fresh medium containing boceprevir or telaprevir at 24 h, and at 24-h intervals thereafter, and secreted GLuc activity was determined 72 h later as described above. The concentration of boceprevir and telaprevir required to reduce the amount of secreted GLuc activity by 50% (antiviral 50% effective concentration [EC50]) was determined using a three-parameter Hill equation (SigmaPlot 10.0).
MD simulation.
Molecular dynamics (MD) simulation was performed for Val55 wild-type and V55A and V55I variants of the NS3 protease, each with (“bound”) or without (“unbound”) the covalently linked ketoamide inhibitor boceprevir (SCH503034). The program package GROMACS (15) and the GROMOS96 53a6 force field (25) were used for the simulations. The X-ray structure 2OC8 (26) from the Protein Databank RCSB PDB (18) served as structural basis for MD. The structure contains an HCV genotype 1a NS3 protease cocrystallized with boceprevir. To obtain starting structures for the variants, the Val55 side chain from the wild-type structure was mutated using the tool IRECS (13). Boceprevir was removed from the corresponding NS3 ligand-binding sites to analyze “unbound” protease structures with empty binding pockets. Topology parameters for boceprevir were calculated by the PRODRG server (31). To describe the covalent linkage between the Ser139 residue (OG atom) and boceprevir (C34 atom), a new residue type (SER2, i.e., deprotonated SER at OG) was defined based on the predefined SER parameters of the GROMOS96 53a6 force field. Appropriate parameters for bond length and angles of the employed force field were added in the topology file to set the covalent bond. Throughout the simulations, periodic boundary conditions were applied. Long-range nonbonded interactions were treated by particle-mesh Ewald (PME) summation. The Berendsen scheme (5) was used to maintain temperature and pressure by weak coupling to an external bath with a temperature coupling relaxation time of 0.1 ps, a pressure coupling constant of 1.0 ps, and a compressibility of 4.5 × 10−5. Bond lengths were constrained to ideal values using the LINCS procedure (14). After steepest-descent energy minimization, the systems were gradually heated from 0 to 300 K over 460 ps. A time step of 1 fs was chosen for the heat-up procedure, and position restraints were applied up to a temperature of 200 K. MD simulations were carried out at 300 K, using a time step of 2 fs and constant pressure of 1 atm for 20 ns. For the “unbound” wild-type structure, the simulation was extended by 10 ns to ensure a steady backbone root mean square deviation (RMSD). Tools of the GROMACS program were employed for analysis of trajectories. Properties (e.g., H bonds) were averaged over the last 6 ns of the trajectories unless otherwise noted. VMD (17) was applied for visualization of central cluster structures from simulated complexes.
RIN.
We used PDB structure 2OC8 to generate a residue-interaction network (RIN) in two dimensions (2D). REDUCE (41) was applied for adding H-atoms to the original X-ray structure. PROBE (40) was used to identify noncovalent residue interactions. The RIN was visualized using Cytoscape (32) and the plugin RINalyzer (8). Protease residues were represented by nodes and corresponding noncovalent interactions by edges. The Cytoscape plugin NetworkAnalyzer (2) was used for calculation of Val55 topology measures within the RIN. We calculated “node degree,” which is the number of edges linked to a specific node within an undirected network, as well as “node connectivity,” which is the number of direct neighbors to a specified node. The “network heterogeneity” was computed to estimate the tendency of the RIN to contain highly connected nodes of putative functional importance (9). The length of a path in the RIN is given by the number of edges forming this path, whereby two nodes are possibly connected via multiple paths. We used the Cytoscape plugin Shortest Path, UCSF Resource for Biocomputing, Visualization, and Informatics (http://www.rbvi.ucsf.edu), to identify the shortest path or distance between two selected nodes. “Network diameter,” which is the maximum length between two nodes in the RIN, was calculated as a reference value for the shortest path analysis. The RIN analysis considered different edge types for all possible interactions, van der Waals contacts, H bonds, overlaps, and main-chain and side-chain interactions. All measures considered weighted edges, if more than one interaction occurred between two nodes.
RESULTS
Genetic diversity of the NS3 protease at Val55.
We performed a comprehensive analysis of NS3 protease sequences from 676 worldwide patient strains of the European HCV Database (euHCVdb) (7), for Val55 variants and correlated substitutions. Val55 was highly conserved throughout all genotypes and subtypes with no variants identified in HCV genotypes 1b, 2, 3, 4, and 6. Two conservative variants were observed in HCV genotype 1a, the V55A and V55I variants. A low variant frequency of four variant strains in 202 isolates was found. No covariant sequence polymorphisms in V55A variant strains were identified, whereas both V55I variant strains showed the covariant sequence polymorphism T54S (EU781818 and EF407443 strains). Two HCV genotype 5 strains with V55L and one genotype 7 strain with V55P were found. Assuming that no sequencing error occurred, the nonconservative change to Pro55 suggests considerable structural differences between the protease domains of HCV genotype 1a and genotype 7. Val55 is located buried at the tip of the β-strand C1 in an antiparallel β-sheet of the genotype 1a structure 2OC8 (37) with the main-chain NH interacting via an H bond with the side chain of Thr54. Different from the wild type, the Pro55 main-chain N atom is not available as an H-bond donor. Furthermore, Gly54 was found as neighboring residue in genotype 7 instead of Thr54, which was found in genotypes 1 to 6. Thus, the Thr54-Val55 H-bond interaction is not present in genotype 7 (38). Since ketoamide compounds are licensed for HCV genotype 1 and no variants were identified for genotype 1b, we subsequently focused on the V55A and V55I variants from HCV genotype 1a.
Ketoamide resistance in Val55 variants.
We introduced the V55A and V55I variants, as well as the V55I variant with its covariant T54S into separate backbones of H77S.3/GLuc2A and determined the resistance level of the HCV genotype 1a RNAs against the ketoamide compound boceprevir. We found boceprevir had antiviral activity in the genotype 1a wild type and both Val55 variants. The V55A variant showed low-level resistance, whereas the V55I variant showed no resistance against boceprevir. Average EC50s were 1,360 nM and 690 nM for the V55A and V55I variants, respectively, corresponding to fold changes of 1.6- and 0.8-fold (Table 1). The V55I variant is more sensitive than the wild type for boceprevir. The T54S V55I double variant has an even lower EC50 of 450 nM, with a fold change of 0.5-fold. Thus, the T54S variant should not confer to ketoamide resistance in the T54S V55I double variant (Table 1).
Table 1.
Impact of the V55A and V55I variants, as well as the T54S V55I double variant on boceprevir resistance
| Variant | IC50 (nM)a | Fold changea | EC50 (nM)b | Fold changeb |
|---|---|---|---|---|
| Single | ||||
| Wild type | 19 | 870 ± 48 | 1 | |
| V55A | 80 ± 18 | 4.2 ± 0.96 | 1,360 ± 82 | 1.6 |
| V55I | NDc | 690 ± 14 | 0.8 | |
| Double | ||||
| T54S V55I | ND | 450 ± 79 | 0.5 |
IC50s previously published by Susser et al. (37). The results shown represent the mean ± standard deviation and fold changes compared to the wild type.
EC50s determined from H77S.3/GLuc2A-transfected cell cultures. The results shown represent the mean ± standard deviation and fold changes compared to the wild type.
ND, not determined.
Viral fitness of Val55 variants in HCV genotype 1a.
To determine the impact of Val55 variants on viral fitness, we measured RNA replication capacity and infectious virus yields for V55A and V55I variants using the H77S.3 cell culture system.
(i) Replication capacity.
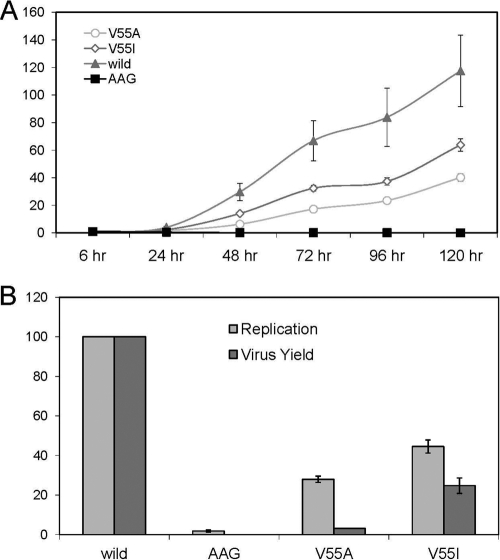
The replication capacity of H77S.3/GLuc2A RNA for the V55A and V55I variants was determined by measuring GLuc activity in supernatant media collected at 24-h intervals following transfection of synthetic RNA. Results were normalized to the activity present at 8 h after transfection, as this represents GLuc expressed directly by the transfected input RNA. Compared to the parental H77S.3/GLuc2A RNA, both variants showed medium-level impairment of their RNA replication capacity, with the GLuc activity generally increasing after 24 h but consistently less than the parental RNA (Fig. 1). Variants showed similar courses in their RNA replication capacity, with slightly higher RNA replication in the V55I variant compared to the V55A variant. The V55I variant showed a steeper increase up to 64% compared to only 40% of parental RNA in the V55A variant. The negative impact on RNA replication correlated well with the fold changes observed in boceprevir resistance for both variants.
Fig 1.
Replication capacity and infectious virus yield of H77S.3 RNAs with V55A and V55I substitutions in NS3. (A) RNA replication capacity reflecting the presence of H77S.3/GLuc2A V55A and V55I variants in Huh7.5 cells. Results are normalized to the 8-h GLuc activity and represent the mean of triplicate samples. (B) Comparison of RNA replication capacity (lightly shaded bars) and infectious virus yields (dark shaded bars). Both are normalized to those obtained with the relevant parental RNA (100%). The wild-type infectious titer is 2,720/ml (± standard deviation [SD] of 198/ml). Data represent the mean ± SD from at least 3 independent experiments. AAG is a replication-defective pH77S/GLuc2A mutant (43).
(ii) Infectious virus yield.
We used H77S.3 RNA lacking the GLuc2A-coding sequence to assess the impact of V55A and V55I variants on the ability to produce infectious virus following transfection into Huh7.5 cells. Cell culture supernatant fluids were collected 72 h after transfection of respective variant RNAs, inoculated onto naïve Huh7.5 cells, and foci of infected cells were detected by immunofluorescence 72 h later. Both Val55 variants produced infectious virus yields lower than that expected from their RNA replication capacity. Thus, both variants directly impair infectious virus assembly or release, as described previously for other resistance-associated NS3 variants located in or near the protease ligand-binding site (34). Infectious virus yields were significantly different between V55I and V55A variants, with 25% and 3% relative infectivity, respectively (Fig. 1). Reductions in the fitness of these particular variants are confined due to defects in viral RNA replication and a significant drop in infectious virus production.
The discordances between RNA replication capacities and yields of infectious virus were higher than expected for conservative variants at a position buried in the NS3 protease domain. The large drop of infectious virus yield observed in the V55A variant resulted in a particularly low viral fitness of this variant compared to the V55I variant.
Network topology analysis for Val55 in the NS3 protease.
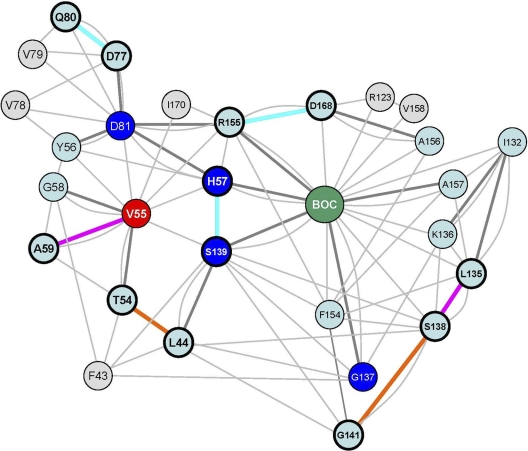
We used a residue-interaction network (RIN) computed from an experimental 3D protease structure to analyze network topology measures and characterize the potential regulatory role of Val55 in the NS3 protease structure. The RIN comprises a total of 206 nodes, referring to residues of the NS3 protease and NS4A, as well as 1,752 edges, referring to noncovalent interactions between residues. The mean number of neighbors for a node in the NS3 protease RIN was calculated as 7.7, with a neighborhood connectivity range from 1 to 17. The majority of nodes showed only few connections with direct neighbors, whereas Val55 was highly connected with a node connectivity of 10. The node degree for Val55 was calculated with 24. The node degree distribution for the NS3 protease RIN identified 167 nodes below 24 (81%), six nodes (3%) with an equal node degree to Val55, and 33 nodes (16%) with a node degree above 24. The network diameter, which is the maximum distance between two connected nodes in the RIN, is 10 edges. Val55 showed direct contacts with 10 residues (Thr54, Tyr56, His57, Gly58, Ala59, Asp81, Ser139, Gly140, Arg155, and Ile170), including residues of the protease catalytic triad and a two-edge distance to the catalytic “oxyanion hole” (Gly137). In addition, Val55 showed van der Waals contacts with two residues of the protease ligand-binding site (Arg155 and Ile170) and indirect interaction via Thr54, Gly58, or Ser139 with Phe43, which participates in the formation of a narrow hydrophobic cavity at the S1′ pocket (38). Val55 resides in an H-bond network with Thr54, Gly58, and Ala59, without direct H-bond interaction to boceprevir. The distance to boceprevir in the RIN is two edges with a path via His57, Ser139, or Arg155 (Fig. 2). Overall, the topology measures suggest a putative structure key role for Val55 in the NS3 protease.
Fig 2.
Residue-interaction network of the NS3 protease from PDB structure 2OC8, with nodes and edges representing protease residues and noncovalent interactions, respectively. The ketoamide compound boceprevir is represented by a single green node (BOC). The residue Val55 is given as a red node. Catalytic residues are given in blue (His57, Asp81, Gly137, and Ser139). H bonds and van der Waals contacts are represented by bold dark gray- or light gray shaded edges, respectively. To reduce complexity, multiple H-bonded or van der Waals contacts were represented by single edges between two nodes, irrespective of the number of pairwise atomic interactions. H bonds and nodes impacted due to V55A and V55I variants and/or boceprevir binding to the ligand-binding site are indicated in bold and color coded. The color coding denotes an H-bond loss upon boceprevir binding in wild-type and variant structures (cyan) and H-bond gain upon boceprevir binding in the wild-type structure (orange), as well as H-bond gain in V55A and V55I variants (magenta).
Molecular dynamics simulation of the Val55 wild type, as well as V55A and V55I variants.
Molecular dynamics (MD) simulation was performed to further investigate the potential structure key role of Val55 in the protease structure. Simulations were carried out for wild-type and variant structures with and without the bound ketoamide boceprevir. Structures without boceprevir are referred as “unbound,” whereas structures with boceprevir are referred as “bound.” The variant structures were obtained by in silico mutation of the X-ray structure from PDB 2OC8 at Val55 with subsequent equilibration and MD simulation. Due to the covalent bond between boceprevir and Ser139, the ligand was kept in the mutated ligand-binding sites during the simulation. The final structures were analyzed for effects of Val55 variants on the H-bond pattern.
(i) Conformational stability.
During simulation, the root mean square deviation (RMSD) serves as a measure for conformational stability and flexibility of the protease structure, with larger RMSDs indicating increasing structural flexibility.
(a) Backbone.
The backbone RMSD of the “unbound” wild-type protease observed over time showed a longer equilibration time than was found in the variant structures. Final values and amplitudes were larger than in the corresponding “unbound” variant backbone RMSDs. In contrast, RMSDs of the “bound” protease were similar for the wild type and variants. RMSDs of “bound” variant structures were not significantly different from respective “unbound” variant structures (Fig. 3A).
Fig 3.
Backbone and side-chain root mean square deviations (RMSDs) and molecular dynamics simulations of the NS3 protease. (A) Backbone RMSDs of “unbound” and “bound” protease wild type and V55A and V55I variants. (B) RMSDs of protease residue His57 in “unbound” and “bound” wild-type and V55A and V55I variant structures. (C and D) Superposition of the “unbound” wild-type and V55I variant protease ligand-binding sites (C), as well as the “unbound” wild-type and V55A variant protease ligand-binding sites (D). Molecular changes in the S1 pocket around Ser139 are shown in the surface representation (wild type in salmon, V55I variant in green, and V55A variant in blue). Wild-type and simulated side-chain orientations of Ser139 upon V55I and V55A mutation are depicted as stick models, with wild-type Val55, the Ile55 variant, and the Ala55 variant as red, green, and blue stick models, respectively.
(b) Single residues.
The analysis of RMSDs from single residues near the protease ligand-binding site provided more detailed information on conformational changes. In the “bound” V55A variant structure, a rearrangement of His57 was observed (Fig. 3B). Similarly, a flip of His57 was found in the “unbound” wild type at the very end of the simulation for only few time steps. In contrast, the “bound” V55A variant structure showed the His57 flip at the beginning of the simulation and maintained this conformation until the end of the simulation. The larger RMSDs reflect higher structural flexibility of residues in the wild-type protease than the corresponding residues in the V55A and V55I variant structures.
(ii) H-bond network.
We explored stabilizing and destabilizing effects on the H-bond pattern in the ligand-binding site for residue pairs selected from the protease residue-interaction network. We evaluated average numbers of H bonds per time frame during simulation.
(a) Wild type versus variants.
The average number of H bonds differed only slightly for “bound” wild-type and variant structures. The total difference relating to all selected residue pairs was smaller than 1.5. This was different in the “unbound” protease structures (Table 2). The average number of H bonds between the residue pairs Leu135-Ser138, Arg155-Asp168, Ser138-Gly141, and Thr54-Leu44 was significantly larger for the “unbound” V55A and V55I variants than for the wild-type structure (Table 2). Additional H bonds in the V55A variant were identified between the catalytic residue pair His57-Ser139, as well as between the residue pair Gln80-Asp77 (Fig. 2). Taking all residue pairs in the ligand-binding site into account, the number of H bonds in the “unbound” variant structures compared to the wild type was increased by four for the V55I variant structure and six for the V55A variant structure.
Table 2.
Difference in the average number of H bonds per time frame for “unbound” variants (V55A and V55I) and wild-type (Val55) structuresa
| Residue pair | Difference in avg no. of H bonds: |
|
|---|---|---|
| V55I − Val55 | V55A − Val55 | |
| X55-Ala59b | 0.2 | 0.6 |
| His57-Ser139 | −0.1 | 0.7 |
| Leu135-Ser138 | 0.7 | 0.7 |
| Gln80-Asp77 | 0.1 | 0.7 |
| Arg155-Asp168 | 0.8 | 0.8 |
| Ser138-Gly141 | 0.9 | 0.8 |
| Thr54-Leu44 | 0.9 | 0.9 |
| Sumc | 3.5 | 5.3 |
A negative sign denotes a loss in H bonds in the variant structure.
X stands for Val (wild type), Ile (V55I variant), or Ala (V55A variant).
Corresponding differences when taking all established residue pairs within the ligand-binding site into account are as follows: V55I − Val55, 4.1; and V55A − Val55, 6.1.
(b) “Bound” versus “unbound” protease structures.
The comparison of “bound” versus “unbound” protease structures reflects the propensity to bind boceprevir based on changes in the H-bond network (Table 3). The comparison between the “bound” and “unbound” wild-type proteases showed an overall increase in H bonds upon boceprevir binding. The bond order for H bonds in the residue pairs Gln80-Asp77 and Arg155-Asp168 was slightly smaller in “bound” than “unbound” wild-type structures. Additional H bonds in the “bound” wild type were observed for the residue pairs Ser138-Gly141 and Thr54-Leu44. The V55A and V55I variant structures showed almost no change in H bonds for the residue pairs Ser138-Gly141 and Thr54-Leu44, while a loss of H bonds was observed for the residue pairs Gln80-Asp77 and Arg155-Asp168. Furthermore, an H bond in the “unbound” but not “bound” V55A variant structure was observed between the catalytic residue pair His57-Ser139 (Fig. 2). Thus, “unbound” variant structures possessed a larger H-bond network than the wild type. Boceprevir binding was shown to disturb rather than stabilize this H-bond network. The “unbound” V55A variant structure showed more H bonds than the V55I variant structure with greater destabilizing impact due to lost H bonds upon boceprevir binding.
Table 3.
Difference in the average number of H bonds per time frame between the “bound” and “unbound” structures for the wild-type (Val55) and variant (V55A and V55I) complexesa
| Residue pair | Difference in avg no. of H bonds |
||
|---|---|---|---|
| Val55 | V55I | V55A | |
| His57-Ser139 | −0.1 | 0.0 | −0.8 |
| Gln80-Asp77 | −0.4 | −1.0 | −1.7 |
| Arg155-Asp168 | −0.4 | −1.4 | −0.6 |
| Ser138-Gly141 | 0.9 | 0.0 | 0.1 |
| Thr54-Leu44 | 0.9 | −0.1 | 0.0 |
| Sumb | 0.9 | −2.5 | −3.0 |
A negative sign denotes a loss of the H bonds upon binding of boceprevir.
Corresponding differences when taking all established residue pairs within the ligand-binding site into account are as follows: Val55, 1.7; V55I, −3.8; and V55A, −4.4.
(iii) Shape of the NS3 protease ligand-binding site.
To estimate the facility for ketoamides approaching the protease ligand-binding site, we analyzed structural changes during MD simulation for a representative pocket in the “unbound” protease structures (S1 pocket). The minimum distances between Ser139 and its next neighbor residues in the S1 pocket were calculated. We found most S1 pocket residues in V55A and V55I variant structures closer to Ser139 compared to the wild-type structure. The overall S1 pocket size was smaller in the variant structures with reduced accessibility to the ligand-binding site. Exceptions were found predominantly for residues involved in the formation of S1′ and S2′ pockets (i.e., Gln41, Thr42, and Lys136).
DISCUSSION
Drug-resistant viral variants are likely to preexist at a low frequency in the replicating viral quasispecies population of the typical HCV-infected patient (29, 39). The V55A variant was previously identified as resistance-associated amino acid variant against the ketoamide compound boceprevir, with Val55 suggested as a regulatory site in the NS3 protease structure (37) involved in resistance development and variant fitness. A recent study found the V55A variant in the long-term follow up of several patients previously treated with boceprevir. Moreover, this variant was identified as dominant baseline strain in one of the patients before treatment. Thus, variations at Val55 are likely to be clinically relevant in patients treated with ketoamide compounds (36). In the present study, we analyzed the natural variation present among 676 sequences from genotypes 1 to 7, collected from geographically diverse sites and deposited in a public database. They likely represent variants present within the dominant quasispecies of the patients from which these sequences were derived (7). We found Val55 highly conserved in all major HCV genotypes and subtypes; however, few conservative variants and one nonconservative variant were identified. Clinically most important, HCV genotype 1a showed four strains from 202 sequences with variations at Val55—two strains with V55A and two strains with V55I—whereas, no variants were found in 335 sequences of genotype 1b HCV.
We used a residue-interaction network approach to characterize the potential key role of Val55 in the NS3 protease structure related to resistance development and viral fitness in Val55 variants. The protease residue-interaction network identified only a few nodes in the network highly connected with their neighboring nodes, while the majority show only a few connections. Val55 is such a highly connected node with connections to protease catalytic residues and the ligand-binding site. The high connectivity points to a potential key role for Val55 with important function in the respective network and protein structure (9), where variants could have an impact on viral fitness and drug resistance development.
To further investigate this role of Val55 and its variants in the NS3 protease structure, we applied molecular dynamics simulation, which is a proven approach to study molecular changes and mechanisms potentially related to drug resistance, already successfully applied to other NS3 protease resistance-associated amino acid variants (38) (S. Schweizer et al., submitted for publication). Analyzing protein backbone RMSDs from the molecular dynamics simulations, we found higher structural flexibility in the wild-type protease than the variant structures. This was further supported by the larger conformational mobility and higher RMSDs of single residues in the wild-type ligand-binding site. Corresponding with that observation, we found fewer H bonds in the local H-bond network for the “unbound” wild type than variant structures. The wild type showed a gain of H bonds upon boceprevir binding, while the variants showed a loss of H bonds, which was most prominent in the V55A variant, with the lowest structural flexibility in that variant. This explains that ketoamides potentially fit easier into the ligand-binding site of the wild type than the Val55 variant structures, particular in that of the V55A variant. Structure simulations are in agreement with resistance data from the cell culture. The V55A variant led to a 1.6-fold increase in EC50 for boceprevir and a 3-fold increase in the telaprevir EC50, which is likely to be clinically significant. The V55I variant showed no fold increase in EC50 for boceprevir and only a 1.4-fold increase in EC50 for telaprevir. Interestingly, the T54S V55I double variant was found for all V55I variant strains but showed no resistance against boceprevir, whereas a clinically significant 7.9-fold increase in the telaprevir EC50 was found, which is much higher than that of the V55I single variant (unpublished data). Accessibility of the ligand-binding site seems to play a crucial role in ketoamide binding. Since it is problematic to calculate absolute volumes of open binding pockets, we used the minimum distance between Ser139 and its next neighbor residues to compute a measure for size and volume of a representative ligand-binding site pocket (S1 pocket). Binding pockets of the “unbound” wild-type structure were superimposed and compared for differences in shape and volume with respective pockets of “unbound” V55I and V55A variant structures (Fig. 3C and D). The variant structures showed most neighboring residues closer to Ser139 with constricted pockets in their ligand-binding sites. Apart from a few exceptions, the minimum distances were smaller than or similar to those obtained for the wild-type pockets. Since Ala55 needs less space than Ile55, we observed overall smaller pockets in V55A than V55I. Molecular changes in the ligand-binding site and effects on the H-bond pattern, together, explain the resistance levels observed in Val55 variants for HCV genotype 1a.
Viral variant fitness is crucial to select resistant variants from the quasispecies population under drug pressure. RNA replication capacity is one measure of the fitness of the virus, and this is dependent on proper processing of the polyprotein by the NS3 protease. V55A and V55I variants showed RNA replication of 28% and 44.5%, respectively, compared to the wild type. Ketoamides mimic the natural substrate of the protease at the site of NS3-NS4A scission, and it is likely that the constricted Val55 variant binding pockets lead to difficulties in protease-substrate interaction and interfere with substrate recognition and cleavage. This is in agreement with the stronger negative impact on RNA replication observed in the V55A variant compared to the V55I variant. Although yields of infectious virus generally correlate well with the RNA replication capacity (34, 39), both Val55 variants were found involved in a drop of infectious virus yield leading to relative infectivity (compared to wild type) of 24.8% for the V55I variant and only 3.1% for V55A. Variants leading to impairments in infectious virus yield are likely to affect NS3 domain-domain interactions between the protease and helicase (3, 4, 20, 34). Corresponding to that, we identified the “unbound” wild-type protease with larger conformational flexibility during molecular dynamics simulation than the Val55 variant structures. The higher structural flexibility might allow the wild type to adapt more efficiently to conformational changes needed for NS3 domain-domain interaction and could serve as a possible explanation for the virus yield drop in Val55 variants with the lowest structural flexibility and corresponding largest virus yield drop in the V55A variant. The steep decline in infectious virus yield is surprising given the fact that the V55A variant was found repeatedly in a public database and as the dominant baseline strain in a patient before ketoamide exposure (36). Furthermore, it was found during long-term follow-up of 5 of 12 patients upon boceprevir treatment (36). Loss of fitness is likely to negatively impact the persistence of NS3 resistance-associated variants and their ability to compete with wild-type virus upon discontinuation of antiviral therapy. Nonetheless, it is possible that they could become fixed in the viral quasispecies by compensatory second-site mutations. Such second-site changes may explain how the V55A variant that negatively impacts replication and infectious virus yield in H77S.3 cell culture could dominate in some treatment-naïve and -experienced patients. Such second-site substitutions could exist within NS3 or outside NS3, so identifying putative compensatory amino acid changes in the same viral strain is a difficult task and beyond the scope of this article.
In summary, we identified preexisting Val55 variants in the NS3 protease, providing a priori for resistance against ketoamides. The topology of Val55 in a residue-interaction network of the NS3 protease indicates a potential structure key role to modulate molecular changes in the ligand-binding site and protease-helicase interaction interface. Val55 variants showed compromised viral fitness due to reduced RNA replication capacity and a distinct drop in their infectious virus yields. Molecular dynamics simulations revealed structural changes in conjunction with fitness costs and drug resistance. Structural flexibility might be important to adapt for NS3 domain-domain interactions involved in particle assembly with lower structural flexibility as a possible explanation for reduced yields in variant viruses.
ACKNOWLEDGMENTS
The present study was supported by a DFG grant (Clinical Research Unit, CRU129, TP3, LE 491/16-2) and a DFG Research Fellowship (WE 4388/3-1) to C.W. S.M.L. was supported by grants and contracts from the National Institute for Allergy and Infectious Diseases and the NIH (U19-AI40035, R21-AI81058, and N01-AI25488). S.S. is grateful for financial support by the CIPSM Gender Support Program. I.A. acknowledges financial support by the CIPSM excellence cluster.
S.M.L. has served as a consultant for Abbott, Hoffmann-LaRoche, Juvaris Biotherapeutics, Merck, Novartis, and Pfizer; research in his laboratory is supported by Merck and Tibotec. The remaining authors disclose no conflicts.
Footnotes
Published ahead of print 17 January 2012
REFERENCES
- 1. Alter HJ, Seeff LB. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17–35 [DOI] [PubMed] [Google Scholar]
- 2. Assenov Y, Ramirez F, Schelhorn SE, Lengauer T, Albrecht M. 2008. Computing topological parameters of biological networks. Bioinformatics 24:282–284 [DOI] [PubMed] [Google Scholar]
- 3. Beran RK, Pyle AM. 2008. Hepatitis C viral NS3-4A protease activity is enhanced by the NS3 helicase. J. Biol. Chem. 283:29929–29937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beran RK, Serebrov V, Pyle AM. 2007. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J. Biol. Chem. 282:34913–34920 [DOI] [PubMed] [Google Scholar]
- 5. Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684 [Google Scholar]
- 6. Chenna R, et al. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Combet C, et al. 2007. euHCVdb: the European Hepatitis C Virus Database. Nucleic Acids Res. 35:D363–D366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doncheva NT, Klein K, Domingues FS, Albrecht M. 2011. Analyzing and visualizing residue networks of protein structures. Trends Biochem. Sci. 36:179–182 [DOI] [PubMed] [Google Scholar]
- 9. Dong J, Horvath S. 2007. Understanding network concepts in modules. BMC Syst. Biol. 1:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fried MW, et al. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975–982 [DOI] [PubMed] [Google Scholar]
- 12. Galtier N, Gouy M, Gautier C. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543–548 [DOI] [PubMed] [Google Scholar]
- 13. Hartmann C, Antes I, Lengauer T. 2007. IRECS: a new algorithm for the selection of most probable ensembles of side-chain conformations in protein models. Protein Sci. 16:1294–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463 [Google Scholar]
- 15. Hess B, Kutzner C, van Der Spoel D, Lindahl E. 2008. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4:1549. [DOI] [PubMed] [Google Scholar]
- 16. Hezode C, et al. 2009. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N. Engl. J. Med. 360:1839–1850 [DOI] [PubMed] [Google Scholar]
- 17. Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38 [DOI] [PubMed] [Google Scholar]
- 18. Kouranov A, et al. 2006. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 34:D302–D305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kwo PY, et al. 2010. Efficacy of boceprevir, an NS3 protease inhibitor, in combination with peginterferon alfa-2b and ribavirin in treatment-naive patients with genotype 1 hepatitis C infection (SPRINT-1): an open-label, randomised, multicentre phase 2 trial. Lancet 376:705–716 [DOI] [PubMed] [Google Scholar]
- 20. Ma Y, Yates J, Liang Y, Lemon SM, Yi M. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 82:7624–7639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Manns MP, et al. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958–965 [DOI] [PubMed] [Google Scholar]
- 22. McHutchison JG, et al. 2009. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N. Engl. J. Med. 360:1827–1838 [DOI] [PubMed] [Google Scholar]
- 23. McHutchison JG, et al. 2010. Telaprevir for previously treated chronic HCV infection. N. Engl. J. Med. 362:1292–1303 [DOI] [PubMed] [Google Scholar]
- 24. Moradpour D, Penin F, Rice CM. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453–463 [DOI] [PubMed] [Google Scholar]
- 25. Oostenbrink C, Villa A, Mark AE, van Gunsteren WF. 2004. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 25:1656–1676 [DOI] [PubMed] [Google Scholar]
- 26. Prongay AJ, et al. 2007. Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3-[2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034). II. Key steps in structure-based optimization. J. Med. Chem. 50:2310–2318 [DOI] [PubMed] [Google Scholar]
- 27. Qiu P, et al. 2009. Identification of HCV protease inhibitor resistance mutations by selection pressure-based method. Nucleic Acids Res. 37:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raney KD, Sharma SD, Moustafa IM, Cameron CE. 2010. Hepatitis C virus non-structural protein 3 (HCV NS3): a multifunctional antiviral target. J. Biol. Chem. 285:22725–22731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rong L, Dahari H, Ribeiro RM, Perelson AS. 2010. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci. Transl. Med. 2:30ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sarrazin C, et al. 2007. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology 132:1270–1278 [DOI] [PubMed] [Google Scholar]
- 31. Schuttelkopf AW, van Aalten DM. 2004. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60:1355–1363 [DOI] [PubMed] [Google Scholar]
- 32. Shannon P, et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shiffman ML, et al. 2007. Peginterferon alfa-2a and ribavirin for 16 or 24 weeks in HCV genotype 2 or 3. N. Engl. J. Med. 357:124–134 [DOI] [PubMed] [Google Scholar]
- 34. Shimakami T, et al. 2011. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology 140:667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Simmonds P, et al. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973 [DOI] [PubMed] [Google Scholar]
- 36. Susser S, et al. 2011. Analysis of long-term persistance of resistance mutations within the hepatitis C virus NS3 protease after treatment with telaprevir or boceprevir. J. Clin. Virol. 52:321–327 [DOI] [PubMed] [Google Scholar]
- 37. Susser S, et al. 2009. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology 50:1709–1718 [DOI] [PubMed] [Google Scholar]
- 38. Welsch C, et al. 2008. Molecular basis of telaprevir resistance due to V36 and T54 mutations in the NS3-4A protease of the hepatitis C virus. Genome Biol. 9:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Welsch C, et al. 7 December 2011, posting date Peptidomimetic escape mechanisms arise due to genetic diversity in the ligand-binding site of the HCV NS3/4A serine protease. Gastroenterology [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Word JM, et al. 1999. Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms. J. Mol. Biol. 285:1711–1733 [DOI] [PubMed] [Google Scholar]
- 41. Word JM, Lovell SC, Richardson JS, Richardson DC. 1999. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 285:1735–1747 [DOI] [PubMed] [Google Scholar]
- 42. Yi M, et al. 2006. Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor: reduced RNA replication fitness and partial rescue by second-site mutations. J. Biol. Chem. 281:8205–8215 [DOI] [PubMed] [Google Scholar]
- 43. Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 103:2310–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zeuzem S, et al. 2004. Peginterferon alfa-2b plus ribavirin for treatment of chronic hepatitis C in previously untreated patients infected with HCV genotypes 2 or 3. J. Hepatol. 40:993–999 [DOI] [PubMed] [Google Scholar]