Abstract
The ClpXP protease is a critical bacterial intracellular protease that regulates protein turnover in many bacterial species. Here we identified a pharmacological inhibitor of the ClpXP protease, F2, and evaluated its action in Bacillus anthracis and Staphylococcus aureus. We found that F2 exhibited synergistic antimicrobial activity with cathelicidin antimicrobial peptides and antibiotics that target the cell well and/or cell membrane, such as penicillin and daptomycin, in B. anthracis and drug-resistant strains of S. aureus. ClpXP inhibition represents a novel therapeutic strategy to simultaneously sensitize pathogenic bacteria to host defenses and pharmaceutical antibiotics.
INTRODUCTION
Caseinolytic proteases (Clp proteases) are intracellular bacterial proteases that regulate protein quality and turnover through controlled proteolysis. Degraded proteins include damaged or nonfunctional proteins as well as transcriptional regulators, rate-limiting enzymes, and proteins tagged during trans translation (10, 13, 16). Clp proteases are comprised of a proteolytic core, ClpP, which is paired with a regulatory ATPase such as ClpX. Clp ATPases recognize, unfold, and transfer the proteins to ClpP for degradation. Orthologs of ClpXP are found in many bacterial species and are often associated with cellular stress, such as heat shock, nutrient deprivation, and oxidative stress (10, 13). ClpX and/or ClpP has also been implicated in virulence of several pathogens, including Listeria monocytogenes, Salmonella, Staphylococcus aureus, and Streptococcus pneumoniae (10, 13). We recently demonstrated that ClpX is critical for the pathogenesis of Bacillus anthracis, a Gram-positive bacterium that is the causative agent of anthrax (20). Loss of ClpX increased susceptibility to innate host defenses, including cationic antimicrobial peptides, and severely attenuated B. anthracis virulence even in the fully pathogenic Ames strain (20).
An alternative strategy in antimicrobial therapy is to target and inactivate bacterial virulence factors rather than directly target growth or survival in the manner of traditional antibiotics (5). Inhibition of virulence factors involved in disease progression should enhance the ability of the host immune system to clear the pathogen. The ClpXP protease is a promising target for pharmacological inhibition due to its conserved nature and its role in the virulence of a wide variety of pathogens. We have identified several inhibitors of ClpXP using a screening system devised in Escherichia coli (8). We targeted the ClpXP protease of B. anthracis Sterne and the Gram-positive human pathogen Staphylococcus aureus using the ClpXP protease inhibitor F2. We found that F2 renders both B. anthracis Sterne and drug-resistant strains of S. aureus more susceptible to host antimicrobial peptides as well as antibiotics that target the bacterial cell envelope, including the cell wall and/or cell membrane.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. anthracis Sterne (pXO1 positive pXO2 negative) was propagated in brain heart infusion (BHI) medium (Sigma), and S. aureus strains were propagated in tryptic soy broth (Hardy). Genetically modified B. anthracis Sterne strains were previously described (20). Clinical daptomycin-nonsusceptible (Dapns) S. aureus clinical strains are described elsewhere (15), and Dapns methicillin-resistant S. aureus (MRSA) strain SA32D was derived through in vitro passage (25).
Synthesis of F2.
3-Chlorobenzoic acid, N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide (EDC), N-hydroxysuccinimide (NHS), triethylamine (TEA), dichloromethane (DCM), and dimethylformamide (DMF) were purchased from Sigma-Aldrich (St. Louis, MO). 1-Propyl-1H-tetrazol-5-amine was purchased from ChemBridge Corporation (San Diego, CA). Silica gel (60 Å, 60 to 200 μm) was purchased from VWR International (Bridgeport, NJ). Thin-layer chromatography (TLC) silica gel (IB-F) plates were purchased from J. T. Baker (Phillipsburg, NJ). 3-Chlorobenzoic acid (250 mg) was added to a round-bottom flask containing a 1:1 solution of DMF-DCM at room temperature, and to this mixture 197 mg EDC, 221 mg NHS, and 619 mg TEA were added. The reaction mixture was stirred for 5 min, a solution of 203 mg 1-propyl-1H-tetrazol-5-amine in DMF-DCM (1:1) was added to the flask, and the reaction mixture was continuously stirred at room temperature. After 12 h, the reaction was stopped and evaporated to dryness. The dry material was dissolved in 10 ml 0.5 M NaHCO3 and extracted with DCM (three times, 20 ml each time). The product was purified via column chromatography on silica gel and eluted with a CH2Cl2-methanol-TEA (9.8:0.1:0.1) solvent system in 65% yield. The product was verified using nuclear magnetic resonance (NMR) and mass spectrometry (MS). 1H NMR (400 MHz, CDCl3) δ 0.93 (t, J = 7.4, 3-H), 1.94 (m, 2-H), 4.28 (t, J = 7.4, 2-H), 7.45 (m, 2-H), 7.90 (m, 2-H), 11.78 (bs, 1-H). 13C NMR (100 MHz, CDCl3) δ 10.6, 21.8, 46.7, 124.8, 127.0, 129.7, 131.1, 134.5, 136.6, 161.1, amd 165.6. MS electrospray ionization m/z C11H12ClN5O: 265.07.
Pulse-chase assays.
The half-life of green fluorescent protein (GFP) with the E. coli transfer-messenger RNA (tmRNA) tag AANDENYALAA (GFP-AA) in vivo was determined using pulse-chase assays (17). E. coli cultures were grown in M9 minimal medium containing no methionine or cysteine at 30°C to an optical density (OD) at 600 nm of 0.6, and GFP-AA expression was induced by the addition of isopropyl-1-thio-β-d-galactopyranoside (IPTG) to 1 mM. At 90 min after induction, cells were labeled with 30 μCi l-[35S]methionine (MP Biomedicals) for 1 min and chased with a mixture of unlabeled l-methionine and l-cysteine at a final concentration of 1.25 mg/ml. At each time point, 450 μl culture was removed and added to 50 μl trichloroacetic acid, and protein was recovered by centrifugation. Protein pellets were resuspended in 50 μl SDS buffer (10 mM Tris-HCl, pH 8.0, 1% SDS, 1 mM EDTA) and separated by gel electrophoresis on 15% SDS-polyacrylamide gels, and the radiolabeled protein bands were visualized and quantified using a Typhoon imager (GE Healthcare Lifesciences). Half-lives were determined by fitting the plots of band intensity versus time to a single exponential. For cells treated with F2, F2 was added to a 100 μM final concentration at the time of induction.
Bacterial survival/growth assays.
Cultures were grown to early log phase and resuspended to an optical density of 0.4 in phosphate-buffered saline. B. anthracis Sterne was diluted 1:10 and S. aureus was diluted 1:100 in assay-specific medium to approximately 1 × 106 CFU/ml and 2 × 106 CFU/ml, respectively, and grown for 24 h at 37°C under static conditions. Survival was measured by serial dilution and plating to enumerate the number of surviving CFU/ml. Growth was measured by determination of the optical density at a wavelength of 600 nm. All experiments were done at least 3 times, and results from individual experiments were combined and presented as mean ± standard error of the mean (SEM).
Antimicrobial peptides.
Cultures were diluted in RPMI 1640 (Gibco) plus 5% Luria-Bertani broth (LB; Acumedia). Assay mixtures contained either F2 or its vehicle control (dimethyl sulfoxide [DMSO]) at the equivalent concentration. Synergy assays had four separate conditions, DMSO, F2, LL-37–DMSO, or LL-37–F2, using the concentrations indicated in the figure legends.
Antibiotics.
Bacterial strains were diluted in BHI medium for penicillin, erythromycin, or ciprofloxacin (all from Sigma) assays; daptomycin (Cubist Pharmaceuticals) assays were carried out in Mueller-Hinton broth II (Difco)–50 μg/ml calcium. Synergy assays had four separate conditions: DMSO, F2, antibiotic (penicillin or daptomycin)-DMSO, or antibiotic-F2 using the concentrations indicated. Assays using Daps and Dapns S. aureus strains were carried out in the presence of either DMSO or F2 plus the indicated amounts of daptomycin.
F2 resistance.
Bacteria were grown overnight in the presence of 0 μM F2 (DMSO control) or 40 μM F2 in RPMI 1640–5% LB. On the next day, the overnight cultures were diluted 1:100 (B. anthracis) or 1:1,000 (S. aureus) in RPMI 1640–5% LB and the indicated amount of F2 was added. Bacteria were then grown for an additional 24 h under static conditions, and the numbers of surviving CFU/ml were enumerated.
Whole-blood killing assays.
Blood collected from healthy donors (use and procedures approved by the University of California, San Diego, Human Research Protections Program) was incubated with 105 CFU B. anthracis and 40 μM F2 or vehicle control (DMSO) in a total volume of 500 μl and rotated at 37°C. At 30 min, a small aliquot was removed, blood was lysed in water, and remaining bacteria were enumerated. Experiments were performed using blood from three individual donors, and results are combined and presented as mean ± SEM.
Cytotoxicity assay.
HeLa cells were grown in Dulbecco's modified Eagle's medium (Difco) with 10% fetal bovine serum (Invitrogen), 1% sodium pyruvate, and 1% penicillin-streptomycin (Difco). Cells (2 × 104 per well) were plated in 96-well plates and grown for 24 h at 37°C in 5% CO2. Fresh medium containing the indicated concentration of F2 was added for an additional 24 h. F2 at 0 μM contained the equivalent vehicle control (DMSO) at the highest concentration. Untreated cells were included as a control. Twenty microliters of MTS tetrazolium [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] from the CellTiter 96 AQueous One Solution cell proliferation assay (Promega) was added to each well and the plates were incubated for 2 h at 37°C in 5% CO2 before quantifying the absorbance at 490 nm. One hundred percent viability was set at the absorbance of the untreated cells. Two independent experiments were combined, and results are presented as mean ± SEM.
RESULTS
Inhibition of the ClpXP protease.
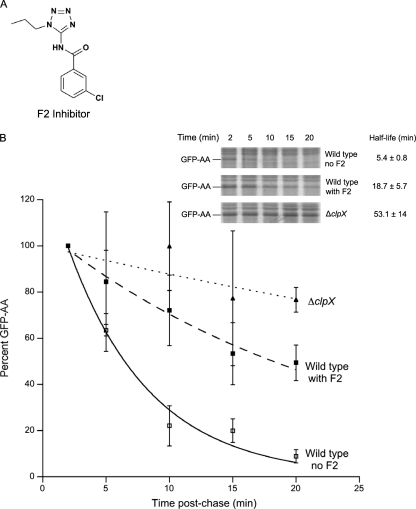
The compound F2 (Fig. 1A) was identified as part of a high-throughput screen for inhibitors of the protein tagging and degradation pathway known as trans translation in E. coli (unpublished data). F2 inhibited proteolysis of ClpXP substrates in vivo using pulse-chase assays that monitored the half-life of GFP with the E. coli tmRNA tag (GFP-AA) (Fig. 1B). The GFP-AA half-life was increased from 5.4 ± 0.8 min to 18.7 ± 5.7 min in the presence of F2. In comparison, bacteria lacking clpX had a protein half-life of 53.1 ± 14 min (average half-life values from 3 experiments). ClpX and ClpP are highly conserved among bacterial species (Fig. 2A and B) (6, 10), and therefore, it is likely that a pharmacological inhibitor may function in multiple species. Genetic deletion of clpX increases the susceptibility of B. anthracis Sterne to LL-37, the human cathelicidin antimicrobial peptide (see reference 20 and Fig. 3A; compare black hatched and light gray hatched bars). Cathelicidins are critical first-line effectors of innate defense against invasive bacterial infection (23). We hypothesized that if F2 could inhibit the ClpXP protease in B. anthracis Sterne, the parental bacteria would be rendered susceptible to cathelicidins in a manner similar to that for the mutant bacteria harboring a genetic deletion of the clpX gene. At a dose of 40 μM, F2 alone did not affect B. anthracis Sterne growth (Fig. 3A; compare black versus dark gray bars). However, in combination with LL-37, growth is inhibited to the same level as it is in the ΔclpX mutant (Fig. 3A; compare light and dark gray hatched bars).
Fig 1.
F2 inhibits the ClpXP protease. (A) Structure of F2; (B) pulse-chase assays to monitor proteolysis of the ClpXP substrate protein GFP-AA in the presence or absence of 100 μM F2; (inset) a representative blot. The graph depicts quantified band intensity of GFP, where the value for wild-type bacteria without F2 is set at 100% and mean results ± standard deviation from 3 experiments are shown.
Fig 2.
Conservation of ClpX/ClpP. Multiple-sequence alignment of ClpP (A) and ClpX (B) sequences from E. coli (Ec), S. aureus (Sa), and B. anthracis (Ba) was performed using the ClustalW program. The residues important for catalytic activity of ClpP are shown in bold. B. anthracis has at least two ClpP genes which are designated 1 and 2. *, amino acids are identical; colons, amino acids have conserved substitutions; periods, amino acids have semiconserved substitutions.
Fig 3.
F2 synergistically interacts with cathelicidin (LL-37) to inhibit B. anthracis survival. (A) Growth of B. anthracis Sterne wild type (WT) (black bars) with vehicle control (DMSO), ΔClpX (light gray bars) with DMSO, or wild type with 40 μM F2 (dark gray bars) after 24 h in the absence (solid bars) or presence (hatched bars) of 1.6 μM LL-37. *, P < 0.05 from untreated wild type; **, P < 0.05 from LL-37-treated wild type by one-way ANOVA, followed by Tukey's post hoc analysis. (B) Survival of wild-type B. anthracis Sterne after 24 h of incubation with DMSO vehicle control (control), 40 μM F2, and/or 1.6 μM LL-37, as indicated. $, no CFU recovered; #, a synergistic interaction (P < 0.05) by two-way ANOVA. (C) Survival of wild-type B. anthracis Sterne after 4 h of incubation with 0.8 μM LL-37 and the indicated amounts of F2. (D) Survival relative to the starting inoculum of wild-type B. anthracis Sterne after 30 min incubation with whole blood treated with vehicle control (DMSO) or 40 μM F2. ***, P < 0.001 by unpaired t test. (E) HeLa cells were incubated with the indicated amount of F2 for 24 h. Cell viability is proportional to the amount of a colored formazan product quantified at an OD of 490 nm. Fifty percent cell viability (50% inhibitory concentration [ID50]) is indicated by the gray line. (F) Survival of wild-type B. anthracis Sterne after incubation with the indicated amount of F2 for 24 h. Bacteria were grown for 24 h before assay in either 0 μM F2 (black bars) or 40 μM F2 (gray bars). $, no CFU recovered.
To examine more precisely the interaction between F2 and LL-37 in killing B. anthracis Sterne, survival of the bacteria was assessed after 24 h of incubation with vehicle control, 40 μM F2 alone, 1.6 μM LL-37 alone, or a combination of 40 μM F2 and 1.6 μM LL-37. Incubation with either compound alone had a negligible effect on survival in comparison to that achieved with the vehicle control; however, when the two compounds were combined, recovery of B. anthracis CFU was below the limit of detection (Fig. 3B). Similar results were seen with murine cathelicidin CRAMP (data not shown). The interaction between F2 and LL-37 was highly significant (two-way analysis of variance [ANOVA], P < 0.0001), indicating a strong synergistic interaction between these two compounds (28). F2 functions in a dose-dependent manner, since increasing amounts of F2 led to decreased bacterial survival when the amount of LL-37 was held constant (Fig. 3C).
In order to test whether the synergistic effect of F2 and antimicrobial peptides on the bacteria could potentially be used in therapeutic treatment of infection, we tested the ability of F2 to augment the bactericidal activity of human whole blood. Treatment of blood with 40 μM F2 significantly enhanced killing of B. anthracis in whole blood compared with that achieved with the vehicle control (Fig. 3D). Cytotoxicity in mammalian cells was not seen until a dose of 500 to 1,000 μM F2 (Fig. 3E).
Since the ClpXP protease plays a role in virulence in a number of pathogens, its inhibition could represent a potential therapeutic target for bacterial species besides B. anthracis. S. aureus is a major human pathogen and a leading cause of skin, soft tissue, and bloodstream infections. Of particular concern is the rise of antibiotic-resistant strains of S. aureus, including MRSA, now epidemic in health care and community settings in the United States and other developed countries. Loss of ClpXP leads to decreased virulence of S. aureus and results in a number of shared phenotypes with B. anthracis, including loss of hemolytic and proteolytic activity (11, 20). We hypothesized that F2 may function in a similar manner in S. aureus, and we tested its activity against several strains, including the methicillin-susceptible strain Newman and the MRSA strain Sanger 252. In both cases, survival of S. aureus was only slightly reduced when the bacteria were incubated for 24 h with either 10 μM LL-37 or 40 μM F2 alone (Fig. 4A and B). However, combining LL-37 and F2 revealed a strong synergistic effect. No colonies were recovered for S. aureus Newman (Fig. 4A), and survival was reduced by at least 10,000-fold for MRSA Sanger 252 (Fig. 4B). F2 exhibits direct antimicrobial activity against B. anthracis Sterne and MRSA Sanger 252 (Fig. 3F and 4C, black bars). Pretreatment of bacteria with 40 μM F2 induces resistance in S. aureus but not in B. anthracis under the same conditions (Fig. 3F and 4C).
Fig 4.
F2 is effective against S. aureus. Survival of S. aureus Newman (A) or MRSA Sanger 252 (B) with vehicle control, 40 μM F2, and/or 10 μM LL-37, as indicated. (C) Survival of MRSA Sanger 252 after incubation with the indicated amount of F2 for 24 h. Bacteria were grown for 24 h before assay in either 0 μM F2 (black bars) or 40 μM F2 (gray bars). $, no CFU recovered; #, a synergistic interaction (P < 0.05) by two-way ANOVA.
Loss of ClpX increases susceptibility to conventional antibiotics.
Deletion of clpX in B. anthracis resulted in increased susceptibility to host defenses such as antimicrobial peptides and lysozyme that target components of the cell envelope, including the cell membrane and/or cell wall (20). Since these bacterial structures are also targets of certain classes of antibiotics, we hypothesized that loss of clpX could increase susceptibility to cell envelope-acting antibiotics. We incubated the parental strain B. anthracis Sterne, the ΔclpX mutant, and the complemented strain ΔclpX pclpX in medium with or without 5 μg/ml of penicillin, a cell wall-acting antibiotic. We found that while all three strains grew well in media without penicillin, growth of the ΔclpX mutant was significantly reduced in the presence of penicillin (Fig. 5A). Incubation with daptomycin, an antibiotic that disrupts membrane potential and causes cell wall stress (22), exerted a similar effect. No viable bacteria were recovered upon incubation of the ΔclpX mutant with daptomycin, whereas the parental and the complemented strains were not significantly affected by daptomycin (Fig. 5B). This effect is not seen with all antibiotics, as no differences in growth were seen upon incubation with either ciprofloxacin, which targets topoisomerase II, or erythromycin, which targets protein synthesis (Fig. 5C and D).
Fig 5.
Loss of the clpX gene increases susceptibility to cell envelope antibiotics. Survival of wild-type B. anthracis Sterne (wild type) plus the empty vector (pEV), ΔclpX plus the empty vector (pEV), or ΔclpX pclpX in the presence of penicillin (A) or daptomycin (B). Survival of wild-type B. anthracis Sterne or the ΔclpX mutant in the presence of erythromycin (C) or ciprofloxacin (D). $, no CFU recovered; *, P < 0.05 from untreated by one-way ANOVA, followed by Tukey's post hoc analysis.
Inhibition of ClpXP increases antibiotic sensitivity.
Since ClpX was important for resistance to penicillin and daptomycin in B. anthracis Sterne, we next tested whether inhibition of the ClpXP protease using F2 would sensitize B. anthracis Sterne to penicillin or MRSA Sanger 252 to daptomycin. Bacteria were incubated with vehicle control, F2 alone, antibiotic alone, or a combination of F2 and antibiotic. Bacterial survival (numbers of CFU/ml) was determined at 24 h. In both cases, neither treatment with F2 nor treatment with antibiotic alone had a significant effect on survival. However, a combination of F2 and penicillin with B. anthracis Sterne (Fig. 6A) or F2 and daptomycin with MRSA (Fig. 6B) significantly reduced bacterial survival in a synergistic manner.
Fig 6.
F2 decreases bacterial resistance to antibiotics. (A) Survival of wild-type B. anthracis Sterne after 24 h of incubation with vehicle control, 40 μM F2, and/or 5 mg/ml penicillin (PEN), as indicated. (B) Survival of MRSA Sanger 252 with vehicle control, 40 μM F2, and/or 1 mg/ml daptomycin (DAP), as indicated. Two daptomycin-susceptible S. aureus parent strains and daptomycin-nonsusceptible strains, 0616/0701 (C) and 32/32D (D), were incubated with the indicated concentration of daptomycin. F2 at 15 to 20 μM was added to 0701 or 32D, as shown, and the surviving numbers of CFU were enumerated 24 h later. #, a synergistic interaction (P < 0.05) by two-way ANOVA; *, P < 0.05 compared to daptomycin-nonsusceptible S. aureus (0701 or 32D) by one-way ANOVA, followed by Tukey's post hoc analysis; $, no CFU recovered.
Although daptomycin is a relatively new antibiotic, resistant strains have already been reported in clinical practice (12, 26). We next asked whether F2 could increase daptomycin susceptibility (Daps) in daptomycin-nonsusceptible (Dapns) S. aureus strains. Strains SA0616 (Daps) and SA0701 (Dapns) were isolated from the bloodstream of a patient with daptomycin treatment failure before and after daptomycin therapy (26). Dapns MRSA strain SA32D was derived by in vitro passage of Daps MRSA strain SA32 (25). Both SA0701 and SA32D were more resistant to daptomycin killing than their parental strains, SA0616 and SA32, respectively (Fig. 6C and D; compare gray and black bars). Also, in both cases, treatment of either SA0701 or SA32D with F2 decreased their resistance to daptomycin (Fig. 6C and D; compare black and white bars), although in neither case did it return daptomycin susceptibility to its original levels (Fig. 6C and D; compare white and gray bars).
DISCUSSION
It is estimated that 20% of newly synthesized proteins are degraded by the proteosome due to transcriptional or translational errors (30) and this number increases under stress conditions such as heat shock (10, 13). Bacterial proteosomes also control the half-life of transcription factors and rate-limiting enzymes, thereby exerting a regulatory effect on gene expression and metabolism. Thus, regulated proteolysis is critical from a quality control as well as a regulatory standpoint, and loss of these intracellular proteases can have detrimental effects (10, 14). During infection, pathogens face numerous stress conditions, including nutrient deprivation, exposure to reactive oxygen species, and temperature and pH changes. Loss of the Clp protease system attenuates virulence in several pathogens, including B. anthracis and S. aureus (10, 13, 20), making the ClpXP protease a potential target for pharmacological intervention.
We have identified a novel inhibitor of the ClpXP protease, F2, which targets bacterial cells with minimal host cell cytotoxicity. While it is unclear exactly how F2 is inhibiting the ClpXP protease, our data indicate that inclusion of F2 decreases the proteolysis of ClpXP substrates in vivo. Similar to a genetic loss of ClpX, cotreatment of B. anthracis with F2 increased susceptibility of the bacteria to cathelicidin antimicrobial peptides. A similar effect was seen with both methicillin-susceptible and methicillin-resistant strains of S. aureus, suggesting that ClpXP also plays a role in cathelicidin resistance in S. aureus. While previous studies have demonstrated the effectiveness of pharmacological interference with ClpP function (2, 3), this is the first study to show that inhibition of ClpXP could result in a synergistic interaction with innate immune defenses. Although we focused upon cathelicidins in this study, we have previously shown that loss of ClpX renders B. anthracis more sensitive to other antimicrobial proteins, including defensins and lysozyme (20). Therefore, pharmacological inhibition of ClpXP may increase susceptibility to multiple host defenses.
F2 also sensitizes B. anthracis and S. aureus to antibiotics such as penicillin and daptomycin, although the synergistic effect between F2 and antibiotics was more pronounced in B. anthracis. This may reflect our assay conditions, or it may indicate that the extent to which ClpXP influences susceptibility to cell envelope antibiotics differs among species. Evidence of a connection between Clp proteases and cell wall-acting antibiotics has been seen in other bacterial species. Loss of ClpP in Streptococcus mutans rendered the bacteria more susceptible to the cell wall-acting antibiotics bacitracin, polymyxin B, and vancomycin, while no effect was seen with non-cell wall-acting antibiotics (7). In Mycobacterium tuberculosis, loss of the ClpCP protease resulted in increased susceptibility to cell wall stress induced by vancomycin or SDS (1). Daptomycin is believed to function by membrane depolarization, but a recent study demonstrated that it also induces the cell wall stress regulon, including Clp family members, in S. aureus (22). The importance of the ClpXP system is further highlighted by our observation that inhibition increased susceptibility to daptomycin in otherwise nonsusceptible strains. However, this suppression of resistance was only partial, indicating that non-Clp-dependent effects also probably contribute to daptomycin resistance.
It is likely that loss of ClpXP has pleiotropic effects on the bacterial cell since the Clp protease regulates a wide range of genes (21, 24). Cell wall-active agents are believed to increase damaged or misfolded proteins and result in induction of genes involved in protein turnover, such as those for chaperones and proteases (22, 29). Loss of ClpXP could hamper this response. The ClpXP protease may also be regulating critical components of the cell wall. In E. coli, ClpXP can degrade FtsZ, a major cytoskeletal protein that is implicated in cell division and cell wall synthesis (4). In Bacillus subtilis, MurAA, an enzyme important in peptidoglycan formation, is degraded by ClpCP (18). It is also possible that cell charge is affected by loss of ClpXP. Daptomycin is an anionic compound that associates with calcium to form a cationic complex similar to an antimicrobial peptide. Resistance to daptomycin and cationic antimicrobial peptides has been linked to mutations in mprF (lysine addition to cell membrane phosphatidylglycerol) and the dltABCD operon (alanylation of cell wall teichoic acids) that result in increased net positive surface charge in both S. aureus and B. anthracis (9, 19, 27, 31). Consistent with this hypothesis, increasing daptomycin resistance in S. aureus was accompanied by increased resistance to cationic antimicrobial peptides such as alpha-defensin HNP-1 and platelet microbicidal proteins (15). The Clp protease could be regulating either directly or indirectly expression of the mprF or the dltABCD operon, although this has not been demonstrated to date.
The ClpXP protease is a promising target for pharmacological intervention. Inclusion of F2 increased the effectiveness of bacterial killing by human whole blood, indicating that this compound can augment innate immune defenses. This therapeutic effect may be magnified at tissue sites of infection where high levels of antimicrobial peptides are produced by cells such as keratinocytes and in patients receiving concurrent antibiotic therapy with cell wall-active agents for which ClpXP inhibition also provides synergism. While we have focused on inhibition of ClpXP, uncontrolled activation of ClpP through a new class of antibiotics, acyldepsipeptides, also has lethal consequences for several bacterial species tested (3). However, as was seen with acyldepsipeptides (3) and other antimicrobial compounds, potential for bacterial resistance to F2 exists. The mechanism behind this resistance is unclear at this point, but it may necessitate that F2 or another pharmacological inhibitor of ClpXP be used in a combination rather than single therapy to limit resistance. Nevertheless, the use of Clp inhibitors can be predicted to contribute to antimicrobial activity on multiple levels, through increased susceptibility to innate immune defenses and decreased resistance to traditional antibiotics, potentially increasing therapeutic effectiveness.
ACKNOWLEDGMENTS
S.M.M. was supported by The Hartwell Foundation and through the Texas Christian University Research and Creative Activities Fund (60595) and the Junior Faculty Summer Research Program. N.S.R., J.N.A., and K.C.K. (GM68720), V.N. (AI052453, AR052728, HD071600), C.Y.O. (GM06852), and G.S. (HD071600) were supported by the National Institutes of Health.
Footnotes
Published ahead of print 17 January 2012
REFERENCES
- 1. Barik S, Sureka K, Mukherjee P, Basu J, Kundu M. 2010. RseA, the SigE specific anti-sigma factor of Mycobacterium tuberculosis, is inactivated by phosphorylation-dependent ClpC1P2 proteolysis. Mol. Microbiol. 75:592–606 [DOI] [PubMed] [Google Scholar]
- 2. Böttcher T, Sieber SA. 2008. Beta-lactones as privileged structures for the active-site labeling of versatile bacterial enzyme classes. Angew Chem. Int. ed. Engl. 47:4600–4603 [DOI] [PubMed] [Google Scholar]
- 3. Brötz-Oesterhelt H, et al. 2005. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 11:1082–1087 [DOI] [PubMed] [Google Scholar]
- 4. Camberg JL, Hoskins JR, Wickner S. 2009. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc. Natl. Acad. Sci. U. S. A. 106:10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. 2008. The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 6:17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chandu D, Nandi D. 2004. Comparative genomics and functional roles of the ATP-dependent proteases Lon and Clp during cytosolic protein degradation. Res. Microbiol. 155:710–719 [DOI] [PubMed] [Google Scholar]
- 7. Chattoraj P, Banerjee A, Biswas S, Biswas I. 2010. ClpP of Streptococcus mutans differentially regulates expression of genomic islands, mutacin production, and antibiotic tolerance. J. Bacteriol. 192:1312–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng L, et al. 2007. Discovery of antibacterial cyclic peptides that inhibit the ClpXP protease. Protein Sci. 16:1535–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fisher N, et al. 2006. The dltABCD operon of Bacillus anthracis Sterne is required for virulence and resistance to peptide, enzymatic, and cellular mediators of innate immunity. J. Bacteriol. 188:1301–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frees D, Savijoki K, Varmanen P, Ingmer H. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 63:1285–1295 [DOI] [PubMed] [Google Scholar]
- 11. Frees D, Qazi SN, Hill PJ, Ingmer H. 2003. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 48:1565–1578 [DOI] [PubMed] [Google Scholar]
- 12. Hayden MK, et al. 2005. Development of daptomycin resistance in vivo in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5285–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ingmer H, Brøndsted L. 2009. Proteases in bacterial pathogenesis. Res. Microbiol. 160:704–710 [DOI] [PubMed] [Google Scholar]
- 14. Jenal U, Hengge-Aronis R. 2003. Regulation by proteolysis in bacterial cells. Curr. Opin. Microbiol. 6:163–172 [DOI] [PubMed] [Google Scholar]
- 15. Jones T, et al. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 52:269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keiler KC. 2008. Biology of trans-translation. Annu. Rev. Microbiol. 62:133–151 [DOI] [PubMed] [Google Scholar]
- 17. Keiler KC, Waller PR, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993 [DOI] [PubMed] [Google Scholar]
- 18. Kock H, Gerth U, Hecker M. 2004. MurAA, catalysing the first committed step in peptidoglycan biosynthesis, is a target of Clp-dependent proteolysis in Bacillus subtilis. Mol. Microbiol. 51:1087–1102 [DOI] [PubMed] [Google Scholar]
- 19. Kraus D, Peschel A. 2006. Molecular mechanisms of bacterial resistance to antimicrobial peptides. Curr. Top. Microbiol. Immunol. 306:231–250 [DOI] [PubMed] [Google Scholar]
- 20. McGillivray SM, et al. 2009. ClpX contributes to innate defense peptide resistance and virulence phenotypes of Bacillus anthracis. J. Innate Immun. 1:494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Michel A, et al. 2006. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188:5783–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Muthaiyan A, Silverman JA, Jayaswal RK, Wilkinson BJ. 2008. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob. Agents Chemother. 52:980–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nizet V, et al. 2001. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414:454–457 [DOI] [PubMed] [Google Scholar]
- 24. Robertson GT, Ng WL, Foley J, Gilmour R, Winkler ME. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 184:3508–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sakoulas G, Alder J, Thauvin-Eliopoulos C, Moellering RC, Eliopoulos GM. 2006. Induction of daptomycin heterogeneous susceptibility in Staphylococcus aureus by exposure to vancomycin. Antimicrob. Agents Chemother. 50:1581–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sakoulas G, et al. 2008. Evaluation of endocarditis caused by methicillin-susceptible Staphylococcus aureus developing nonsusceptibility to daptomycin. J. Clin. Microbiol. 46:220–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Samant S, Hsu FF, Neyfakh AA, Lee H. 2009. The Bacillus anthracis protein MprF is required for synthesis of lysylphosphatidylglycerols and for resistance to cationic antimicrobial peptides. J. Bacteriol. 191:1311–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Slinker BK. 1998. The statistics of synergism. J. Mol. Cell. Cardiol. 30:723–731 [DOI] [PubMed] [Google Scholar]
- 29. Utaida S, et al. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719–2732 [DOI] [PubMed] [Google Scholar]
- 30. Wickner S, Maurizi MR, Gottesman S. 1999. Posttranslational quality control: folding, refolding, and degrading proteins. Science 286:1888–1893 [DOI] [PubMed] [Google Scholar]
- 31. Yang SJ, et al. 2009. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus strains. Antimicrob. Agents Chemother. 53:2636–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]