Abstract
Mupirocin is a topical antibiotic used for the treatment of skin infections and the eradication of methicillin-resistant Staphylococcus aureus carriage. It inhibits bacterial protein synthesis by interfering with isoleucyl-tRNA synthetase activity. High-level mupirocin resistance (MIC of ≥512 μg/ml) is mediated by the expression of mupA (ileS2), which encodes an alternate isoleucyl-tRNA synthetase. In this study, we describe high-level mupirocin resistance mediated by a novel locus, mupB. The mupB gene (3,102 bp) shares 65.5% sequence identity with mupA but only 45.5% identity with ileS. The deduced MupB protein shares 58.1% identity (72.3% similarity) and 25.4% identity (41.8% similarity) with MupA and IleS, respectively. Despite this limited homology, MupB contains conserved motifs found in class I tRNA synthetases. Attempts to transfer high-level mupirocin resistance via conjugation or transformation (using plasmid extracts from an mupB-containing strain) were unsuccessful. However, by cloning the mupB gene into a shuttle vector, it was possible to transfer the resistance phenotype to susceptible S. aureus by electroporation, proving that mupB was responsible for the high-level mupirocin resistance. Further studies need to be done to determine the prevalence of mupB and to understand risk factors and outcomes associated with resistance mediated by this gene.
INTRODUCTION
Mupirocin (pseudomonic acid A), a polyketide antibiotic naturally produced by Pseudomonas fluorescens strain NCIMB 10586, is used topically for the treatment of skin infections, prevention of surgical site infections, and eradication of Staphylococcus aureus carriage. It also exhibits in vitro activity against some Gram-negative organisms, such as Haemophilus and Neisseria spp. (21). The epoxide side chain of mupirocin is structurally similar to that of isoleucine and can bind to the isoleucine-specific binding pocket of isoleucyl-tRNA synthetase, an enzyme that normally promotes the conversion of isoleucine and tRNA to isoleucyl-tRNA. Mupirocin-mediated inhibition of isoleucyl-tRNA synthetase impedes protein and RNA synthesis, ultimately leading to bacterial death (10). The prevalence of mupirocin-resistant S. aureus, particularly among methicillin-resistant S. aureus (MRSA) strains, is increasing (6, 14, 19). Three categories of mupirocin susceptibility have been described for S. aureus (8, 17). S. aureus is considered mupirocin susceptible at a MIC of ≤4 μg/ml. MICs of 8 to 64 μg/ml refer to low-level resistance and are usually due to nonsynonymous changes in the native isoleucyl-tRNA synthetase gene, ileS. Isolates with a MIC of 128 or 256 μg/ml are uncommon and are also considered to demonstrate low-level resistance (17, 19). A MIC of ≥512 μg/ml reflects high-level mupirocin resistance and is mediated by the acquisition of a conjugative plasmid containing mupA (ileS2), which encodes an alternate isoleucyl-tRNA synthetase (11, 23). To our knowledge, there is no other reported mechanism of high-level resistance. Here we report the analysis of isolates that demonstrate high-level mupirocin resistance (MIC of ≥1,024 μg/ml) but are mupA negative by PCR, and we describe a novel gene, mupB, which confers resistance to mupirocin.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The three MRSA strains (designated MUP87, MUP88, and MUP91) included in this study were recovered during surveillance conducted by the Canadian Nosocomial Infection Surveillance Program in 2004 (19, 20). All were highly resistant to mupirocin (MICs of >1,024 μg/ml) but were negative for the mupA gene by PCR (the primers used are described in Table 1). These isolates were from three different patients at the same hospital. Unfortunately, limited clinical information associated with these three isolates (such as prior exposure to mupirocin or other antimicrobials) was available. GP135, a highly mupirocin-resistant strain (MIC, >1,024 μg/ml), was used as a control in PCR assays. The presence of the mupA gene in strain GP135 was confirmed by amplification and sequencing of the entire gene. Mupirocin-susceptible strains NRS1 (tetracycline and gentamicin resistant) and NRS100 (tetracycline resistant) were used as acceptor strains in conjugative assays. Commercial chemically competent Escherichia coli Top10 (Life Technologies), S. aureus strain NRS144, and the pCN48 shuttle vector (2) were used for mupB cloning assays. NRS144 competent cells were also used for transformation assays with plasmids extracted from strains GP135 and MUP87.
Table 1.
Primers used in this study
| Target | Primer | Primer sequence (5′ to 3′) | Amplicon size (bp) | Ta (°C)a | Reference |
|---|---|---|---|---|---|
| ileS | Smr1 | ATAAAGGTAAAAAGCCAGTTTATTGGT | 360 | 55 | 23 |
| ileS-R3 | CAACATACTCCAATTCCTTAC | This work | |||
| Mrm1 | TCCCAGCAGATATGTATTTAGAAGGT | 797 | 55 | 23 | |
| ileS-R2 | TAATGCACGGTTCACATCATC | This work | |||
| mupA | HWmupA-F | ATAAGTGATACTCTAGGAGGC | 447 | 53 | This work |
| HWmupA-R | AGTCCATGTCAACCCAGTATCC | This work | |||
| mupA-Fb | TATATTATGCGATGGAAGGTTGG | 457 | 53 | 1 | |
| mupA-Rb | AATAAAATCAGCTGGAAAGTGTTG | This work | |||
| mupB | mupB-Fb | CTAGAAGTCGATTTTGGAGTAG | 674 | 55 | This work |
| mupB-Rb | AGTGTCTAAAATGATAAGACGATC | This work | |||
| mupBF-BamHIc | GGAAGATGGATCCATTTGGACTTAATAAAAAAGGTC | 3,643 | 53 | This work | |
| mupBR-KpnIc | TAATTTTGGTACCTTAATTTGTAAAGCTAGACATTAAC | This work | |||
| blaZ | blaZ-F | AACTTATTGAGGCTTCAATGAC | 353 | 55 | This work |
| blaZ-R | TGCTTGACCACTTTTATCAGC | This work |
Ta, PCR annealing temperature.
Internal primer for mupA and mupB detection.
Primer for mupB cloning (restriction site is underlined).
Susceptibility testing and molecular typing.
Antibiotic susceptibility testing was performed by Etest according to the manufacturer's recommendations (AB bioMérieux, Marcy l'Étoile, France) and interpreted based on the Clinical and Laboratory Standards Institute breakpoints (4). The genetic relatedness of strains MUP87, MUP88, and MUP91 was assessed by pulsed-field gel electrophoresis (PFGE) of SmaI-digested genomic DNA under previously described conditions (16). DNA profiles were analyzed with BioNumerics software (ver. 3.0; Applied Maths, Austin, TX).
PCR assays and sequencing.
Initial analysis of strain MUP87 targeted the ileS and mupA genes, encoding the native and acquired isoleucyl-tRNA synthetases, respectively. Primers for amplification and sequencing of the ileS gene (Table 1) targeted regions where point mutations conferring mupirocin resistance had been described previously (1, 9, 25). DNA sequencing was performed with BigDye Terminator methodology and a model 3130XL DNA sequence analyzer (Applied Biosystems/Life Technologies, Carlsbad, CA). The nucleotide and deduced amino acid sequences were analyzed with the Vector NTI analysis software package (version 10.3.0; Life Technologies Corp.). Sequence comparisons were performed with the BLAST program, available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/blast/). Multiple-sequence alignments were performed with the ClustalW2 program, available at the European Bioinformatics Institute website (http://www.ebi.ac.uk/Tools/msa/clustalw2).
Conjugative assays.
Overnight cultures of donor and recipient strains were mixed in 500 μl Mueller-Hinton broth (Oxoid) at different donor/recipient ratios (1:1, 1:5, and 1:10). The mixtures were incubated on Mueller-Hinton agar plates (Oxoid) without antibiotic pressure. After 18 h of incubation at 36°C, the cells were collected in 100 μl of Mueller-Hinton broth. Tenfold serial dilutions were plated on selective Mueller-Hinton agar plates supplemented with mupirocin (4 μg/ml) plus tetracycline (4 μg/ml) and/or gentamicin (4 μg/ml), depending on the acceptor cell used, and subsequently incubated at 36°C for 24 h for selection of transconjugants.
Plasmid extraction and transformation.
Plasmids from S. aureus GP135 (harboring the plasmid-mediated mupA gene) and MUP87 (mupB positive) were extracted using a Qiagen plasmid maxiprep kit, with a few modifications. Briefly, 100 μl of 2 mg/ml lysostaphin (Sigma) was added to the resuspended cell pellet and incubated for 1 h at 37°C before adding the lysis buffer. Undigested and digested (HindIII; New England BioLabs) plasmids were analyzed by electrophoresis on 0.7% agarose gels (Tris-acetate buffer) and transferred to a nylon membrane to determine the plasmid location of the mupB gene by Southern hybridization analysis. Specific digoxigenin-labeled mupB, mupA, and blaZ probes, generated by PCR (Table 1), and the hybridization conditions were based on the manufacturer's recommendations (Roche Diagnostics). For transformation assays, NRS144 competent cells were prepared (12) and transformed by electroporation using plasmid extracts from strains GP135 and MUP87. Mueller-Hinton agar plates supplemented with mupirocin (4 μg/ml) were used to select potential transformants.
Whole-genome sequencing.
For identification of the genetic determinant of mupirocin resistance and its genetic background, whole-genome sequencing of S. aureus MUP87 was performed. Genomic DNA was isolated by using a QIAamp DNA miniprep kit (Qiagen), with pretreatment of the strain with 2 mg/ml lysostaphin prior to lysis. Whole-genome sequencing of S. aureus MUP87 was performed with a Roche GS-FLX pyrosequencer using Titanium chemistry (Roche Diagnostics, Laval, Quebec, Canada) (15). De novo assembly of sequencing data was performed with gsAssembler (Roche). Mauve was used to compare these contigs with previously published S. aureus reference genomes (5) and thus to identify genomic and plasmid sequences present only in the MUP87 strain.
Cloning of the mupB gene.
High-quality genomic DNA was used for amplification of the mupB gene by use of Elongase enzyme (Life Technologies) and primers containing BamHI and KpnI restriction sites (Table 1). PCR was performed under the following conditions: 94°C for 1 min and 35 cycles of 94°C for 30 s, 53°C for 30 s, and 68°C for 4 min. Plasmid pCN48, extracted with a Plasmid Midi kit (Qiagen), and mupB amplicons were digested with BamHI and KpnI (New England BioLabs), ligated, and transformed into E. coli Top10 competent cells. Transformed cells were selected with 50 μg/ml carbenicillin. Recombinant plasmids (pCN-mupB) were extracted and sequenced to confirm insert integrity. S. aureus NRS144 competent cells were transformed with pCN-mupB by electroporation (Gene Pulser Xcell; Bio-Rad), and transformants were selected with 4 μg/ml mupirocin.
Nucleotide sequence accession number.
The nucleotide sequence of the mupB gene has been assigned GenBank accession number JQ231224.
RESULTS AND DISCUSSION
Antimicrobial susceptibility testing and PFGE typing.
Strains MUP87, MUP88, and MUP91 displayed identical susceptibility profiles, with the following MICs: erythromycin, ≥256 μg/ml; clindamycin, ≥256 μg/ml; tetracycline, 0.125 μg/ml; gentamicin, 0.38 μg/ml; chloramphenicol, 4 μg/ml; ciprofloxacin, ≥32 μg/ml; linezolid, 0.75 μg/ml; vancomycin, 2 μg/ml; trimethoprim-sulfamethoxazole, 0.047 μg/ml; mupirocin, ≥1,024 μg/ml; and oxacillin, ≥256 μg/ml. The 3 strains had identical banding patterns and were characterized as CMRSA-2 (U.S. pulsotype USA100, sequence type 5) strains by PFGE (3). Based on these phenotypic and genotypic results, we chose one strain (MUP87) for additional molecular studies. Analysis of the MUP87 genome sequence indicates that this strain carries staphylococcal cassette chromosome mec type II (SCCmec II).
mupA detection and identification of missense mutations in the ileS gene.
High-level mupirocin resistance is traditionally associated with acquisition of mupA. However, there is not a perfect correlation, and isolates that are mupA positive by PCR but mupirocin susceptible in vitro have been reported. This discordance is attributed to frameshift mutations that inactivate the MupA product (7). Also, mupA-positive isolates with low-level mupirocin resistance have been identified. In these isolates, the mupA gene was located on the chromosome rather than on a plasmid (18). However, the phenomenon described here, high-level resistance in the absence of mupA, has not been reported previously. To eliminate possible primer mismatch or other technical difficulties associated with PCR-based detection of mupA, we tested two pairs of primers targeting two different regions of the mupA gene (Table 1). However, all PCR attempts with strain MUP87 were negative. We also amplified and sequenced 2 fragments of the chromosomal ileS gene to identify point mutations previously associated with mupirocin resistance (1, 9, 25). Missense mutations (A637G/N213K and A741T/E285K) were identified by this approach, but none was linked to mupirocin resistance.
Characterization of the mupB gene.
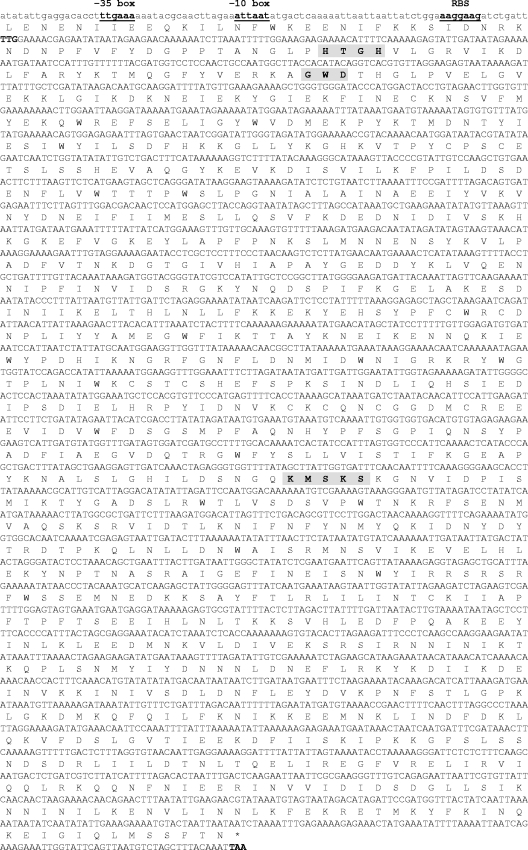
MUP87-specific contigs were identified by comparing the draft genome against MRSA reference sequences. Analysis and preliminary annotation of MUP87-specific sequences identified an ∼19-kb contig harboring an open reading frame (ORF) of 3,102 bp with 65.5% identity to mupA genes (GenBank accession numbers X75439 and GU237136). Only 45.5% identity with ileS (GenBank accession number X74219) was observed. The deduced protein shared 58.1% identity (72.3% similarity) and 25.4% identity (41.8% similarity) with MupA and IleS, respectively. We named the new gene mupB (ileS3). A putative promoter region was identified 67 nucleotides upstream of the start codon: the −35 box (TTGAAA) was separated from the −10 box (ATTAAT) by a 17-bp spacer, and both boxes matched with the consensus sequences TTGACA and TATAAT, respectively (24). A ribosome binding site (AAGGAAG) was found 8 bp upstream of the TTG start codon (Fig. 1). A stem-loop structure described after the stop codon in the mupA gene (GenBank accession number X75439) was not found downstream of mupB. The deduced MupB enzyme contained conserved motifs found in class I tRNA synthetases and required for ATP hydrolysis (HXGH and KMSKS), as well as the GWD motif, involved in amino acid activation and conserved between isoleucyl-tRNA synthetases (Fig. 1) (11). Sequence comparison of mupB-flanking regions with other known prokaryotic ORFs revealed no significant homology (GenBank; December 2011). Overall, this ∼19-kb contig had a 28.4% GC content (for mupB, 29%), compared to ∼33% for S. aureus USA300 (http://www.kazusa.or.jp/codon/).
Fig 1.
Nucleotide sequence of the mupB gene and its promoter region. Start and stop codons are indicated in bold, and protein translation is reported above the DNA sequence. A putative promoter region is indicated, including the −35 box (TTGAAA), the −10 box (ATTAAT), and a ribosome binding site (RBS) (AAGGAAG). Conserved motifs found in class I tRNA synthetases are highlighted in gray.
Mupirocin resistance gene location.
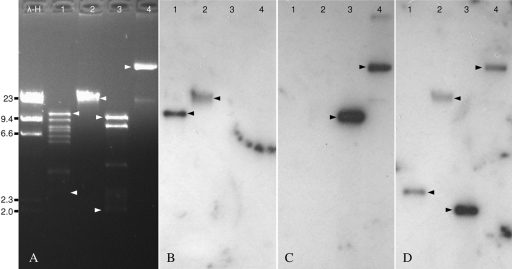
Southern hybridization assays using blaZ (encoding a β-lactamase and commonly found on plasmids from S. aureus), mupA, and mupB probes against plasmid extracts (HindIII digested and undigested) from strains GP135 (mupA+) and MUP87 (mupB+) seemed to be consistent with a plasmid location of mupB (Fig. 2). Cross-reactivity between the mupA and mupB probes was not observed. To analyze the potential horizontal mobility of the mupB gene, the MUP87 isolate was mated with 2 different mupirocin-susceptible S. aureus acceptors. After several attempts, no transconjugants were obtained, suggesting that the gene was carried on a nonconjugative plasmid. Despite multiple attempts using different concentrations of plasmid extract and different batches of competent cells, we were unable to transfer the mupirocin resistance mediated by mupB from strain MUP87 to strain NRS144. In contrast, in control experiments with mupA-containing extracts, mupA transformants were obtained, demonstrating the competency of strain NRS144. Failure to transfer mupB may have been due to some idiosyncrasy of the plasmid carrying the mupB gene. We presume that maintenance of the mupB plasmid is dependent on a specific host factor(s) found in MUP87 but missing in NRS144, affecting plasmid replication in the recipient host after transformation. To confirm this, genetic characterization of strain MUP87, including its whole-genome annotation, is ongoing.
Fig 2.
Southern blot analysis. (A) Plasmids extracted from strains MUP87 (mupB positive) and GP135 (mupA positive) and digested with HindIII (lanes 1 and 3) or left undigested (lanes 2 and 4). Autoradiograms of gel A hybridized with mupB (B), mupA (C), and blaZ (D) probes are shown. Arrows indicate positive bands for the different probes. Lane λ-H, λ phage-HindIII molecular size marker (in kb).
High-level mupirocin resistance is mediated by mupB.
S. aureus NRS144 was transformed with the recombinant plasmid pCN-mupB, carrying mupB plus a 541-bp sequence upstream of the gene (BamHI/KpnI insert). S. aureus NRS144 is a susceptible strain selected for transformability with DNA from E. coli (13). The mupirocin MIC of this strain is <0.064 μg/ml. After transformation, the strain (positive for mupB by PCR) developed high-level resistance to mupirocin (≥1,024 μg/ml). These results confirmed that mupB was the genetic determinant conferring the resistance phenotype.
We designed primers for the specific detection of the mupB gene (Table 1). We tested them against strains MUP87, MUP88, and MUP91 (the 3 clinical MRSA strains highly resistant to mupirocin), strain GP135 (mupA positive), and strain NRS144 (mupA and mupB negative). Amplicons were obtained only with strains MUP87, MUP88, and MUP91, confirming the same mechanism of mupirocin resistance in these 3 clinical isolates.
In summary, we have identified mupB, a new determinant of high-level resistance to mupirocin. The predicted protein exhibits low identity with the previously described MupA and IleS proteins (58.1% and 25.4% identity, respectively) but exhibits motifs common to other isoleucyl-tRNA synthetases. The origin of mupB has not been determined, and sequences flanking the resistance gene do not exhibit homology with any GenBank entries. Southern hybridization results strongly suggest that mupB is located on a presumably nonconjugative plasmid. An epidemiological study of mupirocin-resistant MRSA isolates revealed that the rate of high-level mupirocin resistance increased in Canadian hospitals, from 1.6% in 1995 to 1999 to 7% in 2000 to 2004 (19). In that study, all isolates with mupirocin MICs of >512 μg/ml were mupA positive. These data suggest that the emergence of mupB may have been recent. Recent publications have suggested the presence of non-mupA-mediated high-level mupirocin resistance (17, 22). The identification of mupB would explain those observations. Further studies need to be performed to determine the prevalence of mupB and to understand risk factors and outcomes associated with this novel resistance gene.
ACKNOWLEDGMENTS
We are indebted to A. McGeer for supplying the clinical strains included in this study. We are grateful to N. Tijet, J. Ma, and K. Warren (Public Health Ontario Laboratories, Toronto, Canada) for their technical support. We acknowledge the Network on Antimicrobial Resistance in S. aureus (NARSA) for supplying strains NRS1, NRS100, and NRS144.
Footnotes
Published ahead of print 17 January 2012
REFERENCES
- 1. Antonio M, Mcferran N, Pallen MJ. 2002. Mutations affecting the Rossman fold of isoleucyl-tRNA synthetase are correlated with low-level mupirocin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 46:438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Charpentier E, et al. 2004. Novel cassette-based shuttle vector system for Gram-positive bacteria. Appl. Environ. Microbiol. 70:6076–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Christianson S, Golding GR, Campbell J, Mulvey MR. 2007. Comparative genomics of Canadian epidemic lineages of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 45:1904–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing. 21st informational supplement. M100-S21. CLSI, Wayne, PA [Google Scholar]
- 5. Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss, and rearrangement. PLoS One 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deshpande LM, Fix AM, Pfaller MA, Jones RN, Antimicrobial Surveillance Program Participants Group SENTRY 2002. Emerging elevated mupirocin resistance rates among staphylococcal isolates in the SENTRY Antimicrobial Surveillance Program (2000): correlations of results from disk diffusion, Etest and reference dilution methods. Diagn. Microbiol. Infect. Dis. 42:283–290 [DOI] [PubMed] [Google Scholar]
- 7. Driscoll DG, Young CL, Ochsner U. 2007. Transient loss of high-level mupirocin resistance in Staphylococcus aureus due to MupA polymorphism. Antimicrob. Agents Chemother. 51:2247–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eltringham I. 1997. Mupirocin resistance and methicillin-resistant Staphylococcus aureus (MRSA). J. Hosp. Infect. 35:1–8 [DOI] [PubMed] [Google Scholar]
- 9. Fujimura S, Tokue Y, Watanabe A. 2003. Isoleucyl-tRNA synthetase mutations in Staphylococcus aureus clinical isolates and in vitro selection of low-level mupirocin-resistant strains. Antimicrob. Agents Chemother. 47:3373–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gurney R, Thomas CM. 2011. Mupirocin: biosynthesis, special features and applications of an antibiotic from a Gram-negative bacterium. Appl. Microbiol. Biotechnol. 90:11–21 [DOI] [PubMed] [Google Scholar]
- 11. Hodgson JE, et al. 1994. Molecular characterization of the gene encoding high-level mupirocin resistance in Staphylococcus aureus J2870. Antimicrob. Agents Chemother. 38:1205–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kraemer GR, Iandolo JJ. 1990. High-frequency transformation of Staphylococcus aureus by electroporation. Curr. Microbiol. 21:373–376 [Google Scholar]
- 13. Kreiswirth BN, et al. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712 [DOI] [PubMed] [Google Scholar]
- 14. Liu Q-Z, et al. 2010. Prevalence of clinical meticillin-resistant Staphylococcus aureus (MRSA) with high-level mupirocin resistance in Shanghai and Wenzhou, China. Int. J. Antimicrob. Agents 35:114–118 [DOI] [PubMed] [Google Scholar]
- 15. Margulies M, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:326–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mulvey MR, et al. 2001. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:3481–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patel JB, Gorwitz RJ, Jernigan JA. 2009. Mupirocin resistance. Clin. Infect. Dis. 49:935–941 [DOI] [PubMed] [Google Scholar]
- 18. Ramsey MA, Bradley SF, Kauffman CA, Morton TM. 1996. Identification of chromosomal location of mupA gene, encoding low-level mupirocin resistance in staphylococcal isolates. Antimicrob. Agents Chemother. 40:2820–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simor AE, et al. 2007. Mupirocin-resistant, methicillin-resistant Staphylococcus aureus strains in Canadian hospitals. Antimicrob. Agents Chemother. 51:3880–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simor AE, et al. 2010. Methicillin-resistant Staphylococcus aureus colonization or infection in Canada: national surveillance and changing epidemiology, 1995–2007. Infect. Control Hosp. Epidemiol. 31:348–356 [DOI] [PubMed] [Google Scholar]
- 21. Sutherland R, et al. 1985. Antibacterial activity of mupirocin (pseudomonic acid), a new antibiotic for topical use. Antimicrob. Agents Chemother. 27:495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Swenson JM, et al. 2010. Multicenter study to determine disk diffusion and broth microdilution criteria for prediction of high- and low-level mupirocin resistance in Staphylococcus aureus. J. Clin. Microbiol. 48:2469–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Udo EE, Jacob LE, Mathew B. 2001. Genetic analysis of methicillin-resistant Staphylococcus aureus expressing high- and low-level mupirocin resistance. J. Med. Microbiol. 50:909–915 [DOI] [PubMed] [Google Scholar]
- 24. Voskuil MI, Chambliss GH. 1998. The −16 region of Bacillus subtilis and other gram-positive bacterial promoters. Nucleic Acids Res. 26:3584–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang JA, et al. 2006. Molecular analysis of isoleucyl-tRNA synthetase mutations in clinical isolates of methicillin-resistant Staphylococcus aureus with low-level mupirocin resistance. J. Korean Med. Sci. 21:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]