Abstract
Mammalian macrophage migration inhibitory factor (MIF) is a multifaceted cytokine involved in both extracellular and intracellular functions. Malaria parasites express a MIF homologue that might modulate host immune responses against blood-stage parasites, but the potential importance of MIF against other life cycle stages remains unstudied. In this study, we characterized the MIF homologue of Plasmodium yoelii throughout the life cycle, with emphasis on preerythrocytic stages. P. yoelii MIF (Py-MIF) was expressed in blood-stage parasites and detected at low levels in mosquito salivary gland sporozoites. MIF expression was strong throughout liver-stage development and localized to the cytoplasm of the parasite, with no evidence of release into the host hepatocyte. To examine the importance of Py-MIF for liver-stage development, we generated a Py-mif knockout parasite (P. yoelii Δmif). P. yoelii Δmif parasites grew normally as asexual erythrocytic-stage parasites and showed normal infection of mosquitoes. In contrast, the P. yoelii Δmif strain was attenuated during the liver stage. Mice infected with P. yoelii Δmif sporozoites either did not develop blood-stage parasitemia or exhibited a delay in the onset of blood-stage patency. Furthermore, P. yoelii Δmif parasites exhibited growth retardation in vivo. Combined, the data indicate that Plasmodium MIF is important for liver-stage development of P. yoelii, during which it is likely to play an intrinsic role in parasite development rather than modulating host immune responses to infection.
INTRODUCTION
Macrophage migration inhibitory factor (MIF) is a pleiotrophic factor involved in both innate and adaptive immune responses (reviewed in references 10 and 34). MIF is conserved throughout higher eukaryotes, and homologues have been identified in several disease-causing parasites, including Leishmania (21), several pathogenic worms (47), and Plasmodium (2, 40). Plasmodium species are the etiologic agents of malaria and are responsible for the illness of 300 to 500 million people and the death of 800,000 people annually (49). These parasites are transmitted to a mammalian host during a bite from an Anopheles mosquito, which deposits Plasmodium sporozoites into the skin. The sporozoites migrate to the liver, where they infect hepatocytes. One replicating parasite within a hepatocyte can generate tens of thousands of erythrocyte-infectious forms to initiate blood-stage infection (reviewed in reference 31). Several Plasmodium species, including the human malaria-causing species Plasmodium falciparum and P. vivax and the rodent malarial species P. berghei and P. yoelii, contain MIF homologues (PlasmoDB entries PFL1420w, PVX_124095, PBANKA_144400, and PY05452).
MIF is most commonly thought of as a proinflammatory cytokine and chemokine (38) that can induce the secretion of tumor necrosis factor (TNF), interleukin 18 (IL-18), nitric oxide, IL-6, and other proinflammatory factors (9, 22, 28). It is expressed by multiple cell types, including lymphocytes (4), macrophages (9), and epithelial cells (18). MIF lacks a canonical secretion signal and is released from immune cells in an unknown manner in response to cytokines (e.g., TNF or gamma interferon) (9) and microbial products (5). Hence, it is commonly associated with inflammatory responses during infections such as sepsis (7) and malaria (3, 19, 29). It has been shown that MIF binds to a CD74 and CD44 receptor complex and induces sustained extracellular signal-regulated kinase 1/2 (ERK1/2) mitogen-activated protein kinase (MAPK) activation. This interaction eventually leads to NF-κB activation and the transcription of proinflammatory molecules (27, 37, 41). MIF can also bind to a heterodimer of CD74 and CXCR2/4 to act as a chemokine inducing recruitment of macrophages (6, 38). In addition to its cytokine functions, MIF has multiple intracellular roles. For example, MIF can inhibit the immune regulatory transcription factor AP-1 (23) and downregulates p53 and p53-associated signaling pathways (17, 37). Furthermore, MIF is able to regulate cell division and is necessary for normal progression of the cell cycle (33). MIF further distinguishes itself from other cytokines by concurrently possessing enzymatic capabilities. It contains both an oxidoreductase active site and a tautomerase active site (24, 36), but no substrates for either site have been identified in vivo (14).
Although the protein identity between mammalian and Plasmodium MIF proteins is low (29% for P. falciparum), the structure is well conserved, including the presence of tautomerase and oxidoreductase sites (2, 13). It has been shown that Plasmodium MIF is highly expressed in erythrocyte-stage trophozoites and schizonts, and it is thought that it is released into the host bloodstream during schizont rupture (2, 45). Accordingly, antibodies against parasite MIF have been isolated from sera of both previously and currently infected hosts (11, 45). P. falciparum MIF (Pf-MIF) has also been detected in the blood of infected patients (2, 16, 39). In addition, it was shown that Plasmodium MIF conserves the chemotactic but not cytokine functions of mammalian MIF (2, 45). Interestingly, P. berghei MIF coimmunoprecipitates with human CD74 (13) and can induce MAPK signaling (40). This suggests that Plasmodium MIF can interact with and signal through host MIF receptors in vitro. The in vivo role for Plasmodium MIF during blood stages, however, is still unclear. Interestingly, two groups have shown that artificially increased levels of Plasmodium MIF, induced by either infection with a MIF-overexpressing parasite or injection of recombinant protein, result in a reduction in parasite virulence. This suggests that Plasmodium MIF may be involved in dampening acute infection and allowing prolonged infection (40, 45).
While several groups have examined Plasmodium MIF during blood-stage malaria (2, 11, 40, 45), the importance of MIF during development of preerythrocytic stages has not yet been evaluated. Using the P. yoelii rodent malaria model, we showed that P. yoelii MIF (Py-MIF) is expressed predominantly during blood-stage development and liver-stage development, during which it is confined to the parasite cytoplasm. Furthermore, by utilizing a MIF-deficient P. yoelii knockout parasite, we determined that Plasmodium MIF is required for efficient progression through the liver stage of infection.
MATERIALS AND METHODS
Experimental animals, parasites, and cell lines.
Six- to eight-week-old female BALB/cJ (for in vivo infection studies) or Swiss Webster (SW) (for parasite cycle maintenance) mice were purchased from the Jackson Laboratory (Bar Harbor, ME) or Harlan (Indianapolis, IN). Animal handling was conducted according to Institutional Animal Care and Use Committee-approved protocols. Wild-type (WT) P. yoelii 17XNL (nonlethal strain) clone 1.1 (48) and P. yoelii Δmif parasites were cycled between SW mice and Anopheles stephensi mosquitoes. Infected mosquitoes were maintained on sugar water at 24°C and 70% humidity. Salivary gland sporozoites were extracted from infected mosquitoes on day 14 post-blood meal infection as described before (26, 43). The human hepatoma cell line HepG2-CD81 (42) was used for all in vitro assays and was maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with antibiotics (penicillin and streptomycin) and 10% fetal calf serum (FCS).
Generation of P. yoelii Δmif parasites.
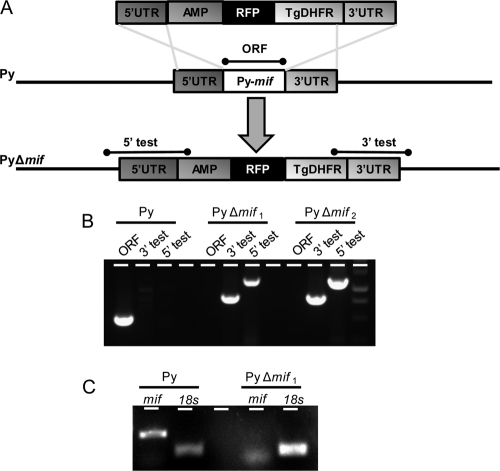
Targeted deletion of Py-mif by double-crossover homologous recombination was achieved by constructing a replacement plasmid in a modified version of the b3D.DT^H.^D targeting vector containing a red fluorescent protein (RFP) cassette (30). Design of this construct was based on the P. berghei mif (Pb-mif) annotation, which was described previously (2) and confirmed in P. yoelii (45). P. yoelii 17XNL genomic DNA (gDNA) was used as a template to amplify a 0.5-kb fragment of the 5′-untranslated region (5′UTR) and a 0.7-kb fragment of the 3′UTR of Py-mif, using oligonucleotide primers PyMIF-1 SacII forward (F), PyMIF-2 ApaI reverse (R), PyMIF-3 ApaI F, and PyMIF-4 NotI R (primer sequences are provided in Table S1 in the supplemental material). PyMIF-2 F and PyMIF-3 R contained a unique ApaI site and overlapping sequences. The 5′UTR and 3′UTR fragments were then combined by sequence overlap extension PCR, and the resulting fragment was inserted into the transfection plasmid between SacII and NotI restriction enzyme sites. The resulting plasmids were digested with ApaI between the 5′UTR and the 3′UTR to linearize the plasmid for transfection. Transfection of P. yoelii 17XNL parasites by use of an Amaxa Nucleofector device (Lonza Cologne GmbH, Germany), resistant parasite selection, and recombinant parasite cloning by serial limiting dilution were all conducted as described elsewhere (20). We obtained two independent P. yoelii Δmif clonal parasite populations (P. yoelii Δmif1 and P. yoelii Δmif2), from two independent transfections. To confirm the targeted deletion and genetic recombination, integration-specific gDNA PCR amplification of the P. yoelii Δmif locus was generated using the specific primer combinations 5′ test (TgF and PyMIFTestF) and 3′ test (PyMIFTestR and TgR). The MIF open reading frame (ORF) was amplified using the primers Sp/PyMIF and Asp/PyMIF (primer sequences are provided in Table S1).
RT-PCR.
Total RNA was extracted using TRIzol reagent (Invitrogen) from P. yoelii salivary gland sporozoites (2 × 106), mixed blood-stage parasites (1 × 107 infected red blood cells were isolated from an infected mouse by heart puncture bleeding and serial limiting dilution of infected blood), or infected livers and then treated with Turbo DNase (Ambion). cDNA synthesis was performed using a SuperScript III Platinum two-step quantitative reverse transcription-PCR (qRT-PCR) kit according to the manufacturer's instructions (Invitrogen). The sequences of specific primers used for amplification from cDNA are listed in Table S2 in the supplemental material. Thirty-five PCR amplification cycles were performed (95°C for 30 s for DNA denaturation and 60°C for 4 min for primer annealing and DNA strand extension). For semiquantitative PCR (QPCR), a standard curve was generated using 1:4 serial dilutions of a reference cDNA sample for PCR amplification of all target PCR products. Experimental samples were compared to this standard curve to give relative transcript abundances. For determination of parasite biomass (see Fig. 4), the average transcript abundance for WT-infected livers was normalized to 1.
Fig 4.
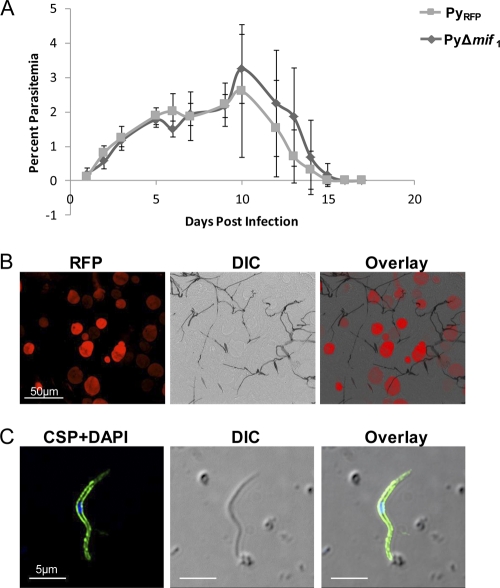
Generation of P. yoelii Δmif parasites. (A) Py-mif was replaced with a selectable drug resistance marker (TgDHFR, the dhfr gene from Toxoplasma gondii [positive selection marker]; and AMP, ampicillin resistance gene for bacterial selection) and a cassette encoding red fluorescent protein by double-crossover homologous recombination using the 5′UTR and 3′UTR regions at the Py-mif locus. The resulting mif-deficient parasite population was fluorescent red. (B) The mixed population of wild-type and transgenic parasites was cloned by serial limiting dilution into SW mice. The genotyping gel shows successful integration on the 3′ (3′ test) and 5′ (5′ test) flanks, as well as the loss of the Py-mif open reading frame, in two independent clones (PyΔmif1 and PyΔmif2). (C) RT-PCR using primers amplifying either mif or 18S rRNA from WT or P. yoelii Δmif1 blood-stage parasites. RT-PCR was performed on cDNA derived from 1 μg of parasite RNA.
IFAs. (i) Salivary gland sporozoites.
Salivary gland sporozoites were isolated and fixed with formalin (Sigma, St. Louis, MO) for 10 min at room temperature. Fixed sporozoites were applied to a polylysine-treated slide, which was housed in a wet chamber at 4°C overnight. Sporozoites were washed twice with phosphate-buffered saline (PBS), permeabilized, and blocked with 2% bovine serum albumin (BSA) plus 0.2% Triton X-100 in PBS for 1 h at 37°C. Immunofluorescence assay (IFA) was performed using murine polyclonal anti-Py-MIF and Alexa Fluor 596-conjugated murine anti-circumsporozoite protein (CSP) antibodies. In other instances, fluorescence staining was achieved with Alexa Fluor-conjugated secondary antibodies specific to rabbit and mouse IgGs. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). The slides were then mounted with FluoroGuard antifade reagent. Fluorescence and differential interference contrast (DIC) images were acquired using an Olympus 1 × 70 Delta Vision microscope equipped with deconvolution software.
(ii) In vitro IFA with HepG2-CD81 cells.
Human hepatoma HepG2 cells expressing the tetraspanin CD81 (HepG2-CD81 cells) were plated on an 8-well chamber slide (Laboratory-Tek Permanox eight-well chamber slide; Nalge Nunc International, Rochester, NY) at a density of 5 × 104 cells/well overnight in DMEM with 10% fetal bovine serum (FBS) at 37°C and 5% CO2. On the second day, salivary gland sporozoites dissected from mosquitoes infected with P. yoelii parasites were incubated with 20% FBS in RPMI 1640 medium for 30 min at room temperature. Next, 3 × 104 sporozoites were added to each well of the 8-well chamber slide cultured with HepG2-CD81 cells, with a subsequent spin at 2,000 rpm for 5 min at room temperature. Cells were incubated at 37°C and 5% CO2 for 24 or 48 h, washed with PBS, and fixed with formalin. Cells were permeabilized and blocked with 2% BSA plus 0.2% Triton X-100 in PBS for 1 h at 37°C. IFA was performed using a rabbit polyclonal antibody (upregulated in infectious sporozoites gene 4 [UIS4] or anti-Py-falstatin antibody) and the anti-Py-MIF antibody. Fluorescence staining was achieved with Alexa Fluor-conjugated secondary antibodies specific to rabbit and mouse IgGs. Nuclei were stained with DAPI. The slides were then mounted with FluoroGuard antifade reagent. Fluorescence and DIC images were acquired using an Olympus 1 × 70 Delta Vision microscope equipped with deconvolution software.
(iii) Liver sections.
To visualize in vivo development of P. yoelii WT or Δmif parasites in livers, 6- to 8-week-old female BALB/cJ mice were injected intravenously with 1 × 106 WT or Δmif sporozoites. Livers from the infected mice were harvested, washed extensively with PBS, and fixed in 4% paraformaldehyde (PFA) at several time points postinfection (p.i.) (2 h, 16 h, 30 h, 44 h, and 50 h). Fifty-micrometer sections were cut from fixed livers by use of a Vibratome apparatus (Ted Pella Inc., Redding, CA). IFAs were performed as previously described (46). Primary antibodies used in IFAs were rabbit polyclonal anti-falstatin or anti-UIS4 antibodies paired with mouse anti-Py-MIF. Fluorescence staining was achieved with Alexa Fluor-conjugated secondary antibodies specific to rabbit and mouse IgGs. Nuclei were stained with DAPI. The slides were then mounted with FluoroGuard antifade reagent. Fluorescence and DIC images were acquired using an Olympus 1 × 70 Delta Vision microscope.
Blood-stage parasite growth curve.
Blood from SW mice infected with P. yoelii WT or Δmif parasites was harvested, diluted with RPMI 1640, and used to infect naïve SW mice for blood-stage growth analysis. One hundred microliters of diluted blood containing 1 × 106 parasites (WT or Δmif) was injected intravenously into each experimental SW mouse (five mice in each group). Parasitemia was tracked by analyzing Giemsa-stained blood smears daily until clearance.
Statistics.
Statistical analyses were performed on two or more independent experiments (unpaired two-tailed Student's t test), as noted in the figure legends or text. Significance indications are as follows: *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; and ***, P < 0.001.
RESULTS
Py-MIF is expressed during liver stages.
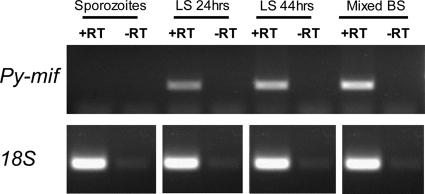
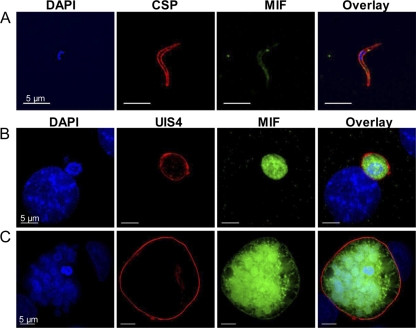
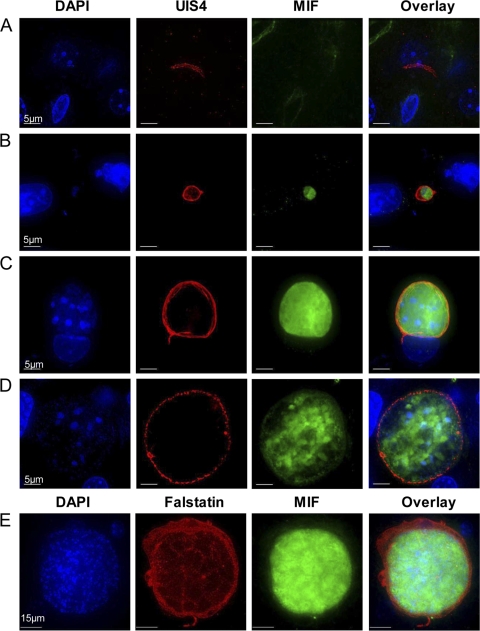
While several groups have determined that Plasmodium MIF is expressed in erythrocyte-stage parasites (2, 45), the expression pattern of MIF during pre-erythrocyte-stage infection had yet to be determined. To establish the expression pattern of Py-mif transcripts in vivo, we monitored Py-mif mRNA in the parasite life cycle by RT-PCR. Sporozoites were isolated from the salivary glands of infected Anopheles stephensi mosquitoes for RNA isolation or were injected into BALB/cJ mice intravenously via the tail vein. Blood or livers from the infected mice were removed after various time points for the isolation of RNA. Py-mif transcripts were detected in mixed blood-stage parasites and 24-h and 44-h liver-stage schizonts but were not observed in infectious salivary gland sporozoites after 35 amplification cycles (Fig. 1) or 45 cycles (see Fig. S1 in the supplemental material). Amplification of parasite 18S rRNA was used as a positive control (Fig. 1; see Fig. S1). We then established the protein expression dynamics and localization of Py-MIF in pre-erythrocyte-stage parasites in vitro by IFA using a polyclonal antibody raised against Py-MIF (45). Py-MIF was detected at low levels in salivary gland sporozoites, but its expression was strong in liver-stage schizonts (Fig. 2A to C). The small amounts of MIF observed in sporozoites (Fig. 2A) were not likely to be unspecific staining, since a similar pattern was observed in parasites expressing a myc-tagged version of Py-MIF by use of an anti-myc antibody (see Fig. S2). At 24 h postinfection (hpi), Py-MIF was distributed uniformly throughout the parasite cytoplasm (Fig. 2B). At 48 hpi, at which point nuclear centers had formed in the late schizonts, Py-MIF appeared to be restricted to the interior of the parasites and was concentrated in globular patterns (Fig. 2C). To confirm this pattern of expression in vivo, BALB/cJ mice were infected with 1 × 106 P. yoelii sporozoites, and their livers were harvested at increasing intervals postinfection. Py-MIF expression was monitored by IFA. We were unable to detect Py-MIF expression at an early time point in vivo (2 hpi) (Fig. 3A); however, Py-MIF was clearly present at later developmental stages (Fig. 3B to E). In 16-h and 30-h liver-stage schizonts, MIF exhibited a uniform distribution throughout the parasite (Fig. 3B and C), while at a later time point (44 hpi) Py-MIF became more compartmentalized and the distribution was focused in the nuclear centers (Fig. 3D). Throughout the exoerythrocytic stages, Py-MIF staining was always observed within the confines of the parasite and never within the host cell, as shown by staining with antibodies against the parasitophorous vacuole membrane (PVM) marker UIS4 (32) (Fig. 2 and 3). However, the parasite plasma membrane is in close proximity to the PVM. Thus, it was difficult to determine whether some MIF was also found in the vacuolar lumen. During blood-stage development, Py-MIF is released or secreted from the parasite during schizont rupture (45). To determine if Py-MIF is released during late liver-stage development, we compared the distribution of Py-MIF to that of Py-falstatin, a protein that is secreted into the PV and released into the host cell during late liver-stage development (35; Y. Pei et al., unpublished data). At 50 hpi, when Py-falstatin was released into the host cell, Py-MIF was still contained within the parasite (Fig. 3E), suggesting that Py-MIF is likely confined to the parasite cytoplasm and not exported beyond the parasite plasma membrane.
Fig 1.
Py-MIF transcript is expressed in liver-stage (LS) and blood-stage (BS) parasites. The expression of Py-mif was determined by RT-PCR on RNA derived from P. yoelii 17XNL. RNA was isolated from salivary gland sporozoites, mixed blood-stage parasites, or mouse livers infected with 1 × 106 sporozoites (isolated at 24 or 44 hpi). Genomic DNA was removed by DNase treatment, and cDNA was generated by reverse transcriptase single-strand DNA synthesis. Either Py-mif or 18S rRNA was amplified from cDNAs via 35 cycles of PCR. Non-RT controls (−RT) were included to confirm that mif amplification was due to cDNA and not to residual genomic DNA.
Fig 2.
Localization of Py-MIF during in vitro liver-stage development. (A) P. yoelii sporozoites were isolated from salivary glands of Anopheles stephensi mosquitoes and stained with polyclonal mouse antibody to Py-MIF (green) and CSP (red). (B and C) HepG2-CD81 cells were infected with P. yoelii salivary gland sporozoites and fixed at 24 (B) or 48 (C) hpi. Cells were stained with a mouse anti-Py-MIF antibody (green), and a polyclonal rabbit anti-UIS4 antibody (red) was used to localize the parasitophorous vacuole membrane. DAPI (blue) was used to stain the nuclei of both parasites and host hepatocytes.
Fig 3.
Localization of Py-MIF in liver-stage parasite mouse infections. One million P. yoelii Δmif sporozoites were injected intravenously into BALB/cJ mice to study liver-stage development in vivo. Liver sections were stained at 2 (A), 16 (B), 30 (C), 44 (D), and 52 (E) hpi with a polyclonal mouse anti-Py-MIF antibody (green). (A to D) A polyclonal rabbit anti-UIS4 antibody (red) was used to localize the parasitophorous vacuole membrane of liver-stage parasites. (E) Cells were stained with a polyclonal rabbit antibody against Py-falstatin (red), a protein that is released into the host cell during late liver-stage development. DAPI was used to stain the nuclei of both parasites and host hepatocytes.
Generation of P. yoelii Δmif parasites.
To determine if Py-MIF is important for liver-stage development, we generated a Py-mif knockout in P. yoelii (P. yoelii Δmif). Using a double-crossover recombination strategy, the mif locus was replaced with a selectable marker (dihydrofolate reductase [DHFR]) and an RFP cassette under the control of a constitutive promoter (30) (Fig. 4A). The recombinant parasites were cloned by serial dilution, and the clonal identity of two parasite lines (P. yoelii Δmif1 and P. yoelii Δmif2 clones), derived from two independent transfections, was confirmed by PCR (Fig. 4B). To verify that Py-mif expression was completely ablated from P. yoelii Δmif parasites, we also monitored Py-mif mRNA expression by RT-PCR. We were unable to detect Py-mif transcripts in the knockout clones (Fig. 4C). The gene knockout was also confirmed by the inability to detect Py-MIF by either IFA or Western blotting using a polyclonal Py-MIF antibody (see Fig. S3 in the supplemental material).
P. yoelii Δmif strain is not attenuated during intraerythrocytic growth and shows normal development of mosquito-stage parasites.
In order to determine if deletion of Py-mif affected the blood-stage development of P. yoelii, we monitored the parasitemia of BALB/cJ mice infected with P. yoelii Δmif parasites. The growth curve was compared to that of an RFP-expressing control parasite in which an RFP expression cassette was inserted into the dispensable S1 locus (19). These parasites were comparable to WT parasites (19) and heretofore are designated control parasites. Mice were injected with 1 × 106 blood-stage parasites, and the resulting parasitemia was determined daily by counting Giemsa-stained parasites in a thin blood smear. No differences were observed between the growth rates of P. yoelii Δmif parasites and the RFP-expressing control parasites (Fig. 5A). Peak parasitemia of mice infected with control parasites was slightly higher than that of mice infected with the knockout strain; however, this difference was not statistically significant (P. yoelii Δmif parasitemia = 2.60% ± 1.93%; control parasitemia = 3.25% ± 1.01%; P = 0.73). Next, we determined if the P. yoelii Δmif strain was defective during mosquito-stage development. Anopheles stephensi mosquitoes were fed on SW mice infected with two independent clones of the P. yoelii Δmif strain or control P. yoelii parasites. Mosquito midguts and salivary glands were harvested and analyzed after 10 and 14 days, respectively. P. yoelii Δmif parasites formed oocysts (Fig. 5B) and generated oocyst sporozoites similarly to WT P. yoelii (Table 1). There were also no significant differences in the number or appearance of salivary gland sporozoites produced in P. yoelii Δmif strain- or WT-infected mosquitoes (Fig. 5C; Table 1).
Fig 5.
mif-deficient parasites develop normally during mosquito or blood stages. (A) Blood-stage growth curve. One million control or P. yoelii Δmif1 blood-stage parasites were injected into BALB/c mice, and parasitemia in the blood was measured daily until all parasites were cleared. (B) P. yoelii Δmif parasites form oocysts. Midguts from mosquitoes infected with the P. yoelii Δmif1 strain were collected at 10 days p.i. and viewed by fluorescence microscopy. P. yoelii Δmif oocysts are fluorescent red. (C) P. yoelii Δmif strain generates salivary gland sporozoites. P. yoelii Δmif1 sporozoites were isolated from mosquito salivary glands and stained with a mouse polyclonal antibody against CSP (green). Parasite DNA was stained with DAPI (blue).
Table 1.
Deletion of Py-mif does not affect sporozoite development in Anopheles mosquitoes
| Genotype | No. of sporozoites per mosquitoa |
|
|---|---|---|
| Oocystsb | Salivary glandsc | |
| RFP-expressing WT | 65,800 ± 26,543 | 17,275 ± 7,864 |
| Δmif1 | 81,500 ± 37,772 | 20,383 ± 9,442 |
| Δmif2 | 64,083 ± 18,304 | 15,016 ± 7,311 |
Values are means ± standard deviations.
Mosquito midguts were removed at 10 days p.i., and oocyst sporozoites were quantified (combination of 3 independent experiments, with >30 mosquitoes per experiment).
Salivary gland sporozoites were dissected at 14 days p.i. and quantified (combination of 4 independent experiments, with >30 mosquitoes per experiment).
P. yoelii Δmif parasites show a liver-stage growth defect.
Since Py-MIF is highly expressed in liver-stage parasites, we hypothesized that it might have an important function in this stage. First, we determined if P. yoelii Δmif parasites were able to complete liver-stage development. BALB/cJ mice were injected intravenously with 1 × 103 or 1 × 104 sporozoites of either control P. yoelii or P. yoelii Δmif1 and P. yoelii Δmif2, and the onset of blood-stage parasitemia was monitored by evaluating thin blood smears. All of the mice infected with the control parasites developed blood-stage patency at approximately 3 days post-sporozoite injection (Table 2). However, less than 50% of mice infected with 1 × 103 P. yoelii Δmif sporozoites had become patent up to the conclusion of the experiment at 20 days postinfection. For mice that became patent, the onset of blood-stage parasitemia was delayed 2 days compared to that with the control, signifying a dramatic reduction in the number of infectious exoerythrocytic merozoites produced by the P. yoelii Δmif strain. Patency was more frequent in mice after injecting 1 × 104 P. yoelii Δmif sporozoites, and the onset of patency was delayed 1 day in mice that did develop blood-stage infection (Table 2).
Table 2.
Delayed patency in BALB/c mice infected with P. yoelii Δmif parasites compared to those infected with control parasites
| Genotype | Dosea | No. of mice injected | % Patent miceb | Mean days to patencyb |
|---|---|---|---|---|
| RFP-expressing WT | 104 | 14 | 100 | 3.0 |
| 103 | 14 | 100 | 3.5 | |
| Δmif1 | 104 | 12 | 83 | 4.4 |
| 103 | 12 | 50 | 5.4 | |
| Δmif2 | 104 | 15 | 47 | 3.8 |
| 103 | 15 | 40 | 5.5 |
Number of sporozoites injected per mouse.
Blood-stage patency was monitored in mice by daily thin smears.
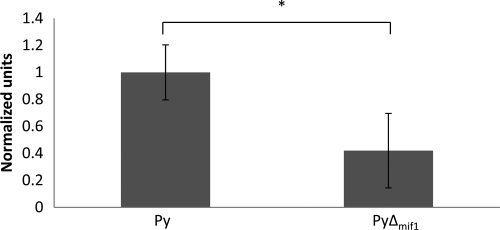
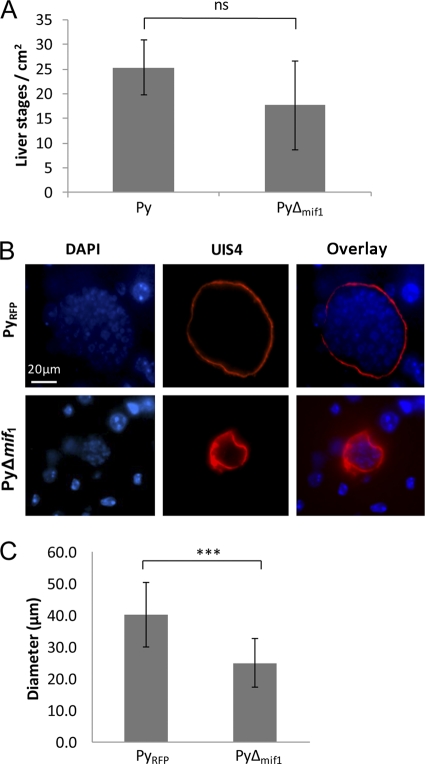
To determine the nature of the liver-stage defect in the P. yoelii Δmif strain, we first quantified the biomass of P. yoelii Δmif parasites in the liver compared to that for control P. yoelii by QPCR. RNA was isolated from the livers of BALB/cJ mice infected with 1 × 106 P. yoelii Δmif or control sporozoites at 44 hpi. The parasite mass was quantified by monitoring the abundance of parasite 18S rRNA compared to total liver RNA (measured by using mouse GAPDH). The P. yoelii Δmif data were further normalized to the abundance levels of the control parasite, which were set to 1. Livers of mice infected with the P. yoelii Δmif strain contained significantly less parasite RNA than those infected with the control parasite (for P. yoelii, 1 ± 0.20 normalized units; for P. yoelii Δmif strain, 0.42 ± 0.28 normalized units), confirming a growth defect of P. yoelii Δmif parasites (Fig. 6). To evaluate if the reduced liver-stage burden observed for P. yoelii Δmif parasites was due to a defect in initial infection of the liver, the parasite-infected livers were examined by IFA on liver sections. Fewer P. yoelii Δmif liver-stage parasites than control parasites were observed in mouse livers; however, these differences were not statistically significant (Fig. 7A). Thus, the initial steps of hepatocyte invasion and dedifferentiation into a trophozoite appeared not to be affected significantly by the deletion of mif. However, further microscopic evaluation of infected livers revealed that the P. yoelii Δmif parasites were significantly smaller than control P. yoelii parasites at 44 hpi (Fig. 7B and C), with the average diameter of a control parasite being 40.1 ± 10.3 μm and the diameter of a mif-deficient parasite being 24.9 ± 7.6 μm (Fig. 7C). We also attempted to evaluate the final phases of liver-stage development, such as merosome formation or merozoite release, in the P. yoelii Δmif parasites. We were unable to detect the presence of merosomes in either P. yoelii Δmif or WT parasites in liver sections (data not shown). However, this absence of merosomes was likely due to the technical difficulty of observing merosome formation in P. yoelii in vivo and not indicative of the ability of P. yoelii Δmif parasites to produce them. It is important to restate here that functional merozoites can be produced by P. yoelii Δmif liver-stage parasites, as some mice infected with the P. yoelii Δmif sporozoites developed blood-stage patency (Table 2).
Fig 6.
Livers from mice infected with the P. yoelii Δmif strain contain less parasite biomass than those infected with WT P. yoelii. BALB/cJ mice were infected with 1 × 106 WT (Py) or mif-deficient clone 1 (PyΔmif1) P. yoelii sporozoites. Livers were harvested at 44 hpi, and the relative amounts of parasite biomass were determined by QPCR for 18S rRNA and normalized to the wild type. Data for a representative sample of two repetitions are shown (n = 5 mice per group). *, P < 0.05.
Fig 7.
Py-mif-deficient liver-stage parasites exhibit a growth defect. (A and B) Livers from mice infected with 1 × 106 P. yoelii WT or Δmif parasites were analyzed at 44 hpi. (A) Quantification of the number of liver-stage parasites per cm2 of liver (for WT, 25 ± 5.60; for P. yoelii Δmif strain, 17.67 ± 8.31; P = 0.09). The data are combinations for 2 experimental replicates. ns, not significant. (B) Liver sections were stained with a rabbit polyclonal antibody against UIS4 (red) to mark the parasitophorous vacuole membrane in infected hepatocytes. DNA is stained with DAPI (blue). Images of a representative sample are shown. (C) Diameters of RFP-expressing P. yoelii control (n = 18) and P. yoelii Δmif (n = 21) parasites were measured using the ruler tool in the Metamorph software package. Three or four diameters were measured for each parasite and then averaged. ***, P < 0.001.
DISCUSSION
MIF is a pleiotropic factor that plays important roles in mammalian immune responses (8). Plasmodium parasites express a MIF homologue; however, the importance of this protein during infection has yet to be delineated clearly (2, 11, 40, 45). Here we characterized a MIF homologue from P. yoelii throughout the parasite life cycle and determined a role for it during liver-stage malaria infection.
During blood-stage replication, Plasmodium MIF is expressed in trophozoites and schizonts and is released during schizont rupture (2, 16, 39). It has been reported that Pf-MIF localizes to Mauer's clefts and is detectable in the cytoplasm of the host erythrocyte (11); however, this localization was not observed with Py-MIF, which was detected only in the parasite cytoplasm (45). P. berghei MIF expression has also been observed in mosquito midgut oocysts and in salivary gland sporozoites (2). Here we show for the first time the expression of Plasmodium MIF in liver-stage parasites. Py-MIF protein was expressed weakly in sporozoites assayed by IFA, but mif transcripts were not detected in salivary gland sporozoites by RT-PCR, as they may have been present in amounts below the limit of detection. Furthermore, we did not detect Py-MIF at 2 h post-hepatocyte invasion in vivo. However, during liver-stage development, Py-MIF was expressed in increasing amounts and was localized within the parasite. During late liver-stage development, Py-MIF exhibited a globular staining pattern within the parasite and was associated with the numerous nuclear DNA masses. At no point did we detect Py-MIF in the host hepatocyte cytoplasm, even during late stages, when secreted proteins such as Py-falstatin are released (Pei et al., unpublished data). To further characterize Py-MIF in liver-stage parasites, we generated a Py-mif knockout strain which showed no difference in blood-stage growth rate or in the generation of mosquito oocyst or salivary gland sporozoites compared to a control parasite line with normal development. However, mif-deficient parasites were defective in liver-stage growth, as shown by the delay or lack of patency observed in mice injected with P. yoelii Δmif sporozoites. Also, mif-deficient parasites were significantly smaller than control parasites at late time points of liver-stage development, suggesting that P. yoelii Δmif parasites do not suffer a defect during initial infection but during subsequent liver-stage growth. In contrast, Augustijn et al. created a mif knockout in P. berghei and stated that it was comparable to the WT throughout the parasite life cycle. However, no data were shown for liver-stage infection (2). It is possible that a liver-stage phenotype may not have been obvious if large numbers of sporozoites were injected into mice; however, it is difficult to speculate due to insufficient data published for the P. berghei system.
Several groups have shown that Plasmodium MIF can modulate host cells by inducing chemotaxis of macrophages or cell signaling or inhibiting apoptosis (2, 11, 40, 45). However, all of these findings are based on experiments with recombinant MIF. We found that Py-MIF is confined within the parasite in P. yoelii liver-stage parasites throughout development in vivo. We did not observe secretion of Py-MIF at any point during liver-stage development, including the late stages, when the PVM breaks down and allows access of proteins in the vacuolar lumen to the host cell cytoplasm. Thus, Py-MIF would not have access to external host receptors or other cell types in the liver on which it could exert cytokine- or chemokine-like functions. This suggests that Py-MIF has parasite-endogenous functions and likely does not interact with external host factors during the liver stage. While it is established that Plasmodium MIF has the intrinsic ability to modulate host cell responses, there is no direct evidence to suggest that MIF interacts physically with host factors in vivo. We cannot rule out the possibility that MIF interacts with host factors during the parasite erythrocytic cycle, as some groups have suggested that Plasmodium MIF is secreted during blood stages (2, 39, 45). However, direct evidence of any such interactions in vivo has yet to be demonstrated.
Mammalian MIF has multiple functions, ranging from cytokine and chemokine properties to regulation of the cell cycle (reviewed in references 10 and 33). It is unlikely that Py-MIF modulates host immune responses through its cytokine or chemokine functions during liver stages because of its intracellular localization in the parasite. However, it is possible that Py-MIF may play a role in parasite survival through its intracellular functions, such as its ability to regulate cell division. In mammalian cells, MIF modulates the cell cycle through its interaction with JAB-1, a protein involved in the proteasomal regulation of cyclins necessary for cell cycle progression. The MIF–Jab-1 interaction is particularly important in the coordination of cell cycle checkpoints, and in MIF-deficient cells, the proteasome is uncoupled from checkpoints in the event of DNA damage (reviewed in reference 33). Thus, MIF-deficient Plasmodium parasites may be unable to efficiently regulate cell division after sustaining DNA damage generated during the normal course of cellular metabolism and may continue through the cell cycle despite DNA damage. This would result in the accumulation of DNA mutations, which may lead to unhealthy or developmentally delayed parasites. The smaller parasites observed in the livers of P. yoelii Δmif parasite-infected mice may have been due to such an accumulation of DNA damage. It would be interesting to determine if P. yoelii Δmif parasites have a higher mutation rate than WT parasites.
In addition to its possible function in regulating the cell cycle, MIF may also be involved in protection from host cell-generated reactive oxygen species (ROS). Cells generate ROS in response to pathogens via the NADPH oxidase (12, 15). These ROS can be delivered directly into pathogen-containing vesicles, and several species, such as H2O2 and nitric oxide, can pass freely through cell membranes. Also, inducible nitric oxide synthase has been observed in P. berghei-infected hepatocytes and may deliver microbicidal nitric oxide directly into the PV (25). MIF possesses a thioredoxin-like oxidoreductase active site. In mammals, thioredoxin-like activity is thought to be important for cellular redox homeostasis, ROS neutralization, and apoptosis inhibition (reviewed in reference 44). Crystal structures for Pb-MIF and Pf-MIF show that the second cysteine (Cys59) of the catalytic CALC motif, thought to be necessary for oxidoreductase activity in mammalian MIF, is not accessible in the parasite homologs (13). However, recent evidence suggests that Pf-MIF may retain oxidoreductase activity through two N-terminal cysteines (Cys3 and Cys4) (1). Thus, another possible role for Py-MIF in liver-stage parasites may be through the antioxidant functions of its oxidoreductase site. Pf-MIF possesses antioxidant functions, as it prevented reactive oxygen species-mediated lipid peroxidation and oxidative damage of DNA in vitro (1). In the absence of MIF, reactive oxygen species or other oxidizing agents may build up in the parasite, inducing DNA damage and/or growth arrest.
Further research remains to determine the specific functions and mechanisms of action of parasite MIF. The importance of MIF in liver-stage parasites determined by our work will help to refocus future experiments to elucidate the role of MIF in the parasite life cycle.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by internal funding from Seattle BioMed.
We thank Jim Burns for providing us with the P. yoelii MIF polyclonal antisera.
Footnotes
Published ahead of print 17 January 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Alam A, et al. 2011. Cysteine-3 and cysteine-4 are essential for the thioredoxin-like oxidoreductase and antioxidant activities of Plasmodium falciparum macrophage migration inhibitory factor. Free Radic. Biol. Med. 50:1659–1668 [DOI] [PubMed] [Google Scholar]
- 2. Augustijn KD, et al. 2007. Functional characterization of the Plasmodium falciparum and P. berghei homologues of macrophage migration inhibitory factor. Infect. Immun. 75:1116–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Awandare GA, et al. 2006. A macrophage migration inhibitory factor promoter polymorphism is associated with high-density parasitemia in children with malaria. Genes Immun. 7:568–575 [DOI] [PubMed] [Google Scholar]
- 4. Bacher M, et al. 1996. An essential regulatory role for macrophage migration inhibitory factor in T-cell activation. Proc. Natl. Acad. Sci. U. S. A. 93:7849–7854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bernhagen J, Calandra T, Cerami A, Bucala R. 1994. Macrophage migration inhibitory factor is a neuroendocrine mediator of endotoxaemia. Trends Microbiol. 2:198–201 [DOI] [PubMed] [Google Scholar]
- 6. Bernhagen J, et al. 2007. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat. Med. 13:587–596 [DOI] [PubMed] [Google Scholar]
- 7. Bozza M, et al. 1999. Targeted disruption of migration inhibitory factor gene reveals its critical role in sepsis. J. Exp. Med. 189:341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Calandra T. 2003. Macrophage migration inhibitory factor and host innate immune responses to microbes. Scand. J. Infect. Dis. 35:573–576 [DOI] [PubMed] [Google Scholar]
- 9. Calandra T, Bernhagen J, Mitchell RA, Bucala R. 1994. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J. Exp. Med. 179:1895–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Conroy H, Mawhinney L, Donnelly SC. 2010. Inflammation and cancer: macrophage migration inhibitory factor (MIF)—the potential missing link. QJM 103:831–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cordery DV, et al. 2007. Characterization of a Plasmodium falciparum macrophage-migration inhibitory factor homologue. J. Infect. Dis. 195:905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Mochel NS, et al. 2010. Hepatocyte NAD(P)H oxidases as an endogenous source of reactive oxygen species during hepatitis C virus infection. Hepatology 52:47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dobson SE, et al. 2009. The crystal structures of macrophage migration inhibitory factor from Plasmodium falciparum and Plasmodium berghei. Protein Sci. 18:2578–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fingerle-Rowson G, et al. 2009. A tautomerase-null macrophage migration-inhibitory factor (MIF) gene knock-in mouse model reveals that protein interactions and not enzymatic activity mediate MIF-dependent growth regulation. Mol. Cell. Biol. 29:1922–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grandvaux N, Soucy-Faulkner A, Fink K. 2007. Innate host defense: Nox and Duox on phox's tail. Biochimie 89:1113–1122 [DOI] [PubMed] [Google Scholar]
- 16. Han C, et al. 2010. Plasma concentration of malaria parasite-derived macrophage migration inhibitory factor in uncomplicated malaria patients correlates with parasitemia and disease severity. Clin. Vaccine Immunol. 17:1524–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hudson JD, et al. 1999. A proinflammatory cytokine inhibits p53 tumor suppressor activity. J. Exp. Med. 190:1375–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Imamura K, et al. 1996. Identification and immunohistochemical localization of macrophage migration inhibitory factor in human kidney. Biochem. Mol. Biol. Int. 40:1233–1242 [DOI] [PubMed] [Google Scholar]
- 19. Jacobs-Lorena VY, Mikolajczak SA, Labaied M, Vaughan AM, Kappe SH. 2010. A dispensable Plasmodium locus for stable transgene expression. Mol. Biochem. Parasitol. 171:40–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Janse CJ, et al. 2006. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol. Biochem. Parasitol. 145:60–70 [DOI] [PubMed] [Google Scholar]
- 21. Kamir D, et al. 2008. A Leishmania ortholog of macrophage migration inhibitory factor modulates host macrophage responses. J. Immunol. 180:8250–8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanai T, et al. 2003. Macrophage-derived IL-18 targeting for the treatment of Crohn's disease. Curr. Drug Targets Inflamm. Allergy 2:131–136 [DOI] [PubMed] [Google Scholar]
- 23. Kleemann R, et al. 2000. Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature 408:211–216 [DOI] [PubMed] [Google Scholar]
- 24. Kleemann R, et al. 1998. Disulfide analysis reveals a role for macrophage migration inhibitory factor (MIF) as thiol-protein oxidoreductase. J. Mol. Biol. 280:85–102 [DOI] [PubMed] [Google Scholar]
- 25. Klotz FW, et al. 1995. Co-localization of inducible-nitric oxide synthase and Plasmodium berghei in hepatocytes from rats immunized with irradiated sporozoites. J. Immunol. 154:3391–3395 [PubMed] [Google Scholar]
- 26. Labaied M, Camargo N, Kappe SH. 2007. Depletion of the Plasmodium berghei thrombospondin-related sporozoite protein reveals a role in host cell entry by sporozoites. Mol. Biochem. Parasitol. 153:158–166 [DOI] [PubMed] [Google Scholar]
- 27. Leng L, et al. 2003. MIF signal transduction initiated by binding to CD74. J. Exp. Med. 197:1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lue H, Kleemann R, Calandra T, Roger T, Bernhagen J. 2002. Macrophage migration inhibitory factor (MIF): mechanisms of action and role in disease. Microbes Infect. 4:449–460 [DOI] [PubMed] [Google Scholar]
- 29. McDevitt MA, et al. 2006. A critical role for the host mediator macrophage migration inhibitory factor in the pathogenesis of malarial anemia. J. Exp. Med. 203:1185–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mikolajczak SA, Aly AS, Dumpit RF, Vaughan AM, Kappe SH. 2008. An efficient strategy for gene targeting and phenotypic assessment in the Plasmodium yoelii rodent malaria model. Mol. Biochem. Parasitol. 158:213–216 [DOI] [PubMed] [Google Scholar]
- 31. Mikolajczak SA, Kappe SH. 2006. A clash to conquer: the malaria parasite liver infection. Mol. Microbiol. 62:1499–1506 [DOI] [PubMed] [Google Scholar]
- 32. Mueller AK, et al. 2005. Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc. Natl. Acad. Sci. U. S. A. 102:3022–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nemajerova A, Moll UM, Petrenko O, Fingerle-Rowson G. 2007. Macrophage migration inhibitory factor coordinates DNA damage response with the proteasomal control of the cell cycle. Cell Cycle 6:1030–1034 [DOI] [PubMed] [Google Scholar]
- 34. Noels H, Bernhagen J, Weber C. 2009. Macrophage migration inhibitory factor: a noncanonical chemokine important in atherosclerosis. Trends Cardiovasc. Med. 19:76–86 [DOI] [PubMed] [Google Scholar]
- 35. Rennenberg A, et al. 2010. Exoerythrocytic Plasmodium parasites secrete a cysteine protease inhibitor involved in sporozoite invasion and capable of blocking cell death of host hepatocytes. PLoS Pathog. 6:e1000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rosengren E, et al. 1996. The immunoregulatory mediator macrophage migration inhibitory factor (MIF) catalyzes a tautomerization reaction. Mol. Med. 2:143–149 [PMC free article] [PubMed] [Google Scholar]
- 37. Salminen A, Kaarniranta K. 2011. Control of p53 and NF-kappaB signaling by WIP1 and MIF: role in cellular senescence and organismal aging. Cell. Signal. 23:747–752 [DOI] [PubMed] [Google Scholar]
- 38. Schober A, Bernhagen J, Weber C. 2008. Chemokine-like functions of MIF in atherosclerosis. J. Mol. Med. (Berlin) 86:761–770 [DOI] [PubMed] [Google Scholar]
- 39. Shao D, et al. 2008. Detection of Plasmodium falciparum derived macrophage migration inhibitory factor homologue in the sera of malaria patients. Acta Trop. 106:9–15 [DOI] [PubMed] [Google Scholar]
- 40. Shao D, et al. 2010. Structural and functional comparison of MIF ortholog from Plasmodium yoelii with MIF from its rodent host. Mol. Immunol. 47:726–737 [DOI] [PubMed] [Google Scholar]
- 41. Shi X, et al. 2006. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity 25:595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Silvie O, et al. 2006. Expression of human CD81 differently affects host cell susceptibility to malaria sporozoites depending on the Plasmodium species. Cell. Microbiol. 8:1134–1146 [DOI] [PubMed] [Google Scholar]
- 43. Tarun AS, et al. 2007. Protracted sterile protection with Plasmodium yoelii pre-erythrocytic genetically attenuated parasite malaria vaccines is independent of significant liver-stage persistence and is mediated by CD8+ T cells. J. Infect. Dis. 196:608–616 [DOI] [PubMed] [Google Scholar]
- 44. Thiele M, Bernhagen J. 2005. Link between macrophage migration inhibitory factor and cellular redox regulation. Antioxid. Redox Signal. 7:1234–1248 [DOI] [PubMed] [Google Scholar]
- 45. Thorat S, Daly TM, Bergman LW, Burns JM., Jr 2010. Elevated levels of the Plasmodium yoelii homologue of macrophage migration inhibitory factor attenuate blood-stage malaria. Infect. Immun. 78:5151–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vaughan AM, et al. 2009. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell. Microbiol. 11:506–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vermeire JJ, Cho Y, Lolis E, Bucala R, Cappello M. 2008. Orthologs of macrophage migration inhibitory factor from parasitic nematodes. Trends Parasitol. 24:355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weiss WR, Good MF, Hollingdale MR, Miller LH, Berzofsky JA. 1989. Genetic control of immunity to Plasmodium yoelii sporozoites. J. Immunol. 143:4263–4266 [PubMed] [Google Scholar]
- 49. WHO 2010. World malaria report. World Health Organization, Geneva, Switzerland [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.