Abstract
In coordination with the ppGpp alarmone, the RNA polymerase regulatory protein DksA controls the stringent response of eubacteria, negatively regulating transcription of translational machinery and directly activating amino acid promoters and de novo amino acid biosynthesis. Given the effects of nitric oxide (NO) on amino acid biosynthetic pathways and the intimate relationship of DksA with amino acid synthesis and transport, we tested whether DksA contributes to the resistance of Salmonella to reactive nitrogen species (RNS). Our studies show that the zinc finger predicted to position DksA in the secondary channel of the RNA polymerase is essential for the resistance of Salmonella enterica serovar Typhimurium to RNS in a murine model of systemic salmonellosis. Despite exhibiting auxotrophies for various amino acids, ΔdksA mutant Salmonella strains regain virulence in mice lacking inducible NO synthase (iNOS). DksA is also important for growth of this intracellular pathogen in the presence of NO congeners generated by iNOS during the innate response of murine macrophages. Accordingly, dksA mutant Salmonella strains are hypersusceptible to chemically generated NO, a phenotype that can be prevented by adding amino acids. The DksA-dependent antinitrosative defenses do not rely on the Hmp flavohemoprotein that detoxifies NO to NO3− and appear to operate independently of the ppGpp alarmone. Our investigations are consistent with a model by which NO produced in the innate response to Salmonella exerts considerable pressure on amino acid biosynthesis. The cytotoxicity of NO against Salmonella amino acid biosynthetic pathways is antagonized in great part by the DksA-dependent regulation of amino acid biosynthesis and transport.
INTRODUCTION
Intracellular pathogens are exposed to a plethora of defense mechanisms within host cells. Salmonella enterica cells residing in macrophages encounter a burst of reactive oxygen and nitrogen species generated through the enzymatic activities of NADPH phagocyte oxidase and inducible nitric oxide (NO) synthase (iNOS) hemoproteins. Macrophages express iNOS in response to lipid A, fimbriae, and porins decorating the Salmonella cell envelope (6, 42, 43). The proinflammatory cytokine gamma interferon (IFN-γ) further enhances the nitrosative potential of Salmonella-infected mononuclear phagocytic cells (28, 41). NO or reactive nitrogen species (RNS) engendered through the reaction of this diatomic radical with molecular oxygen (O2), low-molecular-weight thiols, iron, or superoxide (O2−) modify several bacterial biomolecules, including terminal cytochromes of the electron transport chain, DNA, and DNA binding metalloproteins, as well as [Fe-S] cluster- and thiol-containing enzymes of intermediary metabolism (20, 35). The RNS-mediated inhibition of respiration, central metabolism, and DNA replication is a likely cause of the growth arrest experienced by Salmonella exposed to nitrogen oxides (14). The enzymatic activity of iNOS plays an active role in the anti-Salmonella arsenal of rodent and human mononuclear phagocytes (38, 41) and in host defense against systemic and oral salmonellosis (1, 17, 26).
Salmonella expresses multiple defenses against RNS encountered in the host. The Salmonella pathogenicity island 2 type III secretion-dependent avoidance of iNOS-containing vesicles and DNA damage repair and scavenger systems all increase the antinitrosative defenses of Salmonella (2, 7, 36). In addition, the flavohemoprotein Hmp, which dinitrosylates NO to nitrate, serves as a primary defense against nitrosative stress generated by iNOS (2). Hmp protects the respiratory activity of terminal cytochromes of the electron transport chain (37), increases the resistance of Salmonella against NO congeners produced by human and murine macrophages, and reduces S-nitrosoglutathione formation (9, 38), thereby fostering Salmonella virulence (2). Due to the selective pressure imposed by products of iNOS against Salmonella, this intracellular pathogen must count on a rich regulatory network to fine-tune the expression of antinitrosative defenses. Multiple Salmonella regulators respond to RNS. For example, NsrR and NorR are dedicated sensors of NO, and the transcriptional regulators OxyR, SoxR, fumarate-nitrate reduction (FNR), and Fur, which primarily function as sensors of O2, reactive oxygen species (ROS), and iron, also respond to RNS (10, 12, 15, 18). To sense nitrogen oxides, these regulatory proteins use cysteine residues, non-heme iron centers, and [Fe-S] clusters (10, 12, 15, 18). Nitrosylation of cysteines or iron cofactors in OxyR, SoxR, and NorR activates transcription, whereas the iron-nitrosyl complexes in NsrR and Fur derepress gene expression (12, 13, 15, 18, 39).
Recent evidence indicates that the addition of amino acids to growth media prevents the bacteriostatic actions of NO against Salmonella, suggesting that inhibition of amino acid biosynthesis may be a key target of the anti-Salmonella activity of RNS (35). Work from our laboratory indicates that Salmonella cells undergoing nitrosative stress repress transcription of rRNA, tRNA, and other components of translational machinery, all of which are hallmarks of the stringent response often associated with amino acid deprivation (3). The [Fe-S] cluster in the dihydroxy acid dehydratase of the branch chain amino acid biosynthetic pathway is a prime example of a molecular target of RNS (21), as is the lipoamide-dependent lipoamide dehydrogenase (35). A sudden decrease in the availability of amino acids likely triggers ppGpp synthesis and a stringent response. In addition, RNS could elicit a stringent response by targeting the DksA protein that activates amino acid biosynthetic gene transcription (31). In support of this hypothesis, exposure of Salmonella to acidified nitrite derepresses DksA-inhibited flagellar gene transcription (3, 22). The investigations presented herein tested whether the RNA polymerase binding protein DksA contributes to the antinitrosative defenses of the enteric pathogen S. enterica serovar Typhimurium.
MATERIALS AND METHODS
Bacterial strains.
S. enterica serovar Typhimurium strain ATCC 14028s was used as the wild type and as a background in the construction of mutants according to the method previously described by Datsenko and Wanner (11) (Table 1). PCR amplicons containing kanamycin or chloramphenicol resistance cassettes flanked by the Flp recognition target (FRT) were generated from a pKD13 or pKD3 template, respectively, using AccuPrime Taq high-fidelity polymerase (Invitrogen, Carlsbad, CA) and 60-base-long primers with homology to target genes (Table 2). PCR products were purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA) and were electroporated into S. Typhimurium strain TT22236 harboring the plasmid pTP223, which expresses an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible λ Red recombinase system (34). Mutations were moved into S. Typhimurium strain 14028s via P22-mediated transduction, and pseudolysogens were eliminated by streaking on Evans blue uranine agar. The antibiotic cassette was removed by recombining the flanking FRT sites with the Flp recombinase expressed from the temperature-sensitive pCP20 plasmid (8). In-frame deletions were verified by PCR analysis. To create complementing strains carrying a wild-type or mutated dksA allele, we constructed a template plasmid by cloning an FRT-flanked chloramphenicol cassette from pKD3 into the BamHI and SacI restriction sites of pBluescript SK(+) to generate pSK::cm. A DNA region containing the native promoter and the dksA open reading frame was cloned between EcoRI and BamHI restriction sites of pSK::cm to generate pSK::dksA::cm. dksA variants with a serine substitution at position 114, 117, 135, or 138 were generated by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and were confirmed by DNA sequencing. The ΔdksA mutant strain AV09294 was complemented ectopically by inserting a PdksA-driven, wild-type dksA gene or dksA C114S, C117S, C135S, or C138S variant into the intergenic site downstream of the chromosomal putP locus using the λ Red recombinase system and the pSK::dksA::cm plasmid templates described above (19). The hmp::lacZY transcriptional fusion was transduced from strain AV03005 (27) into the ΔdksA mutant, generating strain AV08174.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Strains | ||
| S. Typhimurium ATCC 14028s | Wild type | ATCC |
| AV03005 | Δhmp::lacZY | 27 |
| AV04068 | Δhmp::FRT | 27 |
| AV08140 | ΔrelA::FRT ΔspoT::FRT | 19 |
| AV08174 | ΔdksA::FRT Δhmp::lacZY | This study |
| AV08261 | ΔdksA::cm ΔrelA::FRT ΔspoT::FRT | This study |
| AV09091 | ΔdksA::cm Δhmp::FRT | This study |
| AV09294 | ΔdksA::FRT | 19 |
| AV10359 | put::dksA::cm ΔdksA::FRT | This study |
| AV10360 | put::dksAC114S::cm ΔdksA::FRT | This study |
| AV10361 | put::dksAC117S::cm ΔdksA::FRT | This study |
| AV10362 | put::dksAC135S::cm ΔdksA::FRT | This study |
| AV10363 | put::dksAC138S::cm ΔdksA::FRT | This study |
| Plasmids | ||
| pKD3 | bla FRT cat FRT oriR6K | 11 |
| pKD13 | bla FRT ahp FRT oriR6K | 11 |
| pCP20 | bla cat cI857 flp pSC101 | 8 |
| pSK | bla pUC ori f1 lacZα | Stratagene |
| pSK::cm | bla cat pUC ori f1 lacZα | This study |
| pSK::dksA::cm | bla cat pUC ori f1 lacZα dksA | This study |
| pSK::dksAC114S::cm | bla cat pUC ori f1 lacZα dksAC114S | This study |
| pSK::dksAC117S::cm | bla cat pUC ori f1 lacZα dksAC117S | This study |
| pSK::dksAC135S::cm | bla cat pUC ori f1 lacZα dksAC135S | This study |
| pSK::dksAC138S::cm | bla cat pUC ori f1 lacZα dksAC138S | This study |
| pCE36 | ahp FRT lacZY+ thisoriR6K | 16 |
Table 2.
Primers used for the construction of strains used in this study
| Mutation | Primer sequencea (5′–3′) |
|---|---|
| dksAC114S | F: TGGAAGATGAAGACTTCGGTTATAGCGAGTCCTGCG |
| R: CGCAGGACTCGCTATAACCGAAGTCTTCATCTTCCA | |
| dksAC117S | F: TATTGCGAGTCCAGCGGGGTGGAGATT |
| R: AATCTCCACCCCGCTGGACTCGCAATA | |
| dksAC135S | F: ACAGCCGATCTGAGCATCGACTGCAAAACGCTGGCT |
| R: AGCCAGCGTTTTGCAGTCGATGCTCAGATCGGCTGT | |
| dksAC138S | F: ACAGCCGATCTGTGCATCGACAGCAAAACGCTGGCT |
| R: AGCCAGCGTTTTGCTGTCGATGCACAGATCGGCTGT | |
| pSK::cm | F: ACGCGGATCCATGGGAATTAGCCATGGTCC |
| R: CTGCAGGAGCTCGTGTAGGCTGGAGCTGCTTC | |
| pSK::dksA::cm | F: ATCGTAGAATTCCGTTGTAGTGGAATAACAGC |
| R: CCGCGGATCCTTAACCCGCCATCTGTTTTT | |
| put::dksA | F: TAGCGATGGGAGAGAGGACACGTTAATTATTCCATTTTAA |
| TAGTGGAATAACAGCCTGATTATTA | |
| R: TACTGCGGGTATTAATGCTGAAAACATCCATAACCCATTGGTGT | |
| AGGCTGGAGCTGCTTC |
Underlined sequences denote SacI, BamHI, or EcoRI restriction sites.
Quantification of NO by polarography.
Wild-type Salmonella cells grown overnight in LB broth were diluted 1:250 in LB broth and treated with 5 mM the NO donor diethylenetriamine (DETA) NONOate, an NO donor with an estimated half-life of 20 h at 37°C, pH 7.4 (Cayman Chemical, Ann Arbor, MI). The amount of NO released into the culture was measured polarographically using an NO-specific probe (World Precision Instruments, Sarasota, FL). The NO concentration was calculated by regression analysis using known standards generated with the NO donor Proli NONOate, which has a half-life of 1.8 s at 37°C and pH 7.4 (Cayman Chemical). Data are represented as μM NO over time.
Susceptibility of Salmonella to NO.
The cytotoxicity of the NO donor DETA NONOate or the polyamine DETA against wild-type and dksA-deficient Salmonella strains was monitored spectrometrically by monitoring bacterial growth on a Bioscreen C microbiology microtiter plate (Growth Curves USA, Piscataway, NJ). Salmonella cultures grown overnight in LB broth were diluted 1:500 in LB broth and treated with 5 mM the NO donor DETA NONOate or the polyamine DETA control. Some cultures were treated with 1 mM DETA NONOate (see Fig. 4). The half-life of DETA NONOate at neutral pH is about 20 h. Bacterial growth was recorded at an optical density at 600 nm (OD600) every 15 min while cultures were shaken at 37°C.
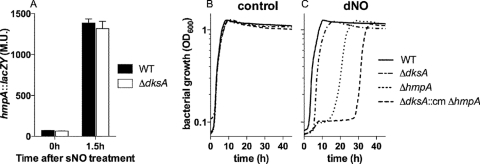
Fig 4.
The antinitrosative role of DksA is independent of Hmp. (A) Transcription of the hmp::lacZY transcriptional fusion 1.5 h after wild-type (WT) and isogenic ΔdksA mutant Salmonella strains were treated with 250 μM spermine NONOate (sNO). The data are presented as Miller units (M.U.) ± standard errors of the means (SEM) of four observations collected in two independent experiments. The addition of 5 mM DETA NONOate (dNO) completely inhibited the growth of hmp-deficient Salmonella for 48 h. Consequently, 1 mM DETA (control) (B) or dNO (C) was instead tested in these experiments. At this concentration of dNO, ΔdksA mutant Salmonella strains were slightly more susceptible than the WT controls. Data represent the means of six independent observations obtained from two separate experiments. The P value for the difference between the ΔhmpA and ΔdksA::cm ΔhmpA mutant strains was <0.0001 as revealed by one-way ANOVA of all data points in the curve followed by Bonferroni's posttest.
β-Galactosidase assay.
LacZ transcriptional activity was quantified spectrometrically from bacterial cultures treated with 250 μM the NO donor spermine NONOate as β-galactosidase activity by monitoring the conversion of o-nitrophenyl-β-d-galactopyranoside to ortho-nitrophenyl compared to that of untreated controls. β-Galactosidase activity is expressed as Miller units calculated according to the equation 1,000 × {[OD420 − (1.75 × OD550)]/[(T min) × (V ml) × OD600]}. Under the experimental conditions used in our assays, the background β-galactosidase activity is ∼1 Miller unit.
Macrophage-killing assays.
The anti-Salmonella activity of macrophages was evaluated as previously described (27). Briefly, peritoneal exudate cells were harvested from C57BL/6 or iNOS-deficient (24) mice 4 days after intraperitoneal (i.p.) inoculation of 1 mg/ml of sodium periodate. The cells were cultured for 36 to 48 h in RPMI 1640 medium (Cellgro, Herndon, VA) supplemented with 10% heat-inactivated fetal bovine serum (BioWhittaker, Walkersville, MD), 15 mM HEPES, 2 mM l-glutamine, 1 mM sodium pyruvate (Sigma-Aldrich, St. Louis, MO), 100 units/ml of penicillin, and 100 mg/ml of streptomycin (Cellgro) (RPMI+). Macrophages washed with media lacking antibiotics were challenged at a multiplicity of infection of 2 with Salmonella that had been opsonized with 10% normal mouse serum for 20 min. After 25 min of infection, the medium was exchanged with prewarmed RPMI+ medium containing 25 μg/ml of gentamicin. Macrophages were lysed with 0.25% deoxycholic acid at the indicated time points after infection, and the surviving intracellular bacteria were enumerated on LB agar plates. Killing is expressed as the fraction of bacteria recovered at the indicated time relative to the bacterial burden isolated after 25 min of internalization.
Mouse infections.
Six- to 8-week-old C57BL/6 or congenic iNOS-deficient (24) mice bred in our animal facility according to Institutional Animal Care and Use Committee guidelines were used to assess the role of DksA in Salmonella virulence. Briefly, individual animals were inoculated intraperitoneally with ∼400 CFU of Salmonella grown overnight to stationary phase in LB broth. Mouse survival was monitored over time. Mice manifesting signs of distress (i.e., low spontaneous activity and ruffled coat) were humanely euthanized by CO2 inhalation followed by cervical dislocation.
Statistical analysis.
Determination of the statistical significances of multiple comparisons was achieved using one-way analysis of variance (ANOVA) followed by Bonferroni's posttest. Data were considered statistically significant when P was <0.05.
RESULTS
DksA mutant Salmonella strains are hypersusceptible to the bacteriostatic effects of NO.
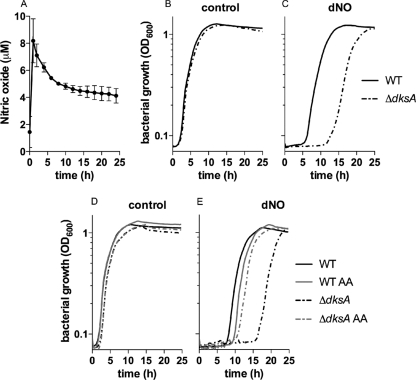
The expression of DksA enhances the antioxidant defenses of Salmonella (19). Due to overlaps in cellular toxicity between ROS and RNS, many defense mechanisms serve a dual role in defending against oxidative and nitrosative stresses (5, 29). Therefore, we evaluated whether DksA contributes to the antinitrosative defenses of Salmonella. The sensitivity of dksA-deficient Salmonella to RNS was tested by assessing bacterial growth in the presence of the NO donor DETA NONOate (Fig. 1). Defects in dksA result in auxotrophies for isoleucine, leucine, valine, glycine, phenylalanine, and threonine, and therefore ΔdksA strains do not grow in minimal media (4). Consequently, the effect of the slow NO releaser DETA NONOate on the growth of dksA mutant Salmonella strains was evaluated in bacteria cultured in LB broth. DETA NONOate has a half-life of 20 h at pH 7. Polarographic determination of the NO generated with 5 mM DETA NONOate under the culture conditions used in our assays indicated an 8 μM NO peak shortly after exposure, followed by a steady 5 μM NO for the duration of the experiment (Fig. 1A). Wild-type and isogenic ΔdksA Salmonella strains exhibited similar growth in LB broth supplemented with 5 mM DETA (Fig. 1B). The ΔdksA mutant Salmonella strain AV09294 was hypersusceptible to the bacterostatic effects of 5 mM DETA NONOate, as shown by its 12-h lag compared to 5 h for the wild-type control (Fig. 1C). Given the inhibitory effects of NO on amino acid biosynthetic pathways and the DksA-dependent regulation of the transcription of amino acid synthesis and transport, we tested whether the addition of amino acids had any effect on the marked susceptibility of ΔdksA mutant Salmonella strains to chemically generated NO. The addition of 0.1% Casamino Acids did not seem to affect the growth of wild-type or ΔdksA mutant bacteria in LB broth (Fig. 1D). However, supplementation of LB broth with 0.1% Casamino Acids abrogated most of the hypersusceptibility of dksA-deficient Salmonella strains to NO (Fig. 1E). These data suggest that DksA contributes to antinitrosative defenses by Salmonella, possibly by fine-tuning the expression of amino acid biosynthetic pathways and transport.
Fig 1.
dksA mutant Salmonella strains are hypersusceptible to the bacteriostatic activity of NO. (A) The concentrations of NO released by 5 mM DETA NONOate (dNO) in LB broth incubated with 2 × 107 CFU/ml of Salmonella Typhimurium strain 14028s were measured polarographically over time at 37°C in a shaker incubator. Data from two independent observations are expressed as means ± standard deviations (SD). (B and C) Wild-type (WT) and the isogenic ΔdksA Salmonella strain AV09294 grown overnight in LB broth were diluted to 2 × 107 CFU/ml in fresh LB broth and treated with 5 mM polyamine DETA (control) (B) or dNO (C). (D and E) Selected control (D) and dNO-treated (E) cultures were supplemented with 0.1% Casamino Acids (AA). Bacterial growth is expressed as the OD600 over time, and the data are represented as the means of 10 independent observations from 2 separate experiments. The P value for the difference between the ΔdksA and wild-type strains was <0.0001 as determined by one-way ANOVA of all data points in the curve followed by Bonferroni's posttest.
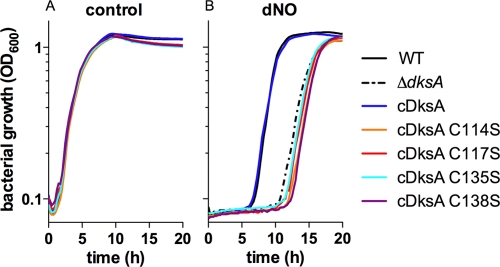
The cysteine residues in the DksA zinc finger are crucial for antinitrosative function.
The DksA protein has a zinc finger in its C terminus coordinated by four cysteine residues (32). Because no function has been ascribed to the zinc finger of DksA and because zinc fingers are notorious for their reactivity with reactive oxygen and nitrogen species, we tested whether the zinc finger is necessary for the antinitrosative defenses associated with DksA. Salmonella strains expressing DksA variants with a single serine substitution at position 114, 117, 135, or 138 were evaluated for susceptibility to NO (Fig. 2B). All four coordinating cysteine residues were found to be critical for DksA-mediated defense against the bacteriostatic effects of NO. All strains showed similar growth in LB broth (Fig. 2A). These data suggest that the integrity of the zinc finger motif is needed for the contribution of DksA to the antinitrosative defenses of Salmonella.
Fig 2.
Cysteine residues in the zinc finger motif are critical for DksA-dependent antinitrosative defense. Wild-type (WT), ΔdksA, or isogenic ΔdksA Salmonella strains complemented with wild-type DksA (cDksA) or a DksA C114S, C117S, C135S, or C138S variant were treated with 5 mM DETA (control) (A) or DETA NONOate (dNO) (B) as described in the legend for Fig. 1. Bacterial growth is expressed as the OD600 over time, and the data are represented as the means of 10 independent observations from 2 separate experiments. The P value for the difference between the ΔdksA and isogenic ΔdksA Salmonella strains complemented with DksA C114S, C117S, C135S, or C138S and the wild-type strain was <0.0001 as determined by one-way ANOVA of all data in the curve followed by Bonferroni's posttest.
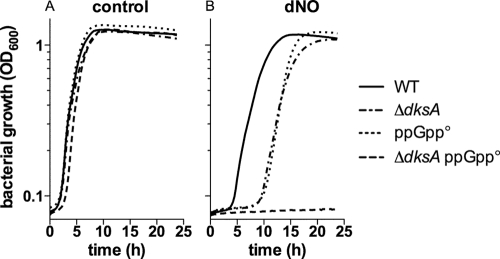
DksA and ppGpp independently increase Salmonella resistance to nitrosative stress.
DksA is thought to regulate RNA polymerase function in conjunction with the alarmone ppGpp (30, 32). The ppGpp synthases RelA and SpoT produce ppGpp in response to nutrient deprivation (33). We examined the effect that NO has on the growth of Salmonella strains lacking both RelA and SpoT ppGpp synthases (ppGpp°) and/or dksA. All strains grew with apparently similar kinetics in LB broth (Fig. 3A). A ppGpp° Salmonella strain and a dksA-deficient Salmonella strain displayed the same increased lag phase after treatment with NO as the wild-type isogenic controls. However, the triple dksA relA spoT mutant did not resume growth even after 24 h of NO treatment (Fig. 3B). Strain AV08261 lacking relA, spoT, and dksA did not lose viability under these experimental conditions (data not shown), indicating that the hypersusceptibility of this triple mutant to NO cannot be rationalized by direct killing by RNS. The marked susceptibility of strain AV08261 to NO could be interpreted as a sign that DksA and ppGpp contribute independently to the antinitrosative defenses of Salmonella.
Fig 3.
DksA and ppGpp independently contribute to the antinitrosative defense of Salmonella. The wild type (WT), the ΔdksA strain, and the ΔrelA ΔspoT-deficient Salmonella strain lacking the ppGpp synthases (ppGpp°) and a triple ΔdksA ppGpp° strain were diluted to 2 × 107 CFU/ml in fresh LB broth from overnight cultures. The cells were treated with 5 mM DETA (control) (A) or DETA NONOate (dNO) (B). Bacterial growth is expressed as the OD600 over time, and the data are represented as the means of 10 independent observations from 2 separate experiments. The P value for the differences between ΔdksA and ΔdksA ppGpp° Salmonella strains was <0.0001 as determined by one-way ANOVA of all data points in the curve followed by Bonferroni's posttest.
Independent contributions of DksA and Hmp to antinitrosative defenses of Salmonella.
ΔdksA mutant bacteria grown 24 h in the presence of 5 mM DETA NONOate became resistant to a subsequent challenge with NO (data not shown). The increased resistance of a ΔdksA mutant Salmonella strain preexposed to NO likely reflects an adaptive response rather than the selection of suppressor mutants, as indicated by the failure of these bacteria to grow in E salts medium (1.66 mM MgSO4, 9.5 mM citric acid monohydrate, 57 mM K2HPO4, 16.7 mM NaHH3PO4) supplemented with 0.4% glucose. Moreover, NO-treated ΔdksA mutant Salmonella strains became hypersusceptible to 5 mM DETA NONOate after overnight growth to stationary phase in fresh LB broth. By detoxifying NO to NO3−, the Hmp flavohemoprotein serves a critical role in the antinitrosative defense of Salmonella (2). Because DksA indirectly regulates gene transcription by interacting with the RNA polymerase, we tested whether the Hmp flavohemoprotein may have played a role in the adaptive response of ΔdksA mutant Salmonella strains pretreated with NO. Toward this end, an hmp::lacZY chromosomal fusion was transduced into the ΔdksA mutant Salmonella strain AV09294. Transcription of the hmp gene was induced ∼20-fold in both wild-type and dksA-deficient Salmonella strains 1.5 h after treatment with 250 μM the NO-donor spermine NONOate (Fig. 4A), suggesting that DksA is not involved in the regulation of hmp transcription. The wild-type, ΔdksA, Δhmp, and ΔdksA::cm Δhmp strains showed similar growth in LB broth (Fig. 4B). Treatment of hmp-deficient Salmonella with 5 mM DETA NONOate resulted in a lag phase of about 48 h (data not shown), making this concentration of DETA NONOate impractical to compare the relative contributions of Hmp and DksA to the resistance of Salmonella to nitrosative stress. Consequently, the concentration of NO was decreased to levels that make the differences between wild-type and ΔdksA mutant Salmonella practically unnoticeable but could still elucidate DksA and Hmp synergism. Salmonella strain AV04064 lacking the flavohemoprotein-encoding hmp gene remained hypersusceptible to 1 mM DETA NONOate, whereas the dksA-deficient strain was slightly more susceptible than its parent, the wild-type control (Fig. 4C). Interestingly, the double ΔdksA::cm Δhmp mutant strain AV09091 showed increased susceptibility to NO compared to single isogenic mutant controls, further supporting the idea that Hmp and DksA independently contribute to the antinitrosative defenses of Salmonella.
DksA fosters intracellular survival of Salmonella within NO-producing macrophages.
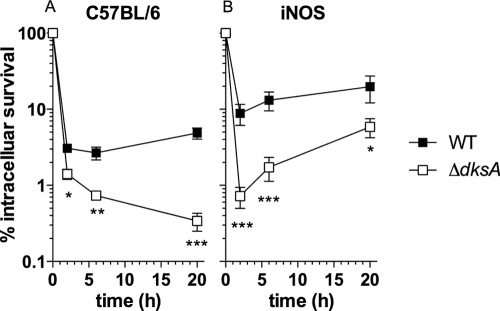
RNS generated by the iNOS hemoprotein plays a role in the innate response of macrophages against Salmonella (41). Therefore, the contribution of DksA to the antinitrosative defenses of Salmonella was evaluated further in primary macrophages expressing a functional iNOS. Compared to the wild-type parent strain, the dksA-deficient Salmonella strain was hypersusceptible to the early phases of the anti-Salmonella activity of macrophages from C57BL/6 mice (Fig. 5A). These findings are consistent with the known hypersusceptibility of dksA mutant Salmonella strains to oxidative products generated by the NADPH phagocyte oxidase-dependent respiratory burst of macrophages (19). Interestingly, later during the course of infection, the viability of the ΔdksA mutant Salmonella strain continued to decline, while the wild-type strain increased in number (Fig. 5A). Together, these findings suggest that in addition to being susceptible to ROS generated by the NADPH phagocyte oxidase, dksA mutant Salmonella strains are hypersusceptible to nitrogen oxides generated by iNOS expressed at later times in infection. To test whether DksA-dependent gene expression antagonizes the anti-Salmonella activity of iNOS-expressing macrophages, we studied the intracellular survival of wild-type and dksA-deficient Salmonella strains in macrophages lacking iNOS (Fig. 5B). iNOS-deficient macrophages showed an even greater difference in the numbers of ΔdksA and wild-type Salmonella cells recovered at early times of infection (Fig. 5B), likely reflecting the higher concentrations of ROS produced by macrophages lacking the iNOS hemoprotein (41). Later during the course of infection, the ΔdksA mutant strain displayed signs of growth in iNOS-deficient macrophages, suggesting that in the absence of NO, dksA-deficient Salmonella can grow intracellularly (Fig. 5B). Collectively, these findings underscore the importance of DksA-mediated antioxidant and antinitrosative defenses for the intracellular fitness of Salmonella.
Fig 5.
Role of DksA in the intracellular survival of Salmonella in iNOS-expressing macrophages. The antimicrobial activities of periodate-elicited peritoneal macrophages isolated from C57BL/6 mice (A) or congenic iNOS-deficient controls (B) were evaluated over time after mice were challenged with the wild-type (WT) or the ΔdksA mutant Salmonella strain. The number of surviving intracellular bacteria was estimated after culture on LB agar plates as described in Materials and Methods. The data represent the mean percent survival ± the SEM of four to eight independent observations from at least two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared to results for WT controls.
DksA antagonizes the cytotoxicity of iNOS in mice.
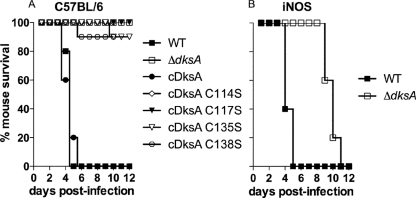
Several groups of investigators have shown that DksA is crucial for Salmonella virulence in mice (19, 40, 44). In agreement with these observations, a ΔdksA mutant Salmonella strain was attenuated in a C57BL/6 murine model of acute systemic infection (Fig. 6A). The ΔdksA mutation could be complemented with a PdksA-dksA wild-type allele expressed ectopically downstream of the putP chromosomal locus but not by PdksA-dksA cysteine variants, suggesting that the integrity of the zinc finger in DksA is critical for Salmonella pathogenesis. Given the hypersusceptibility of the ΔdksA mutant Salmonella strain to the cytotoxic effects of NO and its attenuation in iNOS-sufficient macrophages, we tested whether antinitrosative defenses associated with the expression of a functional DksA promote Salmonella virulence in an acute model of infection. To test this hypothesis, iNOS-deficient mice were challenged intraperitoneally with ∼400 CFU of the wild-type or isogenic ΔdksA mutant Salmonella strain. The dksA-deficient strain regained virulence in iNOS-deficient mice; strain AV09294 killed iNOS-immunodeficient mice 6 days later than the isogenic wild-type controls (Fig. 6B). Since NADPH oxidase contributes to early anti-Salmonella activity, it is possible that the marked susceptibility of ΔdksA mutant bacteria to oxyradicals produced by NADPH phagocyte oxidase may have contributed to the delayed virulence of this strain in iNOS-deficient mice. All Salmonella cells recovered from the spleens of moribund iNOS-deficient mice that had been inoculated i.p. 8 days earlier with ΔdksA mutant bacteria remained auxotrophic for amino acids, as determined by their inability to grow in E salts minimum medium supplemented with 0.4% glucose. Moreover, the bacteria recovered from iNOS-deficient mice contained the ΔdksA mutant allele, as determined by PCR analysis. Together, these findings indicate that in the absence of the amino acid biosynthetic restrictions imposed by nitrosative stress, the phagosomal lumen harbors sufficient amino acids to satisfy the auxotrophies of the ΔdksA mutant Salmonella strain.
Fig 6.
DksA protects Salmonella from nitrogen oxides produced by a functional iNOS hemoprotein in a murine model of salmonellosis. (A) C57BL/6 mice were inoculated intraperitoneally with ∼200 to 400 CFU of either the wild-type (WT), ΔdksA, or ΔdksA mutant Salmonella strain complemented with a wild-type dksA allele (cDksA) or with a dksA C114S-, C117S-, C135S-, or C138S-expressing variant. (B) Congenic iNOS-deficient mice were inoculated intraperitoneally with ∼400 CFU of either the WT or ΔdksA mutant Salmonella strain. The fractions of mice surviving the experimental Salmonella infections were evaluated over time. Data represent 10 mice per group and were obtained from two separate experiments.
DISCUSSION
We previously demonstrated that both oxyradicals and nitrogen oxides produced in the innate response of macrophages activate a stringent DksA-dependent response (3, 19). Furthermore, the RNA polymerase regulatory protein DksA plays a crucial role in the expression of antioxidant defenses of Salmonella (19). Therefore, we evaluated the role of the DksA protein in the antinitrosative defenses of Salmonella. The investigations presented herein indicate that DksA is critical for the ability of Salmonella to resist NO-dependent antimicrobial defenses. DksA increases Salmonella resistance to the bacteriostatic effects of NO, and DksA-mediated antinitrosative defenses support intracellular growth of Salmonella strains within iNOS-sufficient macrophages while fostering virulence in an acute model of infection dependent on the presence of a functional iNOS hemoprotein. The phagosomal lumen is a nutrient-limiting environment. By inhibiting amino acid biosynthetic pathways, NO produced in the innate response of macrophages to Salmonella appears to further restrict the availability of nutrients. According to this model, the regulatory effects of DksA on amino acid biosynthetic synthesis and transport greatly reduce the NO-dependent antimicrobial activity seen in the innate immune response of professional phagocytes. However, in the absence of the iNOS hemoprotein and the consequent burst in nitrosative stress, amino acids must not be a limiting factor in the Salmonella phagosome, since auxotrophic ΔdksA mutant Salmonella strains do in fact replicate in iNOS-deficient macrophages and kill iNOS-deficient mice.
The DksA metalloprotein contains a C-terminal zinc finger coordinated by four cysteine residues (30, 44). Our studies indicate that the DksA zinc finger is critical for the antinitrosative function of the protein. According to a model of the RNA polymerase, ppGpp, and the DksA ternary complex, the globular domain containing the DksA zinc finger is needed for proper positioning of the protein within the secondary channel of the RNA polymerase (32). The zinc finger is likely necessary for the interaction of the acidic residues at the tip of the coiled-coil domain of DksA with the Mg2+ ion bound to the distal diphosphate of the ppGpp molecule inside RNA polymerase (30). Mutations in any of the DksA zinc finger cysteine residues abolish its ability to coordinate antinitrosative defenses, likely due to improper positioning of DksA within the RNA polymerase secondary channel. In accordance with this hypothesis, a mutation in a single cysteine within the Escherichia coli DksA zinc finger results in a relaxed phenotype similar to that of dksA-deficient bacteria (30).
ppGpp° and dksA mutant Salmonella strains exhibit similar susceptibility to NO, suggesting that both stringent-response mediators contribute to the antinitrosative defenses of Salmonella. However, the hypersusceptibility of the triple mutant indicate that ppGpp and DksA may have independent roles in NO defense. In fact, ppGpp and DksA regulate a variety of cellular processes, including motility, adhesion, autoaggregation, and filamentation, independently (22, 25). Moreover, DksA independently activates the PlivJ, Phis, and PthrABC promoters critical for amino acid synthesis and transport (25). The fact that DksA can independently increase amino acid biosynthesis genes suggests that this regulatory protein can modulate RNA polymerase activity without ppGpp. Our data could be alternatively interpreted in light of a model by which ppGpp and DksA may be overexpressed in the ΔdksA and ppGpp° strains, respectively. Overexpression of dksA in ppGpp° E. coli can overcome many of the amino acid auxotrophies associated with the ΔrelA ΔspoT mutant (25). The mechanism by which both ppGpp and DksA add to the antinitrosative defenses of Salmonella awaits further investigation.
The flavohemoprotein Hmp detoxifies NO and protects Salmonella from the iNOS-mediated antimicrobial activity of macrophages (2, 38). Our investigations indicate that DksA does not appear to contribute to hmp transcription, and dksA-deficient and hmp-deficient Salmonella strains display distinct hypersusceptibilities to NO. These findings indicate that DksA protects Salmonella from iNOS-mediated antimicrobial activity independently of the flavohemoprotein-mediated detoxification of NO. Given the NO-mediated targeting of amino acid biosynthetic pathways, it is likely that the DksA-mediated transcriptional regulation of amino acid biosynthesis is crucial for Salmonella to overcome the pressure exerted by RNS on intermediary metabolism.
The addition of amino acids partially abrogated the increased antimicrobial activity exhibited by NO against dksA-deficient Salmonella. DksA-dependent regulation of amino acid biosynthesis and transport may contribute to both the antioxidant and antinitrosative defenses of Salmonella. However, amino acids did not completely reverse the hypersusceptibility of the ΔdksA mutant Salmonella strain to NO. The positive effects of DksA on glutathione biosynthesis and redox homeostasis may also contribute to antioxidant and antinitrosative defenses. Salmonella lacking dksA is auxotrophic for several amino acids, including isoleucine, leucine, valine, threonine, phenylalanine, and glycine. Glycine serves as a substrate in the formation of the tripeptide glutathione, and consequently, dksA-deficient Salmonella contain reduced cytoplasmic glutathione concentrations (19). DksA also maintains proper levels of NAD(P)H-reducing equivalents, which are vital for defense against both oxidative and nitrosative stress (19, 23). The fact that glutathione and the NADPH-dependent glutathione reductase play a critical role in the defense against both ROS and RNS by repairing oxidized and/or nitrosylated thiols raises the interesting possibility that DksA-dependent regulation of low-molecular-weight thiol metabolism could contribute to the antioxidant and antinitrosative defenses of Salmonella.
The ability of Salmonella to survive inside professional phagocytes is a critical aspect in the pathogenesis of this enteric pathogen. Collectively, our investigations demonstrate that DksA helps Salmonella survive the antimicrobial activity of iNOS expressed in the innate immune response of macrophages.
ACKNOWLEDGMENTS
We thank Jessica Jones-Carson, Lin Liu, and James Laughlin for supplying the mice used in the course of these investigations.
This project was supported by National Institutes of Health project AI54959, the Burroughs Wellcome Fund, and Institutional Training Grant T32 AI052066.
Footnotes
Published ahead of print 6 February 2012
REFERENCES
- 1. Alam MS, et al. 2008. Nitric oxide produced in Peyer's patches exhibits antiapoptotic activity contributing to an antimicrobial effect in murine salmonellosis. Microbiol. Immunol. 52:197–208 [DOI] [PubMed] [Google Scholar]
- 2. Bang IS, et al. 2006. Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin hmp. J. Biol. Chem. 281:28039–28047 [DOI] [PubMed] [Google Scholar]
- 3. Bourret TJ, et al. 2008. Nitric oxide antagonizes the acid tolerance response that protects Salmonella against innate gastric defenses. PLoS One 3:e1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown L, Gentry D, Elliott T, Cashel M. 2002. DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol. 184:4455–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bryk R, Griffin P, Nathan C. 2000. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407:211–215 [DOI] [PubMed] [Google Scholar]
- 6. Buzzo CL, et al. 2010. A novel pathway for inducible nitric-oxide synthase activation through inflammasomes. J. Biol. Chem. 285:32087–32095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chakravortty D, Hansen-Wester I, Hensel M. 2002. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J. Exp. Med. 195:1155–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 9. Crawford MJ, Goldberg DE. 1998. Regulation of the Salmonella typhimurium flavohemoglobin gene. A new pathway for bacterial gene expression in response to nitric oxide. J. Biol. Chem. 273:34028–34032 [DOI] [PubMed] [Google Scholar]
- 10. Cruz-Ramos H, et al. 2002. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 21:3235–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. D'Autreaux B, Touati D, Bersch B, Latour JM, Michaud-Soret I. 2002. Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc. Natl. Acad. Sci. U. S. A. 99:16619–16624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. D'Autréaux B, Tucker NP, Dixon R, Spiro S. 2005. A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature 437:769–772 [DOI] [PubMed] [Google Scholar]
- 14. De Groote MA, et al. 1995. Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc. Natl. Acad. Sci. U. S. A. 92:6399–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ding H, Demple B. 2000. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc. Natl. Acad. Sci. U. S. A. 97:5146–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ellermeier CD, Janakiraman A, Slauch JM. 2002. Construction of targeted single copy lac fusions using λ Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153–161 [DOI] [PubMed] [Google Scholar]
- 17. Giacomodonato MN, et al. 2003. Involvement of intestinal inducible nitric oxide synthase (iNOS) in the early stages of murine salmonellosis. FEMS Microbiol. Lett. 223:231–238 [DOI] [PubMed] [Google Scholar]
- 18. Hausladen A, Privalle CT, Keng T, DeAngelo J, Stamler JS. 1996. Nitrosative stress: activation of the transcription factor OxyR. Cell 86:719–729 [DOI] [PubMed] [Google Scholar]
- 19. Henard CA, Bourret TJ, Song M, Vazquez-Torres A. 2010. Control of redox balance by the stringent response regulatory protein promotes antioxidant defenses of Salmonella. J. Biol. Chem. 285:36785–36793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Henard CA, Vazquez-Torres A. 2011. Nitric oxide and Salmonella pathogenesis. Front. Microbiol. 2:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hyduke DR, Jarboe LR, Tran LM, Chou KJ, Liao JC. 2007. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 104:8484–8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lemke JJ, Durfee T, Gourse RL. 2009. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol. Microbiol. 74:1368–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lundberg BE, Wolf RE, Jr, Dinauer MC, Xu Y, Fang FC. 1999. Glucose 6-phosphate dehydrogenase is required for Salmonella typhimurium virulence and resistance to reactive oxygen and nitrogen intermediates. Infect. Immun. 67:436–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. MacMicking JD, et al. 1995. Altered responses to bacterial infection and endotoxic shock in mice lacking inducible nitric oxide synthase. Cell 81:641–650 [DOI] [PubMed] [Google Scholar]
- 25. Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nystrom T. 2007. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J. Bacteriol. 189:5193–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mastroeni P, et al. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J. Exp. Med. 192:237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCollister BD, Bourret TJ, Gill R, Jones-Carson J, Vazquez-Torres A. 2005. Repression of SPI2 transcription by nitric oxide-producing, IFNγ-activated macrophages promotes maturation of Salmonella phagosomes. J. Exp. Med. 202:625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McCollister BD, et al. 2007. N2O3 enhances the nitrosative potential of IFNγ-primed macrophages in response to Salmonella. Immunobiology 212:759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McLean S, Bowman LA, Poole RK. 2010. Peroxynitrite stress is exacerbated by flavohaemoglobin-derived oxidative stress in Salmonella Typhimurium and relieved by nitric oxide. Microbiology 156:3556–3565 [DOI] [PubMed] [Google Scholar]
- 30. Paul BJ, et al. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118:311–322 [DOI] [PubMed] [Google Scholar]
- 31. Paul BJ, Berkmen MB, Gourse RL. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. U. S. A. 102:7823–7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perederina A, et al. 2004. Regulation through the secondary channel—structural framework for ppGpp-DksA synergism during transcription. Cell 118:297–309 [DOI] [PubMed] [Google Scholar]
- 33. Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62:35–51 [DOI] [PubMed] [Google Scholar]
- 34. Price-Carter M, Tingey J, Bobik TA, Roth JR. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183:2463–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Richardson AR, et al. 2011. Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar Typhimurium. Cell Host Microbe 10:33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Richardson AR, et al. 2009. The base excision repair system of Salmonella enterica serovar Typhimurium counteracts DNA damage by host nitric oxide. PLoS Pathog. 5:e1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stevanin TM, et al. 2000. Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo ' or bd, from nitric oxide. J. Biol. Chem. 275:35868–35875 [DOI] [PubMed] [Google Scholar]
- 38. Stevanin TM, Poole RK, Demoncheaux EA, Read RC. 2002. Flavohemoglobin Hmp protects Salmonella enterica serovar Typhimurium from nitric oxide-related killing by human macrophages. Infect. Immun. 70:4399–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tucker NP, et al. 2008. The transcriptional repressor protein NsrR senses nitric oxide directly via a [2Fe-2S] cluster. PLoS One 3:e3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Turner AK, Lovell MA, Hulme SD, Zhang-Barber L, Barrow PA. 1998. Identification of Salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect. Immun. 66:2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vazquez-Torres A, Jones-Carson J, Mastroeni P, Ischiropoulos H, Fang FC. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J. Exp. Med. 192:227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vazquez-Torres A, et al. 2004. Toll-like receptor 4 dependence of innate and adaptive immunity to Salmonella: importance of the Kupffer cell network. J. Immunol. 172:6202–6208 [DOI] [PubMed] [Google Scholar]
- 43. Vitiello M, et al. 2008. Role of mitogen-activated protein kinases in the iNOS production and cytokine secretion by Salmonella enterica serovar Typhimurium porins. Cytokine 41:279–285 [DOI] [PubMed] [Google Scholar]
- 44. Webb C, Moreno M, Wilmes-Riesenberg M, Curtiss R, III, Foster JW. 1999. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol. Microbiol. 34:112–123 [DOI] [PubMed] [Google Scholar]