Abstract
Little is known about the colonization mechanisms of Aeromonas spp. Previous work has suggested that the type IV bundle-forming pilus (Bfp) is an aeromonad intestinal colonization factor. This study provides the first genetic characterization of this structure. To define the role of Bfp in Aeromonas veronii bv. Sobria adherence, a 22-kb locus encoding the bundle-forming pilus was isolated; this contained 17 pilus-related genes similar to the mannose-sensitive hemagglutinin (MSHA) of Vibrio cholerae. Reverse transcriptase PCR (RT-PCR) demonstrated that the locus had two major transcriptional units, mshI to mshF and mshB to mshQ. Transcriptional fusion experiments demonstrated the presence of two strong promoters upstream of mshI and mshB. The locus encoded four putative prepilin proteins, one of which (MshA) corresponded to the N-terminal sequence of the previously isolated major pilin protein. All the pilin genes were inactivated, mutation of each minor or major pilin gene greatly reduced the bacterium's ability to adhere and form biofilms, and complementation of each mutant in trans rescued this phenotype. Mutation of the major pilin MshA and MshB, a minor pilin, resulted in their loss. The position of the mshH gene is conserved within a number of bacteria, and we have shown it is not transcriptionally linked to the other msh genes; moreover, its mutation did not have a dramatic effect on either adhesion or biofilm formation. We conclude that the bundle-forming pilus is required for A. veronii bv. Sobria adherence and biofilm formation; furthermore, both the major and minor pilin proteins are essential for this process.
INTRODUCTION
Mesophilic Aeromonas species are ubiquitous waterborne bacteria that are pathogens of reptiles, amphibians, and fish (1). They can be isolated as part of the fecal flora of a wide variety of other animals, including some used for human consumption, such as cows, sheep, and poultry. In humans, Aeromonas hydrophila belonging to hybridization groups 1 and 3 (HG1 and HG3), Aeromonas veronii bv. Sobria (HG8/HG10), and Aeromonas caviae (HG4) are the main species of the genus that have been associated with disease (9). Aeromonas spp. are an underreported cause of disease in humans and are regarded as emerging pathogens. In humans, they cause diseases ranging from serious wound infections to septicemia (20). However, most are isolated from cases of gastroenteritis that can range from a mild self-limiting diarrhea to a cholera-like illness to dysentery (32).
A range of putative virulence factors has been described for the aeromonads, ranging from the hemolytic toxin aerolysin and cytotonic toxins to capsules and extracellular enzymes (32), and recently aeromonad type III (T3SS) and type VI (T6SS) secretion systems have been described (29, 33).
Adhesion and colonization of host tissue are critical to aeromonad gastrointestinal pathogenesis to allow for effective delivery of toxins and/or invasion that results in disease. However, the adherence process of aeromonads is still poorly understood. A number of factors have been implicated in virulence, such as long/wavy (L/W) type IV pili, outer membrane proteins, lipopolysaccharide O antigen (O-Ag), and the polar flagellum (32). The mesophilic aeromonads are interesting, as most strains express two distinct flagellum systems (4, 22). They have a glycosylated polar flagellum for swimming in liquid and express separate lateral flagella for swarming over surfaces (4, 22, 30).
Type IV pili have been shown to be important for epithelial cell adherence and colonization for several pathogens, including enteric pathogens such as Vibrio cholerae and several types of pathogenic Escherichia coli (5, 27). Previous studies on gastroenteritis-associated Aeromonas spp. has shown that they also encode at least two distinct families of type IV pili, Tap and Bfp, which differ in their N-terminal amino acid sequences and molecular weights (2). The type IV Aeromonas pilus (Tap) was identified following the cloning of the biogenesis cluster (tapABCD) (21). Insertional inactivation of the structural pilin gene tapA had no effect on adherence, as the mutant cells were able to adhere to both biotic and abiotic surfaces as well as the wild-type parental strain (11). The predominant aeromonad type IV pilus that has been isolated is the bundle-forming pilus (Bfp), whose removal by mechanical shearing or blockage using anti-Bfp antibodies reduces the ability of aeromonads to adhere to host cells by over 80% (13, 14). The N-terminal amino acid sequence of Bfp shows closet homology to the mannose-sensitive hemagglutinin (MSHA) of Vibrio cholerae El Tor and that this type IV pilin belongs to the type IVa non-bundle-forming pilus family. Therefore, it appears that Bfp is the major pilus adhesin of mesophilic Aeromonas species, but they have not been genetically characterized. Moreover, the recently published genome sequences of A. hydrophila ATCC 7966T (26) and Aeromonas salmonicida A449 (24) have shown that these strains encode three type IV systems, tap, flp, and msh (Bfp). However, in the psychrophilic species A. salmonicida, the locus encoding the MSHA pilus had a large internal deletion, and the pilus system was therefore thought to be nonfunctional (3). Mutation of the A. salmonicida Flp system made little or no contribution to virulence, but the Tap system was thought to be involved in virulence when tested in the natural host for this species, the Atlantic Salmon (3).
In this study, we present the first data on the genetic characterization of the bundle-forming pilus of mesophilic aeromonads and demonstrate that it has a role in the colonization of both biotic and abiotic surfaces.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids are listed in Table 1. Bacteria were grown aerobically overnight (16 to 20 h), either statically or by shaking, at 37°C. They were grown either in brain heart infusion broth (BHIB) (Oxoid) or on Luria-Bertani agar (LBA), supplemented with the appropriate antibiotics when required. Working stocks of the strains were kept on LBA plates at 4°C for a maximum of 2 weeks. Rifampin (Rif), ampicillin (Amp), streptomycin (Sm), and kanamycin (Km) were used at final concentrations of 50 μg/ml, whereas chloramphenicol (Cm) and tetracycline (Tc) were used at 25 μg/ml.
Table 1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Genotype/phenotypea | Source/reference |
|---|---|---|
| Strains | ||
| Aeromonas veronii bv. Sobria | ||
| BC88 | Wild type, dysenteric isolate | 13 |
| BC88R | BC88, spontaneous Rifr | This study |
| BC88mshA | BC88RmshA::Kmr | This study |
| BC88mshB | BC88RmshB::Kmr | This study |
| BC88mshC | BC88RmshC::Kmr | This study |
| BC88mshD | BC88RmshD::Kmr | This study |
| BC88TapD | BC88RmshD::Kmr | This study |
| BC88MshH | BC88RmshH::Kmr | This study |
| Escherichia coli | ||
| CC118λpir | Δ(ara,leu)7697 araD139 ΔlazX74 glaE glaK phoA20 thi-1 rspE rpoB(Rfr) argE(am) recA1 λpir+ | 8 |
| S17-1λpir | hsdR pro recA, RP4-2 in chromosome, Km::Tn7 (Tc::Mu) λpir Tpr Smr | 19 |
| XL1-Blue | endA1 recA1 hsdR17 supE44 thi-1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tcr)] | Stratagene |
| BL21(DE3) | F−ompT gal dcm lon hsdSB (rB−mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | Novagen |
| Staphylococcus epidermidis | ||
| NCTC11047 | Wild-type strain, laboratory culture collection | |
| Plasmid | ||
| pUC19 | High-copy-no. cloning vector, MCS, Ampr | Gibco BRL |
| pUC4-KIXX | Source of Tn5-derived nptII gene (Kmr) | Pharmacia |
| pBBR1MCS | Broad host range, IncP, -W, -Q, ColE1 and p15A compatible, containing pBluescript IIKS-lacZα-polylinker, Cmr | 15 |
| pKNG101 | oriR6K mobRK2 strAB sacBR, 6.8 kb, Smr | 10 |
| pZErO2.1 | Cloning vector Kmr | Invitrogen |
| pET28a(+) | E. coli overexpression vector for N-terminal His6-tagged proteins, Kmr | Novagen |
| pKAGb4(−) | Broad-host-range vector ori1600 carrying promoterless lacZ gene, Cmr | (M. S. Thomas) |
| pTB037 | 78 bp, encoding MshA N-terminal region in pZErO2.1 | This study |
| pETMshA | pET28a(+) expressing MshA as an N-terminal His6-tagged protein, Kmr | This study |
| pETMshB | pET28a(+) expressing MshB as an N-terminal His6-tagged protein, Kmr | This study |
| pBBRMshA | mshA of A. veronii bv. Sobria BC88 in pBBR1MCS, Cmr | This study |
| pBBRMshB | mshB of A. veronii bv. Sobria BC88 in pBBR1MCS, Cmr | This study |
| pBBRMshC | mshC of A. veronii bv. Sobria BC88 in pBBR1MCS, Cmr | This study |
| pBBRMshD | mshD of A. veronii bv. Sobria BC88 in pBBR1MCS, Cmr | This study |
| pBBRMshH | mshH of A. veronii bv. Sobria BC88 in pBBR1MCS, Cmr | This study |
| pBBRTapD | tapD of A. veronii bv. Sobria BC88 in pBBR1MCS, Cmr | This study |
| pKAG-PB | Promoter probe suicide vector pKAGb4 with BC88 mshB promoter region ligated upstream of the promoterless lacZ gene, Cmr | This study |
| pKAG-PA | Promoter probe suicide vector pKAGb4 with BC88 mshA promoter region ligated upstream of the promoterless lacZ gene, Cmr | This study |
| pKAG-PC | Promoter probe suicide vector pKAGb4 with BC88 mshC promoter region ligated upstream of the promoterless lacZ gene, Cmr | This study |
| pKAG-PD | Promoter probe suicide vector pKAGb4 with BC88 mshD promoter region ligated upstream of the promoterless lacZ gene, Cmr | This study |
| pKAG-PI | Promoter probe suicide vector pKAGb4 with BC88 mshI promoter region ligated upstream of the promoterless lacZ gene, Cmr | This study |
Amp, ampicillin; Cm, chloramphenicol; Km, kanamycin; Nal, nalidixic acid; Sm, streptomycin; MCS, multiple cloning site.
Protease, DNase, and hemolysin detection.
For protease plates, the method described by Sokol et al. (28) was used. Dialyzed brain heart infusion (D-BHI) milk medium was prepared by dissolving 18.5 g of BHI powder (Difco) in 50 ml of water that was then dialyzed against 1 liter of distilled water (dH2O) for 18 h at 4°C. Agar (Difco) was added to the dialysate to a concentration of 3% (wt/vol). A 3% (wt/vol) solution of skim milk was prepared, and the solutions were autoclaved separately. Equal volumes of the two sterile solutions were mixed at 60°C, and the mixture was dispensed into petri plates. The bacterial strains were cultured on the plate and incubated at 37°C for 24 h. The presence of clear zones of hydrolysis on the D-BHI milk medium was considered a positive result.
For DNase detection, appropriate Aeromonas strains in addition to a Staphylococcus epidermidis wild-type strain were grown on a DNase agar plate (Oxoid) at 37°C overnight for approximately 16 h. The plate was then flooded with 1 M HCl to precipitate the DNA and make the plate opaque. A positive reaction was indicated by the disappearance of the opaqueness and an area of clearing around the colony of bacterial growth. For hemolysin detection, Aeromonas strains were streaked onto a Columbia blood agar plate containing horse blood (Oxoid). Plates were incubated at 37°C overnight for approximately 16 h and examined for β-hemolysis.
Pilin purification.
The pilins MshA and MshB were overexpressed and purified as N-terminal His6-tagged proteins. Both mshA and mshB, encoding the corresponding proteins minus their signal sequences, were amplified using primers, including NdeI/XhoI and NdeI/EcoRI restriction sites, respectively, to facilitate cloning in frame into the overexpression vector pET28a (Novagen). The proteins were overexpressed in E. coli BL21λ(DE3) according to the manufacturer's instructions and then purified using a HisTrap HP column (GE Healthcare).
Preparation of antibody.
Approximately 200 μg of purified pilin was emulsified with 1 ml of Freund complete adjuvant and inoculated subcutaneously into dwarf lop-eared rabbits. Booster injections of the 100-μg pilin protein in 0.5 ml Freund complete adjuvant were administered 4 and 6 weeks later. Antibodies were obtained by bleeding 10 days after the second booster injection.
Whole-cell protein preparation, SDS-PAGE, and immunoblotting.
Aeromonas strains were grown statically overnight in BHIB at 37°C. Equivalent numbers of cells were harvested by centrifugation, and the cell pellet was either boiled in SDS-PAGE loading buffer for whole-cell proteins or was resuspended in 1 ml of phosphate-buffered saline (PBS), which was subsequently sheared of pili by passing the bacterial suspension through an 18-gauge needle 20 times. Bacteria were pelleted by centrifugation and discarded; 100 μl of the supernatant was added to 100 μl of SDS-PAGE loading buffer and boiled for 5 min. Protein samples were separated on SDS-polyacrylamide gels (12% [vol/vol]). For immunoblotting, proteins were transferred onto a Hybond-C (GE Healthcare) nitrocellulose membrane. Following transfer, membranes were blocked with 5% (wt/vol) powdered skim milk and probed with a polyclonal rabbit anti-pilin antibody (1:500). The unbound antibody was removed by five washes in PBS, and a goat anti-rabbit peroxidase-conjugated secondary antibody (1:1,000) was added. The unbound secondary antibody was washed away with PBS as described for the primary antibody. The conjugate was then detected using the ECL detection system (GE Healthcare).
Adherence assay.
Tissue culture was maintained as described by Thornley et al. (31). The HEp-2 cells (ATCC CCL23, derived from human epidermoid larynx carcinoma) were grown in basal Eagles medium containing newborn calf serum. The cell line was grown until confluent at 37°C in air with 5% CO2, diluted to 50 to 100 cells/mm2, and seeded in 1-ml amounts into 24-well tissue culture plates. The HEp-2 cells were used at semiconfluence in duplicate for each separate experiment, with monolayer age being kept as constant as possible. The HEp-2 cells were 24 h old after the final seeding. Bacteria were grown statically in BHIB at 37°C, harvested by gentle centrifugation (1,600 × g, 5 min), and resuspended in PBS (pH 7.2) at approximately 107 CFU/ml (A600 of ∼0.07). The HEp-2 cells were infected with 1 ml of the bacterial suspension for 90 min at 37°C in 5% CO2. Following infection, the nonadherent bacteria were removed from the monolayer by three washes with PBS. The remaining adherent bacteria and the monolayers were then fixed in 100% methanol for 5 min. Methanol was removed by washing with PBS, and the HEp-2 cells with the adherent bacteria were stained for 45 min in 10% (vol/vol) Giemsa stain (BDH, United Kingdom) prepared in Giemsa buffer. The coverslips were air dried, mounted, and viewed by oil immersion under a light microscope at ×1,000 magnification. Twenty HEp-2 cells/coverslip were randomly chosen, and the number of bacteria adhering/HEp-2 cell was recorded. Assays were carried out in duplicates or triplicates.
Biofilm assay.
Biofilm formation in borosilicate glass tubes (10 by 75 mm) was assessed by the method of O'Toole and colleagues with slight modifications (12). In brief, the glass tubes were inoculated with 300 μl of a 1:100 dilution of bacteria from brain-heart infusion broth (BHIB) cultures grown overnight (16 to 18 h). These were then lightly covered with foil and incubated for up to 30 h at 37°C without shaking. To detect and quantify biofilm formation, the tubes were rinsed thoroughly and vigorously with water, and the remaining cells were stained with crystal violet solution (0.5% [wt/vol], 15 min at room temperature [RT]). The crystal violet-stained biofilm was then solubilized by the addition of 100% ethanol (600 μl, 10 min, RT), and the optical density at 570 nm (OD570) of 200 μl of the resultant suspension was measured using a microplate reader (Bio-Rad, Hercules, CA). Replicates of three to six tubes per organism were examined in each experiment, and a broth control that was not inoculated was included in each experiment. The growth rates and motility of the pilus mutant strains were assessed by measuring OD600 and swimming in motility agar (22), respectively, and were shown not to differ significantly from that of the wild-type strain (data not shown).
Statistical analysis.
The differences in adherence to cell lines between the wild-type and mutant strains and the mutant strains versus the complemented strains were analyzed using GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA). Data are given as means ± standard deviations (SD). Statistical significance was compared to the wild-type by one-way analysis of variance (ANOVA).
DNA techniques.
Plasmid DNA was isolated by alkaline lysis or by using the Qiagen miniprep DNA purification system (Qiagen). A. veronii bv. Sobria chromosomal DNA isolation was carried out according to standard techniques (25). DNA restriction digestions and T4 ligations were carried out according to the manufacturer's instructions. DNA samples were separated on 0.8% (wt/vol) agarose gels; when required, extraction of DNA from gels was carried out using the QIAquick gel extraction kit (Qiagen).
PCR.
Reactions were performed using Pfx DNA polymerase (Invitrogen) at 2.5 mM MgCl2 in a Techne thermal cycler. Initial DNA denaturation was carried out for 2 min, and amplification reactions were carried out for 30 cycles with denaturation at 95°C for 30 s, primer annealing at 55°C for 1 min, and elongation at 72°C for 2 min.
Southern and dot blot hybridizations.
Southern and dot blot hybridizations were performed by capillary transfer (25). Probe labeling, hybridization, and detection were carried out using the enhanced chemiluminescence (ECL) labeling and detection system (GE Healthcare) according to the manufacturer's instructions.
Nucleotide sequencing and sequence analysis.
Double-stranded DNA sequencing was performed by using the Sanger dideoxy-chain termination method with the Abi Prism dye terminator cycle sequencing kit (Perkin Elmer company). DNA fragments were ligated into pUC19 and sequenced using an ABI Prism 377 DNA sequencer (Perkin Elmer company). The M13 universal primers were employed in sequencing the ends of the DNA inserts. Following the first sequencing reaction and whenever required, primers were designed until the insert sequences were complete. Primers used for DNA sequencing were purchased from Eurogentec. For chromosomal walking to extend the sequence into flanking regions, direct genomic sequencing was used. Custom 24-mer primers were designed to a known nucleotide sequence and were used with sheared A. veronii bv. Sobria genomic DNA in a 99-cycle polymerase reaction using the BigDye Terminator mix according to the manufacturer's instructions (Perkin Elmer company). The DNA sequence was translated in all six frames, and all open reading frames (ORFs) greater than 100 bp were inspected. Deduced amino acid sequences were compared with those of DNA translated in all six frames from nonredundant GenBank and EMBL databases by using the BLAST network service at the National Center for Biotechnology Information and analyzed as previously described (6, 22).
Construction of defined insertion mutants.
Mutants were created by the insertion of the Tn5-derived kanamycin resistance cartridge (nptII) from pUC4-KIXX (Pharmacia). This cartridge contains an outward reading promoter that drives the transcription of downstream genes when inserted in the correct orientation. For each mutant, the 1.4-kb SmaI-digested kanamycin resistance cartridge was inserted into a convenient restriction site within the middle of the gene. If a convenient site was not present, one was created by spliced overlap extension (SOE) PCR. Constructs containing the mutated genes were ligated into the suicide vector pKNG101 (10) and transferred into Aeromonas by conjugation. Conjugal transfer of the recombinant plasmids from E. coli S17-1λpir to A. veronii bv. Sobria BC88 was performed using a filter mating technique. Bacterial conjugation was allowed to proceed for 6 to 8 h at 37°C on sterile nitrocellulose filters (0.45-mm pore size) placed onto an LBA plate. Serial dilutions of the mating mix were then plated on LBA supplemented with Rif and Km, the latter added in order to select for recombination. Colonies that were kanamycin resistant (Kmr) and streptomycin sensitive for pKNG101 derivatives (those not likely to have retained the vector) were purified and probed for the kanamycin cartridge and absence of any plasmid sequences by Southern hybridization, thus demonstrating a double recombination event and allelic exchange.
Construction of lacZ transcriptional fusions.
The mobilizable broad-host-range lacZ promoter probe plasmid pKAGb-4(−) was used in this study (K. Agnoli and M. Thomas, unpublished data). The putative promoter regions of the four A. veronii bv. Sobria pilus genes and mshI gene were amplified by PCR, and the resulting fragments were directionally ligated separately into pKAGb-4(−), giving the plasmids pKAG-PB, pKAG-PA, pKAG-PC, pKAG-PD, and pKAG-PI for the mshBACD promoters and the mshI promoter, respectively. These plasmids were introduced separately by conjugation into the wild-type strain. Activity of the four putative promoters was measured as a function of β-galactosidase activity. A. veronii bv. Sobria cultures were grown in triplicate to an optical density at 600 nm (OD600) of 0.5 to 0.8 and were then chilled on ice for 15 min. Duplicate assays were performed at 30°C on 200 μl of cells for each culture in a total volume of 1 ml following permeabilization of the cells with chloroform and sodium-dodecyl sulfate; values are presented in Miller units (MU) (18).
RT-PCR.
Reverse transcriptase PCR (RT-PCR) was performed as described by Tabei et al. (30).
Nucleotide sequence accession number.
The nucleotide sequence of the genes described here have been assigned the following GenBank accession number: JQ361815.
RESULTS
Cloning of the bfp pilus locus.
At the outset of this project, the genome sequence of Aeromonas veronii bv. Sobria was not available. The N-terminal sequence of the major pilin purified from A. veronii bv. Sobria strain BC88 (13) was used to design the degenerate forward and reverse primers P019 (ATGACIYTIATHGARYTIGT) and P022 (TTIARRAAYTTIGGIGCIGC) that corresponded to amino acids 1 to 7 and 26 to 20, respectively. This enabled the amplification of a 78-bp DNA fragment by PCR, which was subsequently cloned into pZErO2.1, generating pTB037. Upon nucleotide sequencing, the translated sequence of the insert exactly corresponded to the N-terminal sequence of the Bfp pilin protein. The DNA sequence both upstream and downstream of this locus was then determined by direct genomic sequencing of the BC88 chromosomal DNA using custom-designed primers. This strategy extended the sequence by over 20 kb.
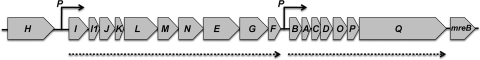
Sequence analysis revealed 19 ORFs (Fig. 1A, Table 2). The arrangement and encoded protein sequences of the ORFs in this region closely resemble those of the type IV pilus mannose-sensitive hemagglutinin (MSHA) proteins of V. cholerae El Tor (17) and have been designated mshA to mshQ, accordingly. The locus encoded four pilin/pseudopilin proteins and 14 proteins that have been linked with pilus biogenesis in other bacteria; in addition, there was a hypothetical protein (MshF) that is encoded at the same position in A. hydrophila ATCC 7966T and a diguanylate cyclase and phosphodiesterase domain-containing protein (MshH). Downstream of the locus was the gene mreB that encodes a protein involved in the determination of cell shape. Recently, the genome sequences of the related Aeromonas species, A. hydrophila ATCC 7966T, A. salmonicida A449, and A. veronii B565, have been completed (16, 24, 26), and these also contained the 22-kb msh locus, with the analogous genes designated AHA0383 to AHA0399, ASA3949 to AHA3938, and B565_3672 to B565_3654, respectively. Moreover, the corresponding gene cluster is present in many Vibrio, Photobacterium, and Shewanella strains. All 18 genes that are thought to be required for the biogenesis of the bundle-forming pilus are present in A. hydrophila, but A. salmonicida A449 has a large deletion between the mshN and mshD genes (3). Unusually for MSHA, the A. veronii bv. Sobria MshI protein was encoded by two genes, mshI and mshI1. This was also true in A. hydrophila ATCC 7966T (26), A. salmonicida (24), and A. caviae Sch3 (J. Shaw, unpublished data).
Fig 1.
Genetic organization of the A. veronii bv. Sobria BC88, mannose-sensitive hemagglutinin (MSHA) type IV pilus locus. The predicted ORFs were named after the msh homologues in other species and are indicated by gray arrows, which indicate the direction of transcription. The right-angled arrows with solid lines and a P depict the promoter regions, whereas the arrows with dotted lines show the linked transcriptional units.
Table 2.
Characteristics of the A. veronii bv. sobria BC88 msh locus-encoded proteins
| msh locus gene product | Mol mass (kDa) | Similarity to equivalent V.cholerae El Tor N16961 MSHA protein | Predicted function |
|---|---|---|---|
| MshH | 74.2 | 5.6e−104 | GGDEF/EAL domain-containing regulatory protein |
| MshI | 31.3 | 4.9e−25 | Bitopic inner membrane protein, GspL-like N-terminal domain of MshI |
| MshI1 | 22.3 | 6.2e−23 | Bitopic inner membrane protein, GspL-like C-terminal domain of MshI |
| MshJ | 24.3 | 3.4e−26 | Bitopic inner membrane protein, GspM-like |
| MshK | 11.4 | 3.13e−09 | Periplasmic protein, GspC-like |
| MshL | 59.9 | 1.4e−125 | Outer membrane secretin, GspD-like |
| MshM | 33.6 | 2.7e−49 | Membrane protein with ATPase domain, GspA-like |
| MshN | 39.5 | 2.1e−23 | Tetratricopeptide repeat protein, PilF-like |
| MshE | 63.2 | 9.0e−161 | ATPase, GspE-like |
| MshG | 44.9 | 7.3e−111 | Polytopic integral membrane protein |
| MshB | 22.4 | 5.7e−24 | Prepilin |
| MshA | 14.3 | 0.77 | Prepilin |
| MshC | 14.7 | 9.1e−7 | Prepilin |
| MshD | 15.4 | 7.6e−13 | Prepilin |
| MshO | 16.1 | 4.6e−8 | Pseudopilin, GspG-like |
| MshP | 16.7 | 7.4e−15 | Periplasmic type IV pilus protein PilX |
| MshQ | 144.5 | 5.0e−144 | Pilin adhesin, PilY1-like, lectin-like domain |
Transcript mapping of the pilus locus.
RT-PCR was employed to determine which genes were cotranscribed with their immediate partners downstream. Primer pairs that overlapped different pilus genes (from mshH to mreB) within the locus were designed toward the 3′ end of the upstream gene and near the 5′ end of the downstream gene. This was in order to amplify the intergenic region between the two genes that would be expressed if the two genes were cotranscribed. RT-PCR products of the expected sizes were detected (data not shown) for every gene pair from mshI to mshF and from mshB to mshQ. This suggested the presence of two major transcriptional units, one incorporating mshI to mshF and the other mshB to mshQ in the pilus locus, with the two major promoters being located upstream of mshI and mshB, respectively (Fig. 1). It also suggested that both mshH and mreB were not transcriptionally linked to the other msh genes.
The four pilin genes (mshBACD) were located adjacent to each other in the same transcriptional orientation, with relatively small intergenic regions between the end of one gene and the start of another. These data along with the RT-PCR information suggested that they were part of a polycistronic operon. To support this, the putative promoter regions of the pilin genes mshB, mshA, mshC, and mshD were transcriptionally fused to the promoterless reporter gene lacZ in pKAGb-4(−). DNA fragments of around 500 bp were amplified by PCR from BC88 wild-type strain chromosomal DNA. These represented the 5′ ends of the genes and their putative promoter regions. The PCR fragments were ligated into the broad-host-range lacZ expression vector pKAGb-4(−) in an orientation to allow expression of the lacZ gene. The constructs were separately introduced into the A. veronii bv. Sobria wild-type strain BC88. Quantitative assays on cells grown in BHI for β-galactosidase activity showed that in the A. veronii bv. Sobria wild-type parental strain, the putative mshA, mshC, and mshD promoters had very low intrinsic β-galactosidase activities equivalent to that of the vector-only background control (212 to 300 MU). In contrast, a 20-fold increase in β-galactosidase activity (6,082 MU) was detected for the mshB promoter (Table 3), suggesting that the mshBACD genes comprise a single transcriptional unit.
Table 3.
β-Galactosidase activities of putative msh promoters in A. veronii bv. Sobria BC88
| Plasmid | Construct | Activity (MU) |
|---|---|---|
| pKAGb4 | Vector | 300 ± 54 |
| pKAG-PB | mshBp-lacZ | 6,082 ± 564 |
| pKAG-PA | mshAp-lacZ | 212 ± 45 |
| pKAG-PC | mshCp-lacZ | 233 ± 44 |
| pKAG-PD | mshDp-lacZ | 233 ± 45 |
| pKAG-PI | mshIp-lacZ | 8,415 ± 653 |
Similarly, the mshII1JKLMEGF genes are located adjacent to each other in the same transcriptional orientation and appear to be part of a single transcriptional unit under the control of a promoter upstream of mshI. Therefore, the 500-bp region upstream of mshI was also amplified by PCR and ligated upstream of lacZ in pKAGb-4(−). When introduced into A. veronii bv. Sobria wild-type cells, this promoter repeatedly showed a higher activity (8,415 MU) than the mshB promoter (Table 3), suggesting that the expression of mshII1JKLMEGF is controlled by a promoter upstream of mshI.
The A. veronii bv. Sobria msh pilin genes are critical for adherence to epithelial cells.
The msh locus contained four putative pilin/pseudopilin genes. As stated above, the N-terminal sequence of the purified pilin exactly matched that of the derived N terminus (minus cleaved signal sequence) of the mshA gene product (13). The major aim of this study was to create isogenic mutants in the major and minor pilin genes that would allow the first determination of the aeromonad bundle-forming pilus in adhesion. A kanamycin resistance cassette was inserted in the same transcriptional orientation as the target gene. The presence of an outward reading promoter on the cassette ensures expression of downstream genes, thereby reducing any polar effects. However, such insertions may alter the regulation of the downstream genes. The construction of all mutants was verified by Southern hybridization of chromosomal DNA and by PCR (data not shown).
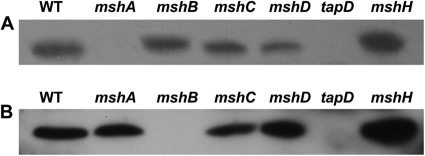
Furthermore, both the MshA and MshB proteins as examples of both major and minor pilins were overexpressed as N-terminal His-tagged proteins in E. coli and purified. They were subsequently used to raise polyclonal antibodies against MshA and MshB in rabbits. Western blotting demonstrated that the MshA or MshB pilins were absent from their respective mutants, mshA and mshB, but present in the wild-type mshC, mshD, and mshH mutants (Fig. 2A and B). A mutant in the prepilin N-peptidase TapD was also created (see section below), and both pilins (MshA and MshB) were absent in the TapD mutant (Fig. 2A and B).
Fig 2.
Msh-pilin immunoblotting of whole-cell proteins of the wild-type strain (WT) and isogenic mutants. The mutation is indicated above the lane. (A) MshA Western immunoblot; (B) MshB Western immunoblot. Proteins were obtained from bacteria grown at 37°C in BHIB and were analyzed by SDS-PAGE (12%). They were transferred onto nitrocellulose membranes and immunoblotted with anti-MshA or anti-MshB antibody (1:1,000).
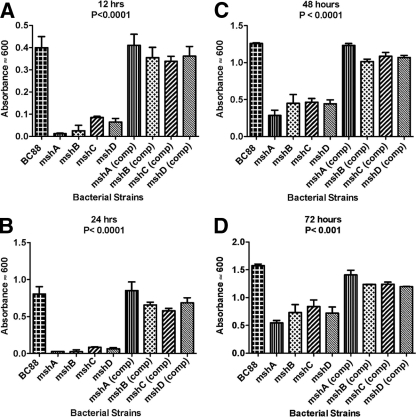
To determine if the msh locus encoding the A. veronii bv. Sobria BC88 bundle-forming pilus had a true role in adhesion, the wild-type and mutant strains were assessed by adherence to HEp-2 cells and by biofilm formation on borosilicate tubes. The wild-type strain BC88 adheres to HEp-2 cells with approximately 21 bacteria per cell, whereas in the mutants where each pilin gene had been individually knocked out, bacterial adherence was significantly (P < 0.001) reduced by over 90% to between 1 to 2 bacteria per cell (Fig. 3). To confirm these results and rule out any polar effects, complementation analysis was performed. Each of the individual pilin genes was amplified by PCR and cloned into the broad-host-range vector pBBR1MCS in an orientation to allow expression from the lac promoter. When such vectors were mobilized into the corresponding mutant strains, the adherence phenotype was rescued and near-wild-type levels of adhesion were observed (P < 0.001) (Fig. 3).
Fig 3.
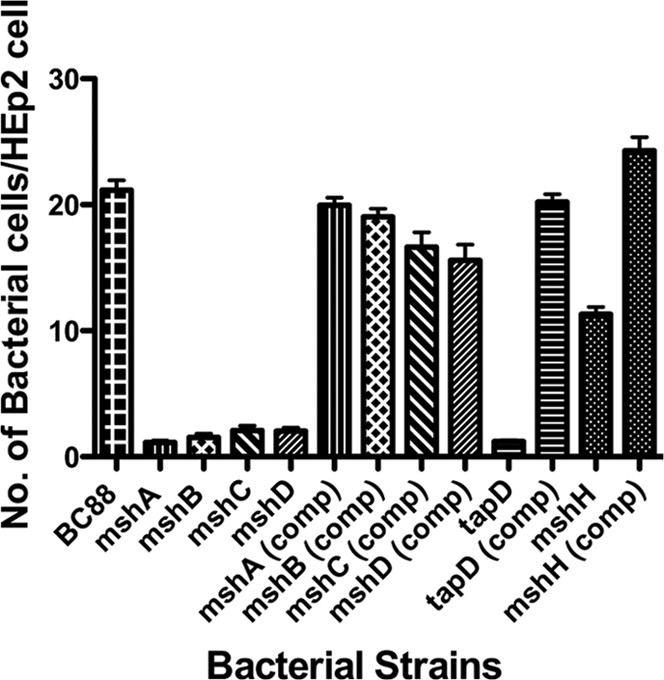
Adherence of A. veronii bv. Sobria strains to HEp-2 cells. Bacteria were grown statically in BHIB at 37°C, and adherence assays were carried out. Assays were carried out in triplicate on three separate occasions, and the mean number of adherent bacteria per HEp-2 cell was recorded. The error bars represent one standard deviation. Strains are indicated under the x axis, and significance of the mutants versus the wild-type and the complemented strains was by ANOVA (P < 0.001).
The A. veronii bv. Sobria pilin genes are essential for biofilm formation.
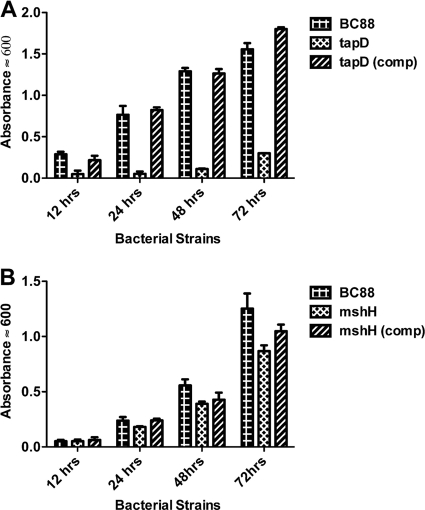
In previous studies investigating the role of A. caviae flagella in biofilm formation, borosilicate glass gave more consistent results (12). Here, we used the same assay to investigate the role of the pilin mutants in biofilm formation over time (Fig. 4). Mutation of any of the pilin genes resulted in a dramatic reduction in the ability to form a biofilm after 12 h of incubation (Fig. 4A). At this time point, biofilm absorbance values were 10 to 20% of the wild-type values. At 24 h, biofilm formation by the mutant strains was again approximately 10% of that of the wild type (Fig. 4B), and by 48 and 72 h the mutant biofilm was between 30 and 50% of the wild type (Fig. 4C and D). Reductions in biofilm formation in the mutants at all time points were significant (P < 0.001). Complementation with the individual wild-type pilin genes expressed in pBBR1MCS rescued the ability to form biofilms; this was especially obvious at 12 and 24 h (P < 0.001) (Fig. 4A and B).
Fig 4.
Biofilm development on borosilicate glass (37°C) over time by the A. veronii bv. Sobria wild-type, Msh-pilin mutant, and complemented strains. Values shown are the means of results from three replicate tubes ± standard deviations, and significance of the mutants versus the wild-type and complemented strains was by ANOVA (P < 0.001).
The TapD prepilin peptidase is essential for epithelial cell adherence and biofilm formation.
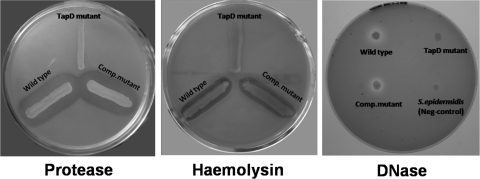
In each of the published aeromonad genome sequences, there is only one copy of the prepilin peptidase gene (tapD) that is located in the tapABCD locus that encodes part of another type IV pilus system (11). TapD is required for the processing and removal of the signal sequences from both pilins and pseudopilins. Therefore, TapD should be required not only for the assembly and functionality of the Tap type IV pilus but also for the MSHA (Bfp) type IV pilus and the type II secretion system (T2SS). Its mutation should also affect the adhesion of all A. veronii bv. Sobria type IV pilus systems. The tapABCD gene cluster has been previously isolated and sequenced from A. veronii bv. Sobria BC88 (11). Therefore, tapD was amplified by PCR and mutated using the method employed for the pilin genes, and its phenotype was investigated. To confirm the phenotype of the tapD mutant, its ability to secrete DNase, protease, and hemolysin was determined. As expected, the tapD mutant was negative for DNase production on DNase plates, negative for hemolysin production, and negative for protease production (Fig. 5). The wild-type copy of the tapD gene was amplified by PCR and cloned into the broad-host-range vector pBBR1MCS in an orientation to allow expression from the lac promoter. When this construct was mobilized into the corresponding tapD mutant strain for complementation analysis, the secreted enzyme production was restored (Fig. 5).
Fig 5.
Analysis of secreted enzyme production of A. veronii bv. Sobria wild-type, tapD mutant, and complemented tapD mutant strains. Bacterial strains were plated onto protease agar, blood agar, or DNase agar plates, and a positive reaction was assessed due to the zones of clearing around the areas of growth.
Based on the absence of another potential prepilin peptidase (26), we predicted that TapD would be required for the production of both the Msh and Tap type IV pilus systems, and its mutation resulted in a significant reduction of adherence (P < 0.001), with around 1 bacterium per tissue culture cell (Fig. 3). Mutation of TapD also had a great effect on biofilm formation, resulting in approximately 20% of that of the wild type (Fig. 6A). Complementation restored both adherence levels and biofilm formation to around those of the wild type (P < 0.001) (Fig. 3, Fig. 6A).
Fig 6.
Biofilm development on borosilicate glass (37°C) over time by the A. veronii bv. Sobria wild-type, the TapD mutant, and complemented strains (A) and the MshH mutant and complemented strains (B). Values shown are the means of results from three replicate tubes ± standard deviations, and significance of the mutants versus the wild-type and the complemented strains was by ANOVA (P < 0.001).
Role of MshH in epithelial cell adherence and biofilm formation.
Upstream of mshI is a gene, mshH, that is conserved in the MSHA loci of a number of bacteria, including Photobacterium, Shewanella spp., and Vibrio spp., and has been annotated as an MSHA biogenesis gene. However, Marsh and Taylor (17) and work in this report by RT-PCR showed that this gene was not coupled or transcribed with other genes in the msh locus. We therefore aimed to determine if mutation of mshH would result in a phenotype similar to those observed for the other msh genes with regard to adhesion and biofilm formation. The mshH mutant strain showed a 48% reduction in adhesion to HEp-2 cells compared to that of the wild-type strain, resulting in approximately 11 bacteria per HEp-2 cell versus over 21 bacteria per cell (P < 0.001) (Fig. 3). Complementation using a wild-type copy of mshH in pBBR1MCS rescued the adherence phenotype to wild-type levels (Fig. 3). However, mutation of mshH did not appear to have a great effect on biofilm formation, as the levels of the mutant were close to those of the wild type at all time points, and any large decreases that had been observed for the pilin or tapD mutants were absent (Fig. 6B).
Prevalence of the msh locus among the aeromonads.
Using a 1-kb PCR-generated probe that corresponded to the 3′ end of the mshB gene, the 5′ end of the mshC gene, and the complete mshA gene, we investigated the distribution of the msh locus among 31 aeromonads by Southern hybridization. The strains investigated included A. hydrophila, A. veronii bv. Sobria, A. caviae, A. salmonicida, and several type strains for the aeromonad hybridization groups. The probe bound to all strains tested except for the four strains of A. salmonicida (Table 4).
Table 4.
Sources of reference and test strains tested for hybridization with the 3′mshB-mshA-5′mshC probe
| Strain | Source | Aeruginosa species | Hybridization |
|---|---|---|---|
| BC88 | Stool | A. veronii bv. Sobria | + |
| ATCC 7966T | Canned milk | A. hydrophila (HG1) | + |
| CDC 9533-76 | Fish | A. bestiarum (HG2) | + |
| CDC 0862-83 | Freshwater | A. salmonicida (HG3) | − |
| ATCC 15468T | Guinea pig | A. caviae (HG4) | + |
| CDC 0862-83 | Fish | A. media (HG5A) | + |
| ATCC 23309 | Freshwater | A. eucrenophila (HG6) | + |
| CIP 7433T | Fish | A. sobria (HG7) | + |
| CDC 0437-80 | Stool | A. veronii bv. Sobria (HG8) | + |
| CDC 0787-80 | Stool | A. jandaei (HG9) | + |
| ATCC 35624T | Sputum | A. veronii bv. Veronii (HG10) | + |
| AH-1 | Fish | A. hydrophila | + |
| AH-3 | Fish | A. hydrophila | + |
| Sch13 | Stool | A. hydrophila | + |
| Sch27 | Stool | A. hydrophila | + |
| AH/V101 | Stool | A .hydrophila | + |
| BC96 | Stool | A. veronii bv. Sobria | + |
| FA132 | Stool | A. veronii bv. Sobria | + |
| BC96 | Stool | A. veronii bv. Sobria | + |
| CA110 | Stool | A. veronii bv. Sobria | + |
| Sch22 | Stool | A. veronii bv. Sobria | + |
| Sch39 | Stool | A. veronii bv. Sobria | + |
| F 1965 | Stool | A. veronii bv. Sobria | + |
| Sch3 | Stool | A. caviae | + |
| Sch18 | Stool | A. caviae | + |
| Sch29 | Stool | A. caviae | + |
| Sch46 | Stool | A. caviae | + |
| CA195 | Stool | A. caviae | + |
| NCMB 1102 | Fish | A. salmonicida | − |
| NB8601 | Fish | A. salmonicida | − |
| MT1326 | Fish | A. salmonicida | − |
DISCUSSION
Our previous work has demonstrated that some strains of Aeromonas species possessed at least two type IV pilus structures, the Tap and bundle-forming pilus (Bfp) (3). The published genome sequences of A. hydrophila and A. salmonicida confirmed this by showing the genes for three distinct type IV pilus systems, Tap, Msh (Bfp), and a third as-yet-uncharacterized system, Flp, present on the chromosome (3, 26). Previous studies have suggested that the bundle-forming pilus of A. veronii bv. Sobria is a major adherence factor for diarrheagenic strains (14). However, until now genetic studies on this system in Aeromonas species have not been undertaken. Here, we have isolated the gene cluster encoding the complete A. veronii bv. Sobria bundle-forming type IV pilus system. Homology searches demonstrated that the system is similar to the mannose-sensitive hemagglutinin (MSHA) of Vibrio cholerae El Tor (17), and hence the Aeromonas genes were also designated msh. The N-terminal amino acid sequence of the protein encoded by the aeromonad mshA gene matched that of the previously reported major pilin sequence for both A. veronii bv. Sobria BC88 and the N-terminal sequences of other pili isolated from Aeromonas strains, placing it in the classical type A type IV pilus group (13). The gene organization of the A. veronii bv. Sobria msh locus matched that previously reported for V. cholerae (17), with the exception that there are two mshI genes in the Aeromonas chromosome, mshI and mshI1, that encode proteins equivalent to the N-terminal and C-terminal domains of the V. cholerae MshI protein, respectively. Database searches show that this arrangement can also be found in A. hydrophila and A. salmonicida as well as Shewanella species, whereas the single MshI arrangement is prevalent in most Vibrio and Photobacterium species.
The major pilin gene mshA was found in a gene cluster along with three other minor pilin genes, mshB, mshC, and mshD, and RT-PCR and lacZ fusions showed that these were transcribed in a single transcriptional unit from a promoter located upstream of mshB. The second promoter of the msh gene cluster was shown to be upstream of mshI, and this promoter gave a higher level of activity than the mshB promoter, which is similar to the observations in V. cholerae with regard to activity and arrangement (17). As the major and minor pilins are produced from the same mRNA, this would suggest that rate of translational initiation must vary between the separate msh pilin genes to give rise to the different amounts of pilin proteins. As in previous studies, we have detected only the major pilin protein (MshA) from purified pilin filaments and not the other minor pilin proteins (13).
The major pilin gene was mutated, resulting in a 10-fold or more decrease in adherence to both HEp-2 cells and biofilm formation to borosilicate glass (at 12 h). Furthermore, other mutations in the minor pilin proteins also had a great effect on biotic and abiotic adherence, suggesting that all the pilin proteins are required for a functional pilus structure for optimal adherence and that the pilins are not able to compensate for the loss of one of the others. Mutation of the mshA or mshB gene resulted in the absence of MshA or MshB protein on Western blots, further supporting the argument that both the major and minor pilin proteins are required for adherence.
The marked effect seen by mutating the bundle-forming pilus genes in A. veronii bv. Sobria BC88 is supported by our previous work that demonstrated that using the purified pili could block adhesion to both HEp-2 and Henle 407 cells and that anti-Bfp (MshA) antibodies also blocked adhesion to cell lines (14). Mutation of tapD had a dramatic effect on adhesion that was equivalent to the reduction observed for the mutations of the pilin structural genes. Furthermore, in the TapD mutant, we were unable to detect the presence of either MshA or MshB on Western blots, demonstrating the importance of this protein in MSHA biogenesis. The published aeromonad genomes to date all show one copy of the prepilin peptidase required for both maturation of the type IV pilin proteins and the pseudopilins required for the type II secretion system (T2SS). Therefore, as expected, mutation of tapD had a pleiotropic effect by inhibiting the secretion of various enzymes through the T2SS and by effecting aeromonad adherence and biofilm formation. The latter observations are possibly due to effects on the maturation of the Msh pilins or the Flp or Tap pilins.
The mshH gene has been annotated as part of the MSHA pilus system for a number of bacterial species, including Photobacterium, Shewanella, and Vibrio spp. Here, we have shown that the gene is not transcriptionally linked to the other pilin genes. The gene encodes a protein that has GGDEF and EAL domains that are involved in di-guanylate cyclase synthase and phosphodiesterase activity, which are involved in making and breaking down c-di-GMP, respectively. The intracellular concentration of c-di-GMP modulates bacterial lifestyle choices from motile to sessile or vice versa and is important for adhesion and the formation of biofilms (7). Even though we see a slight decrease in the A. veronii bv. Sobria mshH mutant adhesion to HEp-2 cells, this is not supported by an inhibition in biofilm formation on borosilicate glass (Fig. 6), and this reduction is not like the greater than 80 to 90% drop in adhesion and biofilm formation that we observed for the other aeromonad MSHA mutants. Therefore, the mshH mutation is most likely having a more pleiotropic affect through the modulation of c-di-GMP concentrations, as reductions in c-di-GMP in another strain of A. veronii bv. Sobria have been shown to reduce adhesion and biofilm formation (23). This modulation could be affecting many other aeromonad factors associated with adhesion, including flagellar motility and outer membrane proteins (12, 22). Furthermore, the c-di-GMP network appears to be complex in the aeromonads, as the genome sequence of A. hydrophila demonstrated the presence of 13 proteins with GGDEF/EAL domains, 31 with only a GGDEF domain, and 9 with only an EAL domain (23, 26). Therefore, the mutation of MshH could possibly be compensated for by one of the many other GGDEF or EAL domain proteins.
Moreover, a homologue of mshH is found in enteric bacteria, such as Serratia, Enterobacter, and Yersinia, that do not possess an MSHA pilus system. Furthermore, the gene directly downstream of mshH in these bacteria is mreB, which is found downstream of the msh locus in Aeromonas, Photobacterium, Shewanella, and Vibrio spp. This suggests that the MSHA locus has been lost or deleted from this site in the enteric bacteria or that an ancestor of the closely related bacteria that possess this pilus system inserted it between mshH and mreB. A similar observation was also made by Marsh and Taylor, who suggested that the MSHA in Vibrio cholerae had been inserted into this particular locus due to the presence of a 7-bp direct repeat that flanks the locus (17). However, this evidence could further support the hypothesis that mshH is not a gene directly required for function of this type IV pilus.
Interestingly, the gene probe that encompassed part of mshB and all of mshA hybridized with all aeromonad mesophilic strains but no psychrophilic strains (A. salmonicida) tested, suggesting that the msh locus is present in all mesophilic strains but not in psychrophilic strains. This supports the evidence presented by Boyd et al. (3), who demonstrated that in A. salmonicida A449, there is a deletion in the msh locus between mshN and mshD, thus the area that the probe would have hybridized to has been deleted; this appears to be the case in other A. salmonicida strains also. The probe was able to demonstrate that the msh locus is present in mesophilic strains, whereas previously we were unable to do this, as the anti-Bfp antibody did not cross-react with 104 other strains of Aeromonas when tested (13), even though all previously isolated aeromonad long/wavy pili had very similar N-terminal amino acid sequences of the classical type A, type IV pilus group (13).
In conclusion, here we have undertaken the first genetic characterization of the mesophilic aeromonad bundle-forming pilus system, and through the use of defined mutants we have demonstrated the importance of this structure for adherence to tissue culture cells and biofilm formation. This supports our earlier findings that this structure is an important factor in aeromonad adherence.
Footnotes
Published ahead of print 6 February 2012
REFERENCES
- 1. Austin B, Adams C. 1996. Fish pathogens, p 197–243 In Austin B, et al. (ed), The genus Aeromonas. John Wiley and Sons, Chichester, West, Sussex, United Kingdom [Google Scholar]
- 2. Barnett TC, Kirov SM, Strom MS, Sanderson K. 1997. Aeromonas spp. possess at least two distinct type IV pilus families. Microb. Pathog. 23:241–247 [DOI] [PubMed] [Google Scholar]
- 3. Boyd JM, et al. 2008. Contribution of type IV pili to the virulence of Aeromonas salmonicida subsp. salmonicida in Atlantic salmon (Salmo salar L.). Infect. Immun. 76:1445–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gavin R, et al. 2002. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol. 43:383–397 [DOI] [PubMed] [Google Scholar]
- 5. Giron JA, Levine MM, Kaper JB. 1994. Longus: a long pilus ultrastructure produced by human enterotoxigenic Escherichia coli. Mol. Microbiol. 12:71–82 [DOI] [PubMed] [Google Scholar]
- 6. Gryllos I, Shaw JG, Gavin R, Merino S, Tomás JM. 2001. Role of flm locus in mesophilic Aeromonas adherence. Infect. Immun. 69:65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 8. Herrero M, de Lorenzo V, Timmis KN. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. J. Bacteriol. 172:6557–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Janda JM, Abbott SL. 2010. The genus Aeromonas: taxonomy, pathogenicity and infection. Clin. Microbiol. Rev. 23:35–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaniga K, Delor I, Cornelis GR. 1991. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137–141 [DOI] [PubMed] [Google Scholar]
- 11. Kirov SM, Barnett TC, Pepe CM, Strom MS, Albert MJ. 2000. Investigation of the role of type IV Aeromonas pilus (Tap) in the pathogenesis of Aeromonas gastrointestinal infection. Infect. Immun. 68:4040–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kirov SM, Castrisios M, Shaw JG. 2004. Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect. Immun. 72:1939–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kirov SM, Sanderson K. 1996. Characterization of a type IV bundle-forming pilus (SFP) from a gastroenteritis-associated strain of Aeromonas veronii biovar Sobria. Microb. Pathog. 21:23–34 [DOI] [PubMed] [Google Scholar]
- 14. Kirov SM, O'Donovan LA, Sanderson K. 1999. Functional characterization of type IV pili expressed on diarrhea-associated isolates of Aeromonas species. Infect. Immun. 67:5447–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kovach ME, Phillips RW, Elzer PH, Roop RM, II, Peterson KM. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800–802 [PubMed] [Google Scholar]
- 16. Li Y, Liu Y, Zhou Z, Huang H, Yren Zhang Y, Li G, Zhou Z, Wang L. 2011. Complete genome sequence of Aeromonas veronii strain B565. J. Bacteriol. 193:3389–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marsh JW, Taylor RK. 1999. Genetic and transcriptional analyses of the Vibrio cholerae mannose-sensitive hemagglutinin type 4 pilus gene locus. J. Bacteriol. 181:1110–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 19. Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parker JL, Shaw JG. 2011. Aeromonas spp. clinical microbiology and disease. J. Infection. 62:109–118 [DOI] [PubMed] [Google Scholar]
- 21. Pepe CM, Eklund MW, Strom MS. 1996. Cloning of an Aeromonas hydrophila type IV pilus biogenesis gene cluster: complementation of pilus assembly functions and characterization of a type IV leader peptidase/N-methyltransferase required for extracellular protein secretion. Mol. Microbiol. 194:857–869 [DOI] [PubMed] [Google Scholar]
- 22. Rabaan AA, Gryllos I, Tomas JM, Shaw JG. 2001. Motility and the polar flagellum are required for Aeromonas caviae adherence to HEp-2 cells. Infect. Immun. 69:4257–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rahman M, et al. 2007. The role of c-di-GMP signalling in an Aeromonas veronii biovar Sobria strain. FEMS Microbiol. Lett. 273:172–179 [DOI] [PubMed] [Google Scholar]
- 24. Reith ME, et al. 2008. The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics 9:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26. Seshadri R, et al. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 188:8272–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sharma DP, Stroeher UH, Thomas CJ, Manning PA, Attridge SR. 1989. The toxin-coregulated pilus (TCP) of Vibrio cholerae: molecular cloning of genes involved in pilus biosynthesis and evaluation of TCP as a protective antigen in the infant mouse model. Microb. Pathog. 7:437–448 [DOI] [PubMed] [Google Scholar]
- 28. Sokol PA, Ohman DE, Iglewski BH. 1979. A more sensitive plate assay for detection of protease production by Pseudomonas aeruginosa. J. Clin. Microbiol. 9:538–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suarez G, et al. 2008. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44:344–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tabei SMB, et al. 2009. An Aeromonas caviae genomic island is required for both O-antigen lipopolysaccharide biosynthesis and flagellin glycosylation. J. Bacteriol. 191:2851–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thornley JP, Shaw JG, Gryllos IA, Eley A. 1996. Adherence of Aeromonas caviae to human cell lines HEp-2 and Caco-2. J. Med. Microbiol. 45:445–451 [DOI] [PubMed] [Google Scholar]
- 32. Thornley JP, Shaw JG, Gryllos IA, Eley A. 1997. Virulence properties of clinically significant Aeromonas species: evidence for pathogenicity. Rev. Med. Microbiol. 8:61–72 [Google Scholar]
- 33. Vilches S, et al. 2004. Complete type III secretion system of a mesophilic Aeromonas hydrophila strain. Appl. Env. Microbiol. 70:6914–6919 [DOI] [PMC free article] [PubMed] [Google Scholar]