Abstract
The angiosperm genus Silene exhibits some of the most extreme and rapid divergence ever identified in mitochondrial genome architecture and nucleotide substitution rates. These patterns have been considered mitochondrial specific based on the absence of correlated changes in the small number of available nuclear and plastid gene sequences. To better assess the relationship between mitochondrial and plastid evolution, we sequenced the plastid genomes from four Silene species with fully sequenced mitochondrial genomes. We found that two species with fast-evolving mitochondrial genomes, S. noctiflora and S. conica, also exhibit accelerated rates of sequence and structural evolution in their plastid genomes. The nature of these changes, however, is markedly different from those in the mitochondrial genome. For example, in contrast to the mitochondrial pattern, which appears to be genome wide and mutationally driven, the plastid substitution rate accelerations are restricted to a subset of genes and preferentially affect nonsynonymous sites, indicating that altered selection pressures are acting on specific plastid-encoded functions in these species. Indeed, some plastid genes in S. noctiflora and S. conica show strong evidence of positive selection. In contrast, two species with more slowly evolving mitochondrial genomes, S. latifolia and S. vulgaris, have correspondingly low rates of nucleotide substitution in plastid genes as well as a plastid genome structure that has remained essentially unchanged since the origin of angiosperms. These results raise the possibility that common evolutionary forces could be shaping the extreme but distinct patterns of divergence in both organelle genomes within this genus.
Keywords: chloroplast, indels, inversions, organelle genome architecture, positive selection, substitution rate
Introduction
Plants and other photosynthetic eukaryotes share the distinction of having plastids, an endosymbiotically derived organelle that coexists with mitochondria in the cytoplasm (Gould et al. 2008; Kim and Archibald 2009). There are clear parallels in the long-term evolution of the genomes of these two organelles. For example, both have experienced massive gene loss (Adams and Palmer 2003; Timmis et al. 2004), which appears to be a universal pattern in obligately intracellular symbionts (Andersson and Kurland 1998; Moran and Wernegreen 2000). Organelle gene loss has generally been associated with transfer of genetic control to the nucleus, so most of the genes required for organelle function are located in the nuclear genome, including virtually all of those responsible for the maintenance of organellar DNA (Day and Madesis 2007; Sloan and Taylor 2012). Interestingly, many of the plant genes involved in DNA replication and repair in one organelle genome have related paralogs that function in the other (Zaegel et al. 2006; Shedge et al. 2007; Cappadocia et al. 2010). Moreover, the products of many nuclear genes are targeted to both organelles, including a disproportionate fraction of genes associated with DNA synthesis and processing (Carrie et al. 2009). Therefore, the evolution of DNA replication and repair machinery in organelles involves a complex history of gene transfer, co-option, duplication, retargeting, and replacement (reviewed in Sloan and Taylor 2012).
Despite sharing components of their DNA replication and repair machinery, mitochondrial and plastid genomes differ greatly in their structural organization and evolution. For example, seed plant plastid genomes are gene-dense and exhibit a high degree of syntenic conservation (Raubeson and Jansen 2005). In contrast, seed plant mitochondrial genomes contain an abundance of noncoding sequence and experience rapid rates of rearrangement among and even within species (Mower et al. forthcoming). Mitochondrial and plastid genomes also exhibit different rates of nucleotide substitution, which are believed to reflect underlying differences in mutation rate. Rates of synonymous substitutions are typically two to four times faster in plastid DNA than mitochondrial DNA in seed plants (Wolfe et al. 1987; Palmer and Herbon 1988; Drouin et al. 2008). However, a handful of seed plant lineages exhibit dramatic increases in mitochondrial substitution rates, reaching levels that are more typical of fast-evolving animal mitochondrial genomes (Cho et al. 2004; Parkinson et al. 2005; Bakker et al. 2006; Mower et al. 2007; Sloan et al. 2008, 2009; Ran et al. 2010).
Observed cases of rate acceleration in plant mitochondrial DNA are often characterized as mitochondrial-specific phenomena because sequenced nuclear and plastid genes show little or no correlated increase in substitution rate (e.g., Cho et al. 2004; Mower et al. 2007; Sloan et al. 2008). Nevertheless, limited evidence suggests that some of these dramatic changes in mitochondrial rate may not be entirely independent of evolution in the plastid genome. For example, the Geraniaceae, which has experienced a series of extreme changes in mitochondrial substitution rate (Parkinson et al. 2005), also exhibits abnormally high rates of structural evolution in the plastid genome and accelerated substitution rates in a subset of plastid genes (Chumley et al. 2006; Guisinger et al. 2008, 2011; Blazier et al. 2011). Likewise, gnetophytes exhibit elevated rates of evolution in both plastid and mitochondrial genomes (Mower et al. 2007; McCoy et al. 2008; Wu et al. 2009). However, comparisons based on complete genomes from both the mitochondria and the plastids are not available in these cases to assess the potential evolutionary parallels between the organelle genomes.
We recently reported the complete mitochondrial genome sequences of four Silene species with highly divergent mitochondrial substitution rates (Sloan et al. 2012). Two of these species (S. noctiflora and S. conica) exhibit nearly 100-fold increases in synonymous substitution rates, whereas the other two (S. latifolia and S. vulgaris) have low rates that are more typical of other angiosperm mitochondrial genomes. The accelerated rates in S. noctiflora and S. conica are associated with major expansions in mitochondrial genome size, resulting in the largest known mitochondrial genomes, as well as numerous other changes in mitochondrial gene and genome architecture.
To assess the relationship, if any, between mitochondrial and plastid genome evolution in Silene, we sequenced and analyzed the complete plastid genomes from these same four Silene species. The species with fast-evolving mitochondrial genomes (S. noctiflora and S. conica) do not show evidence of comparable genome-wide increases in plastid synonymous substitution rates. However, they do exhibit substantial rate accelerations in a subset of plastid genes, particularly at nonsynonymous sites, suggesting that altered selection pressures are acting on specific plastid pathways or functions in these species. In addition, the S. noctiflora and S. conica plastid genomes have experienced rapid structural evolution. In contrast, the S. latifolia and S. vulgaris plastid genomes are highly conserved relative to other angiosperms. These results provide an example of recent and correlated accelerations in mitochondrial and plastid genome evolution among closely related species, but the specific patterns of sequence and structural change differ between the two organelle genomes in many respects. We discuss the possibility that shared forces are acting, either directly or indirectly, on both mitochondrial and plastid genomes in Silene.
Materials and Methods
Source Material and Plastid DNA Extraction
For each of four Silene species (S. latifolia Poir., S. vulgaris [Moench] Garcke, S. noctiflora L., and S. conica L.), approximately 200 g of fresh tissue was collected from multiple individuals from a single maternal family. The maternal families and collection methods correspond to those previously described for mitochondrial genome sequencing (Sloan, Alverson, et al. 2010; Sloan et al. 2012). Intact chloroplasts were isolated using a combination of differential centrifugation and separation on a sucrose step gradient (Palmer 1986; Jansen et al. 2005). Chloroplasts were then lysed, and DNA was purified by phenol:chloroform extraction. These preparations yielded between 4 and 20 μg of DNA per species. The purity of plastid DNA was confirmed by restriction digestion.
Roche 454 and Illumina Sequencing
For each plastid DNA sample, shotgun libraries were constructed with multiplex identifier (MID) tags following standard protocols for sequencing on a Roche 454 GS-FLX platform with Titanium reagents. MID-tagged libraries were sequenced as part of a larger pooled sample with each of the four species constituting the equivalent of 2.5% of a full 454 plate. All 454 library construction and sequencing were performed at the Genomics Core Facility in the University of Virginia’s Department of Biology.
Multiplex barcoded libraries were also prepared for paired-end sequencing on an Illumina GAII sequencing platform as described previously (Sloan et al. 2012). For S. noctiflora, plastid DNA was amplified with GenomiPhi V2 (GE Healthcare, Piscataway, NJ) to produce sufficient starting material for Illumina library construction. All other Illumina libraries (and all 454 libraries) were generated without whole-genome amplification. The barcoded libraries were sequenced as part of a larger pooled sample in a single Illumina lane on a 2 × 85 bp paired-end run with each species representing 8% of the pool. Illumina sequencing was performed at the Biomolecular Research Facility in the University of Virginia’s School of Medicine.
Genome Assembly and Annotation
Shotgun 454 sequencing produced between 3.7 and 7.1 Mb of sequence data for each species. These reads were assembled with Roche’s GS de novo Assembler v2.3 (“Newbler”) using default settings. Initial assembly produced complete or nearly complete plastid genome sequences. Sequencing coverage in single-copy regions for each of the four species ranged from 21 to 46 ×. As expected, roughly twice those coverage levels were obtained for the inverted repeat (IR). The assemblies for each species contained as many as three gaps, but these generally reflected uncertainty regarding the length of long homopolymer (i.e., single-nucleotide repeat) regions. These regions were combined and then corrected with Illumina data (see below) to produce finished genomes.
Roche 454 data are known to have high insertion and deletion error rates associated with long homopolymer regions. To correct errors in the 454 assembly, paired-end Illumina reads were mapped to the genome using SOAP v2.20 (Li et al. 2009) as described previously (Sloan et al. 2012). After quality trimming and removal of multiplex barcode sequences, the Illumina run produced between 40 and 259 Mb of sequence with an average read length between 60 and 65 bp for each species. This data set provided deep coverage for the entirety of all four genomes with an average read depth between 297 and 1400×. The Illumina mapping results were used to identify and correct between 50 and 96 sequencing errors per genome, the vast majority of which were associated with homopolymer lengths.
Protein, transfer RNA (tRNA), and ribosomal RNA (rRNA) gene content in each of the finished genomes was annotated using DOGMA (Wyman et al. 2004). The annotated genome sequences were deposited in GenBank (accessions JF715054–JF715057).
Analysis of Genomic Inversions and Indels
To reconstruct the history of large inversions in Silene plastid genomes, gene order and orientations in each genome were compared with the inferred ancestral state for angiosperms (Raubeson and Jansen 2005) using GRIMM v2.0.1 (Tesler 2002). In addition, all four Silene plastid genomes were aligned with the outgroup Spinacia oleracea using MAUVE v2.3.1 (Darling et al. 2010).
To identify and quantify the number of indels in each plastid genome, syntenic blocks of sequence for all four Silene species and the outgroup Spinacia oleracea were aligned using MUSCLE v3.7 (Edgar 2004). Intergenic regions containing inversion breakpoints were not included in this analysis. Large indels (>100 bp) were identified by manual inspection of the sequence alignments. In many cases, the size, number, and polarity of smaller indel events were ambiguous because multiple indels often overlapped in structurally variable regions. Therefore, to estimate the relative frequency of smaller indels (<100 bp) in each species, we restricted our focus to the subset of events that are unique to a single species within the aligned data set and show no overlap with structural variants in the other four species. A custom Perl script was used to identify all indels meeting these criteria.
Phylogenetic Analysis and Substitution Rate Variation
To assess the phylogenetic relationships among the four Silene species, nucleotide sequences from all Silene protein genes and introns were aligned with the corresponding sequences from the closest available outgroup, Spinacia oleracea, as well as Arabidopsis thaliana (for protein-coding sequences only). Alignments were performed using MUSCLE v3.7 (Edgar 2004) and adjusted manually. Phylogenetic analyses were performed with RAxML v7.0.4 on three different concatenated data sets: 1) all protein genes except accD, clpP, ycf1, and ycf2 (which were excluded because of extreme sequence and/or structural divergence in S. noctiflora and S. conica), 2) all protein genes in the photosynthesis-related atp, pet, ndh, psa, and psb complexes, and 3) all introns. RAxML analyses were performed with the following parameters: -f d, -b 1, -p 1, -#1,000, and -m GTRGAMMA.
The relative rates of sequence divergence in the four Silene genomes (and the outgroups Spinacia and Arabidopsis) were analyzed using both codon- and nucleotide-based models of evolution in PAML v 4.4 (Yang 2007) as described previously (Sloan et al. 2009; Sloan, Alverson, et al. 2010). Because the phylogenetic relationships among the four Silene species are not confidently resolved, all PAML analyses implemented a constrained topology with the four Silene species radiating from a single polytomy. Protein-coding sequences were analyzed with codon-based models to quantify the rates of synonymous and nonsynonymous substitution, whereas RNA genes and intronic sequences were analyzed with nucleotide-based models. Analyses were performed on the following data sets: 1) a concatenation of all protein genes except accD (see below); 2) separate concatenations of each of the following protein gene sets: atp, pet, ndh, psa, psb, rpl, rpo, and rps; 3) each of the following individual protein genes: ccsA, cemA, clpP, matK, rbcL, ycf1, ycf2, ycf3, and ycf4; 4) a concatenation of all rRNA genes; and 5) a concatenation of all introns. The accD gene is too structurally divergent in S. noctiflora and S. conica to produce a useful alignment that includes both species. However, a large portion of accD from each of these species can be separately aligned against the remaining species in the analysis. Therefore, two separate analyses of accD sequence divergence were performed, one involving S. noctiflora and one involving S. conica.
To test for evidence of positive selection acting on individual genes or sets of genes, all loci with estimated dN/dS ratios greater than one in any Silene species were reanalyzed with the dN/dS ratio constrained to a value of one for that species. Likelihood ratio tests were performed to compare the constrained and unconstrained analyses and determine whether the estimated dN/dS ratios significantly exceed one (Yang 1998). Because we performed a total of 72 rate analyses in Silene protein genes (18 genes or gene sets for each of four Silene species), we applied a Bonferroni correction factor of 72 to account for multiple comparisons.
RNA Editing
In land plants, mitochondrial and plastid messenger RNA transcripts undergo systematic conversion of cytidines to uridines (C-to-U editing), restoring conserved codons (Knoop 2011). RNA editing sites were previously identified by cDNA sequencing for a subset of plastid genes in all four of the Silene species analyzed in this study (Sloan, MacQueen, et al. 2010). To predict editing sites in other plastid genes, we aligned Silene genes against all protein-coding sequences from A. thaliana, Nicotiana tabacum, and Zea mays that are known to undergo RNA editing. Editing data for these three species were obtained from REDIdb (Picardi et al. 2007) and other published sources (Tillich et al. 2005; Chateigner-Boutin and Small 2007). Any site that is edited in one or more of these outgroups was predicted to be edited in Silene species that have a C at the corresponding genomic position (supplementary table S1, Supplementary Material online). A number of editing sites appear to have been lost in one or more Silene species as a result of C-to-T substitutions at the genomic level. For any site that was predicted to vary in its editing status among the four Silene species, cDNA sequencing was performed as described previously (Sloan, MacQueen, et al. 2010) to confirm editing in at least one species. The results of cDNA sequencing confirmed editing in all cases except for rps14 (nucleotide position 80). This site was predicted to be edited in S. latifolia, S. vulgaris, and S. conica but to have been lost by a genomic C-to-T substitution in S. noctiflora. However, cDNA sequencing in S. latifolia found no evidence of editing. Therefore, this site was excluded from the counts shown in table 1 and supplementary table S1 (Supplementary Material online).
Table 1.
Summary of Silene Plastid Genomes
| S. latifolia | S. vulgaris | S. noctiflora | S. conica | |
| Genome size (bp) | 151,736 | 151,583 | 151,639 | 147,208 |
| IR | 25,906 | 26,008 | 29,891 | 26,858 |
| Large single-copy region | 82,704 | 82,258 | 79,475 | 80,129 |
| Small single-copy region | 17,220 | 17,309 | 12,382 | 13,363 |
| G + C content (%) | 36.43 | 36.25 | 36.51 | 36.12 |
| Protein genesa | 77 | 77 | 77 | 77 |
| tRNA genesa | 30 | 30 | 30 | 30 |
| rRNA genesa | 4 | 4 | 4 | 4 |
| Intronsa | 20 | 20 | 16 | 16 |
| RNA editing sitesb | 25 | 26 | 24 | 24 |
Gene and intron counts exclude putative pseudogenes and duplicate copies in the IR.
Editing site counts are from supplementary table S1 (Supplementary Material online) and include predicted sites that have not been confirmed by cDNA sequencing (see Materials and Methods).
Results
Gene Content in Silene Plastid Genomes
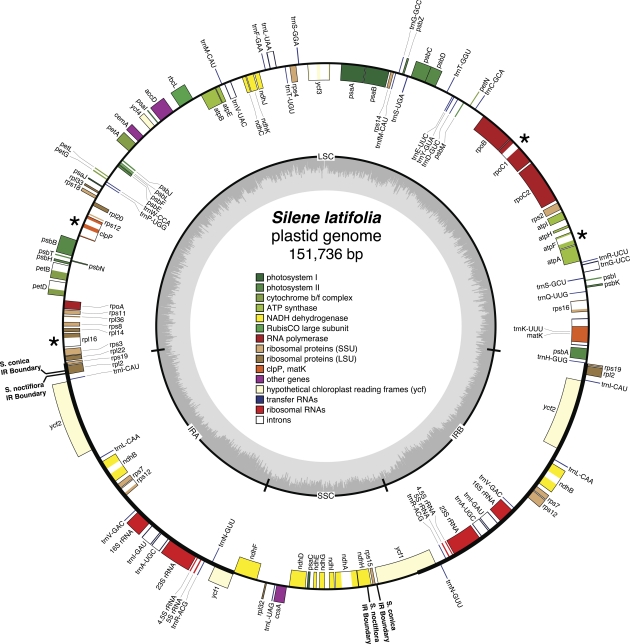
All four Silene plastid genomes are typical in size relative to other angiosperms and exhibit a classic circular genome map with a pair of large IRs separating two single-copy regions (fig. 1, supplementary fig. S1, Supplementary Material online and table 1). The four genomes share a gene complement that encodes 77 proteins, 30 tRNAs, and 4 rRNAs. Genes coding for the translation initiation factor A (infA) and the ribosomal protein subunit L23 (rpl23) appear to be present only as pseudogenes in the genomes of all four species and are not included in the totals above. These two genes have been lost independently in multiple angiosperm lineages, including other species within the Caryophyllales (Zurawski and Clegg 1987; Millen et al. 2001; Funk et al. 2007; Logacheva et al. 2008). The infA gene has been subject to repeated functional transfers to the nucleus (Millen et al. 2001), whereas there is evidence that rpl23 has been functionally replaced by its cytosolic counterpart in other species (Bubunenko et al. 1994). In addition to the functional loss of infA and rpl23, it is possible that some annotated genes in the Silene plastid genomes are pseudogenes. For example, as reported previously (Sloan et al. 2009), the intron-encoded open reading frame (ORF) matK contains an internal frameshift indel in S. conica. The gene encoding the RNA polymerase α subunit (rpoA) also contains a frameshift indel approximately 200 bp upstream of the normal stop codon position in three of the four Silene species. Silene latifolia and S. noctiflora share a 7 bp deletion in rpoA, whereas S. vulgaris has a 10 bp deletion in the same region. In both matK and rpoA, frameshift indels have occurred in homopolymer regions and have introduced premature stop codons. Finally, multiple genes are highly divergent in sequence and/or structure in S. conica and S. noctiflora (see below) and could be pseudogenes. Most notably, the entire 3′ half of the accD gene is missing in S. noctiflora.
FIG. 1.—
Plastid genome map for Silene latifolia. Boxes inside and outside the circle correspond to genes on the clockwise and anticlockwise strand, respectively. The inner circle depicts GC content. The positions of the IR are labeled on the inner circle and noted with thicker black lines on the outer circle. All differences >100 bp in IR boundary positions among the four sequenced Silene plastid genomes are labeled on the outer circle. Asterisks indicate genes that have lost introns in S. noctiflora and/or S. conica. Maps of all four Silene plastid genomes are provided as supplementary material (supplementary fig. S1, Supplementary Material online). This figure was generated with OGDraw v1.2 (Lohse et al. 2007).
Rapid Structural Evolution in the Plastid Genomes of S. noctiflora and S. conica
The S. latifolia and S. vulgaris plastid genomes show nearly perfect syntenic conservation with Spinacia (fig. 2) and other angiosperms including Amborella trichopoda (data not shown), suggesting that these two Silene species have maintained the ancestral angiosperm genomic structure (Raubeson and Jansen 2005). In contrast, the two species with fast-evolving mitochondrial genomes (S. noctiflora and S. conica) have experienced numerous changes in plastid genome structure in just the few million years since the divergence of these four Silene species. These changes include multiple inversions, intron losses, large indels, and shifts in the IR boundaries.
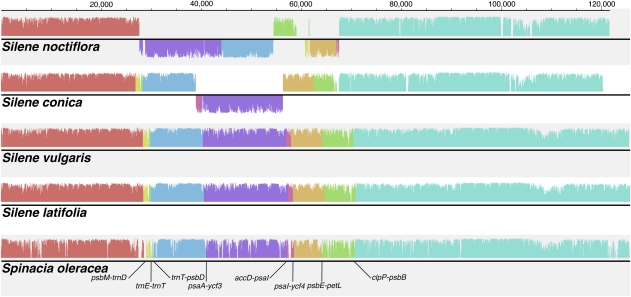
FIG. 2.—
Structural alignments of Silene and Spinacia plastid genomes. The coloring identifies collinear sequence blocks shared by all five genomes. Bars drawn below the black line indicate sequences found in inverted orientation. The height of each bar reflects sequence similarity. The eight inversion breakpoints identified by GRIMM are labeled at the bottom. Only one copy of the IR is shown for each genome, and the orientation of the small single-copy region was reversed relative to its conventional presentation to minimize complexities associated with changes in the IR boundaries. This figure was generated with MAUVE v2.3.1 (Darling et al. 2010).
The rearranged gene order in the S. noctiflora plastid genome suggests that it has experienced four inversions involving six breakpoints found within or between the following gene pairs: psbM-trnD, accD-psaI, psbB-clpP, petL-psbE, psbD-trnT, trnT-trnE (fig. 2). The S. conica genome appears to have experienced a single inversion with a pair of breakpoints (psaA-ycf3 and psaI-ycf4) that are distinct from any of those involved in the S. noctiflora rearrangements (fig. 2). At least some of the inversions are likely the result of recombination between short IRs, as has been observed in other angiosperms (Knox et al. 1993; Haberle et al. 2008). All four Silene species have a pair of divergent IRs (ca. 170 bp and 80% sequence identity) that coincide with the breakpoints for the S. conica inversion. Outside of Silene, this sequence is widely conserved in seed plant plastid genomes but found only as a single copy. In addition, S. noctiflora has a unique pair of IRs (154 bp, 99% sequence identity) corresponding to the petL-psbE and psbD-trnT breakpoints. However, any repeats that may have been associated with other inversion events in S. noctiflora are not readily identifiable. Interestingly, the S. conica inversion interrupts a genomic fragment that was previously sequenced and analyzed in a number of species within the Sileneae including S. conica (Erixon and Oxelman 2008a). This earlier study did not detect the inversion found in our analysis of the S. conica plastid genome, which could indicate that it is polymorphic within the species. However, the 2008 study was based on polymerase chain reaction (PCR) and Sanger sequencing of individual fragments, so artifacts involving PCR-mediated recombination (Alverson et al. 2011) might also explain the earlier finding of a noninverted genome conformation in S. conica.
The S. latifolia and S. vulgaris plastid genomes share an identical complement of 19 group II introns (including the trans-splicing first intron of rps12; Koller et al. 1987) as well as a single group I intron in the trnL-UAA gene. The S. noctiflora and S. conica genomes each lack four of these introns. Both S. noctiflora and S. conica have lost the rpoC1 intron as well as both introns in the fast-evolving clpP gene (Erixon and Oxelman 2008b). In addition, S. noctiflora has lost the rpl16 intron and S. conica has lost the atpF intron. All five of these introns have been lost independently in other angiosperm lineages (Downie et al. 1996; Campagna and Downie 1998; Jansen et al. 2007). Like other members of the core Caryophyllales, all four Silene species lack the rpl2 intron found in other plant lineages (Downie et al. 1991; Logacheva et al. 2008). In every case, missing introns have been precisely excised at their normal splicing boundaries.
In addition to the deletions associated with intron loss, the S. noctiflora and S. conica plastid genomes have also experienced a total of 10 and 11 large indels of >100 bp in size, respectively. Ten of these indels were found in protein-coding sequences (all in the highly divergent accD, ycf1, and ycf2 genes), whereas the remaining 11 are in noncoding sequences (10 in intergenic regions and one in the trnL-UAA intron). In contrast, no indel greater than 100 bp was found in either S. latifolia or S. vulgaris. The S. noctiflora and S. conica plastid genomes also have a higher frequency of small indels. Alignments of all four Silene species with the outgroup Spinacia oleracea identified a total of 18, 46, 107, and 151 unique nonoverlapping indels of <100 bp in size for S. latifolia, S. vulgaris, S. conica, and S. noctiflora, respectively.
The structures of the S. noctiflora and S. conica plastid genomes have also been altered by shifts in the boundaries between their IRs and single-copy regions. Although the precise boundaries of the IR in angiosperms are subject to frequent shifts (Goulding et al. 1996), they are generally found within the rps19 and ycf1 genes. These boundary positions appear to be the ancestral state for most angiosperm lineages, including the genus Silene based on their presence in both S. latifolia and S. vulgaris (fig. 1, supplementary fig. S1, Supplementary Material online) as well as in Spinacia oleracea, the closest outgroup with a sequenced plastid genome (Schmitz-Linneweber et al. 2001). Silene latifolia and S. vulgaris share identical boundary positions between the IR and the large single-copy region and differ only slightly (<100 bp) in the positions of their boundaries between the IR and the small single-copy region. In contrast, the IR in S. noctiflora and S. conica has contracted at the boundary with the large single-copy region and expanded at the boundary with the small single-copy region (fig. 1). As a result, the IR in S. noctiflora and S. conica does not contain any portion of rps19 and lacks a substantial fraction of rpl2. In addition, the IR now includes the entirety of ycf1 in both species as well as rps15 and a portion of ndhH in S. noctiflora. Interestingly, Fagopyrum esculentum, another member of the Caryophyllales with a fully sequenced plastid genome, also has an expanded IR that contains a full copy of ycf1 and which is shared with related species in the Polygonaceae and Plumbaginaceae (Logacheva et al. 2008, 2009). However, the absence of this expansion in other families within the noncore Caryophyllales (Logacheva et al. 2009) suggests that it evolved multiple times independently, including at least once within the genus Silene and another time in a common ancestor of the Polygonaceae and Plumbaginaceae.
Although the directions of the inferred IR boundary shifts are the same in both S. noctiflora and S. conica, the magnitudes of the expansions and contractions differ (fig. 1). Differences in IR boundary positions account for 0.7 kb of the observed 3 kb difference in IR length between S. noctiflora and S. conica (table 1). The remaining 2.3 kb difference in IR length between these species is the result of indels within the IR, particularly in the coding sequences of ycf1 and ycf2.
Elevated and Variable Substitution Rates in the Plastid Genomes of S. noctiflora and S. conica
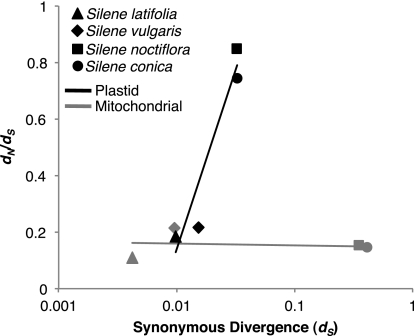
Silene noctiflora and S. conica exhibit increased substitution rates in plastid genes. However, the observed rate accelerations differ in two important respects relative to the dramatic rate increases in the mitochondrial genomes of these species. First, the elevated plastid rates are primarily driven by a disproportionate increase in the frequency of nonsynonymous substitutions (dN) with only a modest 2- or 3-fold change in synonymous divergence (dS). In contrast, mitochondrial dN and dS values have each increased by nearly two orders of magnitude in these species, resulting in virtually no change in the dN/dS ratio (fig. 3). Second, whereas the mitochondrial rate accelerations in S. noctiflora and S. conica appear to be genome-wide phenomena (Sloan et al. 2012), plastid rates differ substantially among genes (fig. 4 and supplementary fig. S2, Supplementary Material online).
FIG. 3.—
Synonymous (dS) and nonsynonymous divergence (dN) in Silene mitochondrial and plastid genomes as estimated with PAML. Plastid data are based on an analysis of all protein genes except accD. Mitochondrial data are from table 1 in Sloan et al. (2012). The solid lines represent best fit trend lines for the plastid (black) and mitochondrial (gray) data.
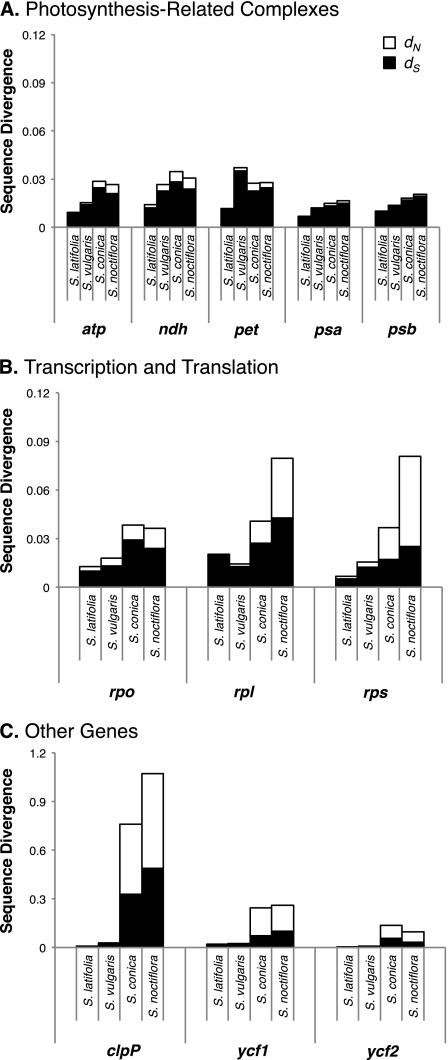
FIG. 4.—
Sequence divergence in Silene plastid genes as measured by the estimated number of substitutions per site in the terminal branch for each species. The black and white portions of each bar represent the amount of synonymous and nonsynonymous divergence, respectively. (A) Genes in major photosynthesis-related complexes. (B) Genes coding for RNA polymerase subunits and ribosomal proteins. (C) Three other highly divergent genes (note the 10-fold change in scale). Additional plots are available as supplementary material (supplementary fig. S2, Supplementary Material online) for individual protein genes, rRNA genes, and introns.
Plastid genes in five major complexes associated with photosynthesis show little rate increase in S. noctiflora and S. conica (fig. 4A), whereas informational protein genes including RNA polymerase subunits (to some extent) and ribosomal proteins show larger increases (fig. 4B). Other plastid genes have experienced even greater rate changes, including the large ORFs ycf1 and ycf2, which are known to be essential for cell survival but are otherwise uncharacterized (Drescher et al. 2000), as well as the protease subunit clpP (fig. 4C), which was previously found to have highly accelerated substitution rates in multiple species within the tribe Sileneae, including S. conica (Erixon and Oxelman 2008b). The accD gene, which is required for fatty acid biosynthesis, shows some evidence of substitution rate acceleration (supplementary fig. S2, Supplementary Material online) and has also undergone rapid structural evolution, including large deletions in both S. noctiflora and S. conica.
Phylogenetic Analysis of Silene Plastid DNA
An analysis of multiple concatenated data sets did not provide a clear consensus on the phylogenetic relationships among the four Silene species in this study (supplementary fig. S3, Supplementary Material online). A concatenated data set of all plastid protein genes except accD, clpP, ycf1, and ycf2 supported a sister relationship between the fast-evolving S. noctiflora and S. conica lineages (supplementary fig. S3A, Supplementary Material online). However, this support disappeared when the analysis was restricted to photosynthesis-related genes (supplementary fig. S3B, Supplementary Material online), which do not exhibit a history of major rate accelerations in S. noctiflora and S. conica (fig. 4A). These genes supported a sister relationship between S. latifolia and S. conica (supplementary fig. S3B, Supplementary Material online). Analysis of shared intron sequences provided weak support for yet another topology with S. latifolia sister to S. noctiflora (supplementary fig. S3C, Supplementary Material online). In all three analyses, internal branch lengths were very short, indicating a rapid radiation of these four Silene lineages.
Discussion
Recent and Correlated Changes in Mitochondrial and Plastid Genome Evolution
Sequencing of the S. noctiflora and S. conica mitochondrial genomes revealed that they are exceptional, even when compared with the already complex mitochondrial genomes of most flowering plants. These mitochondrial genomes exhibit extreme changes in genome size, structure, and rate of sequence evolution (Sloan et al. 2012). In this study, we have shown that the plastid genomes in these species have also experienced recent and rapid divergence that distinguishes them from the plastid genomes of most angiosperms, including other members of the same genus. Although comparisons of complete mitochondrial and plastid genome sequences have not been performed in other angiosperm species with accelerated mitochondrial substitution rates, there is some evidence to suggest that similar correlated increases in the rate of sequence and/or structural evolution in both organelle genomes have occurred in lineages such as the Geraniaceae and gnetophytes (Parkinson et al. 2005; Chumley et al. 2006; Mower et al. 2007; Guisinger et al. 2008, 2011; McCoy et al. 2008; Wu et al. 2009; Blazier et al. 2011).
Although these cases constitute relatively few independent data points, they nevertheless raise the possibility of a shared mechanism affecting both organelle genomes. In angiosperms, the mapping and sequencing of plastid genomes have far outpaced progress on mitochondrial genomes. As a result, there are numerous angiosperm lineages, such as the Campanulaceae (including the Lobeliaceae), Fabaceae, Goodeniaceae, Oleaceae, Passifloraceae, and Ranunculaceae, that have been identified as having accelerated and/or rearranged plastid genomes, but for which we have little or no mitochondrial data (Jansen et al. 2007, 2008). Many of these lineages contain plastid genomes that are far more divergent and rearranged than those found in Silene and, therefore, represent a natural starting point for generating additional mitochondrial genome sequences. It is unlikely that there is any simple or absolute relationship between the patterns of evolution in mitochondrial and plastid genomes. For example, note that some of the most divergent plastid genomes in the Geraniaceae (Blazier et al. 2011; Guisinger et al. 2011) occur in genera with only moderately accelerated mitochondrial substitution rates (Parkinson et al. 2005). Nevertheless, a more comprehensive comparison of organelle genomes across angiosperms may help identify mechanisms that jointly affect mitochondrial and plastid genome evolution.
The idea that rates of sequence evolution might be correlated between mitochondrial and plastid genomes is not new. In fact, there are many factors expected to affect rates and patterns of evolution at an organismal level (Ohta 1992; Whittle and Johnston 2002; Smith and Donoghue 2008). Therefore, one of the intriguing elements of this study is not necessarily that the mitochondrial and plastid genomes are both highly divergent in S. noctiflora and S. conica but that they have diverged in such different ways (table 2). Our findings raise the question of what evolutionary mechanisms could generate these correlated, yet distinct patterns of divergence between the mitochondrial and the plastid genomes. There are many potential answers to this question (including simple coincidence), but one intriguing possibility involves modification of nuclear genes encoding dual-targeted protein products. For example, homologs of the bacterial recA gene are known to play an important role in plant organelle genome stability, and the Arabidopsis genome contains three characterized recA homologs with one targeted to plastids, one to mitochondria, and one to both organelles (Shedge et al. 2007; Rowan et al. 2010). Modification of the dual-targeted gene RECA2 (Shedge et al. 2007) could affect the evolution of both genomes but in potentially different ways given the possibility that the gene product serves different functional roles in the two organelles or maintains different levels of redundancy with other members of the gene family.
Table 2.
Contrasting Patterns of Mitochondrial versus Plastid Genome Divergence in Silene noctiflora and Silene conica
| Mitochondrial | Plastid | |
| Sequence | ||
| Major genome-wide increase in synonymous substitution rate | Yes | No |
| Large increases in dN/dS in a subset of protein genes | No | Yes |
| Large decrease in the frequency of RNA editing | Yes | No |
| Structural | ||
| Genomic expansion | Yes | No |
| Multichromosomal genome structure | Yes | No |
| Gene duplication | Yes | No |
| Elevated indel rate | Yes | Yes |
| Intron losses | Only one | Yes |
| Inversions | N/Aa | Yes |
| Shifts in IR boundaries | N/A | Yes |
Rates of genome rearrangement between and even within species are so high in angiosperm mitochondrial genomes that estimating the number or rate of inversions in any given lineage is not feasible.
The discovery and history of the bacterial mutS homolog MSH1 may also be informative with respect to correlated patterns of evolution between mitochondrial and plastid genomes. This nuclear locus was originally named CHM (for chloroplast mutator) because mutants exhibited a variegated leaf phenotype and modifications in plastid morphology that could subsequently be inherited maternally (Redei 1973). Therefore, it was predicted that disruptions of this nuclear gene destabilize the plastid genome. Subsequent work, however, found that the MSH1 gene product is targeted to mitochondria, where it regulates recombinational activity and genome reorganization, and a direct role of MSH1 in plastid genome stability became uncertain (Martinez-Zapater et al. 1992; Abdelnoor et al. 2003; Shedge et al. 2007; Arrieta-Montiel et al. 2009). The documented effects of MSH1/CHM on plastids may be partially mediated through indirect physiological pathways linking these two organelles. Because mitochondria and plastids maintain a high degree of functional interdependence, (Roussell et al. 1991; Woodson and Chory 2008; Yoshida and Noguchi 2011), it is possible that perturbation of one organelle genome will have evolutionary consequences for the other. More recent findings indicate that MSH1 is also targeted to plastids and may play a direct role in plastid genome stability as originally predicted (Xu et al. 2011). Therefore, this dual-targeted gene highlights the potential for both direct and indirect mechanisms to link the evolution of mitochondrial and plastid genomes. MSH1, RECA, and other gene families known to be involved in plant organelle genome stability (e.g., Zaegel et al. 2006; Cappadocia et al. 2010) represent important candidates for further investigation in Silene.
Causes of Substitution Rate Variation Among Plastid Genes
The pattern of mitochondrial substitution rate acceleration in S. noctiflora and S. conica has been attributed to genome-wide increases in the mutation rate (Mower et al. 2007; Sloan et al. 2009, 2012). However, a similar interpretation is inconsistent with the observed substitution patterns in the plastid genomes. The magnitude of rate accelerations in S. noctiflora and S. conica vary markedly across plastid genes. Some of this variation might be explained by “localized hypermutation” as has been proposed in cases of gene-specific rate accelerations in both plastid (Magee et al. 2010) and mitochondrial (Sloan et al. 2009) genomes. However, even a model with a diverse range of localized gene-specific mutation rates could not explain the disproportional increases in dN found in many plastid genes in S. noctiflora and S. conica. Instead, the observed increases in dN/dS suggest a history of relaxed purifying selection and/or increased positive selection acting on a subset of plastid genes in S. noctiflora and S. conica. Increased rates of sequence and structural evolution might be associated with the process of functional gene transfer to the nucleus (Magee et al. 2010). In addition, some loci exhibit dN/dS ratios that are significantly greater than one when averaged across the entire length of the gene (table 3), strongly suggesting at least some role for positive selection in the rate accelerations observed in these species.
Table 3.
Positive Selection on Silene Plastid Genes
| Gene/Complex | S. noctiflora | S. conica |
| accD | 2.20 | 0.98 |
| cemA | 1.21 | 1.48 |
| clpP | 1.19 | 1.31 |
| rps (concatenated) | 2.23 (0.002) | 1.17 |
| ycf1 | 1.6* | 2.33 (6 × 10−6) |
| ycf2 | 1.87 (0.02) | 1.39* |
Note.—All genes (or sets of concatenated genes belonging to a single complex) with estimated dN/dS values greater than 1 are shown. Estimates that are significantly greater than 1 are shown in bold with Bonferroni-corrected P values in parentheses.
Values that are significant based on an uncorrected P value of 0.05 but not after Bonferroni correction are marked with an asterisk.
The differences in substitution rate and dN/dS across functional classes of plastid genes (fig. 4, supplementary fig. S2, Supplementary Material online) suggest that changes in selection pressure may be associated with specific biochemical pathways or functions rather than the entire genome. Interestingly, the patterns of rate variation among genes in S. noctiflora and S. conica exhibit some clear parallels with the evolution of plastid genomes within the Geraniaceae, which have experienced a longer and more extreme history of genome rearrangement (Chumley et al. 2006; Guisinger et al. 2008, 2011; Blazier et al. 2011). For example, both lineages show a high degree of sequence conservation in genes directly involved in photosynthesis and greater levels of divergence in other genes such as ribosomal proteins. Furthermore, the most divergent genes in S. noctiflora and S. conica, in particular, accD, clpP, ycf1, and ycf2, have been completely lost from the plastid genome in multiple lineages within the Geraniaceae (Guisinger et al. 2008, 2011). The parallels are not perfect, however. Some of the highest levels of divergence in the Geraniaceae are found in genes coding for RNA polymerase subunits (Guisinger et al. 2008), which show only modest accelerations in S. noctiflora and S. conica (fig. 4). In addition, one clade within the Geraniaceae appears to have lost all functional copies of its ndh genes (Blazier et al. 2011), but these genes are highly conserved in Silene.
The evolution of plastid genomes in nonphotosynthetic angiosperms provides some insight into the patterns of selection acting on Silene plastid genes. Not surprisingly, evolution of a nonphotosynthetic lifestyle is generally associated with plastid genome reduction and gene loss (Wolfe et al. 1992; Delannoy et al. 2011). Nevertheless, nonphotosynthetic angiosperms retain a plastid genome, demonstrating that the functional importance of plastids extends beyond photosynthetic pathways (Barbrook et al. 2006; Benning et al. 2006). Most of the genes retained in the plastid genomes of nonphotosynthetic plants are required for plastid gene expression. For example, of the 42 functional genes identified in the highly reduced plastid genome of the parasitic eudicot Epifagus virginiana, only 4 (accD, clpP, ycf1, and ycf2) are not involved in plastid gene expression (Wolfe et al. 1992). The nonphotosynthetic orchid Rizanthella gardneri, which has the smallest sequenced plastid genome of any land plant, has independently converged on a remarkably similar set of genes (including accD, clpP, ycf1, and ycf2) (Delannoy et al. 2011).
Strikingly, these are the same four genes that exhibit the greatest accelerations in the rate of sequence and/or structural evolution in S. noctiflora and S. conica, suggesting that there have been significant changes in selection pressures acting on nonphotosynthetic pathways in plastids in both Silene species. Although these four genes are widely retained in land plants (Delannoy et al. 2011), each has been lost from the plastid genome of some lineages, including multiple angiosperms (Katayama and Ogihara 1996; Knox and Palmer 1999; Chumley et al. 2006; Haberle et al. 2008; Guisinger et al. 2011). There is also evidence for positive selection acting on these genes in other land plant lineages (Erixon and Oxelman 2008b; Greiner et al. 2008). Knockout experiments in tobacco have shown that all four are essential (Drescher et al. 2000; Shikanai et al. 2001; Kuroda and Maliga 2003; Kode et al. 2005). The protein encoded by clpP is a component of a complex multimeric protease with broad substrate specificity within the plastid (Peltier et al. 2004; Stanne et al. 2009), whereas accD codes for a subunit of the acetyl-CoA carboxylase, which is involved in fatty acid biosynthesis (Kode et al. 2005). Despite the essential nature of ycf1 and ycf2 (Drescher et al. 2000), the functions of these genes have not yet been identified. Silene and other lineages with a history of extreme divergence in accD, clpP, ycf1, and ycf2 may provide an opportunity to better understand the more general role of these genes and their related pathways in plants.
Single or Multiple Origins of Accelerated Organelle Genome Evolution in Silene?
The clear similarities between S. noctiflora and S. conica in the evolution of both mitochondrial and plastid genomes raise the obvious question of whether these lineages form a monophyletic group that experienced shared ancestral changes associated with the organelle genomes. Although it is tempting to assume that commonalities between these species (e.g., shared intron losses in clpP and rpoC1) reflect common ancestry, phylogenetic analyses in other angiosperms have shown that such patterns can occur in parallel across independent evolutionary lineages (e.g., Guisinger et al. 2011). Therefore, an independent phylogenetic estimate of the relationships between these Silene lineages is necessary. However, such analyses have generally failed to resolve relationships among the major lineages of Silene subgenus Behenantha, which includes the species examined in this study (Erixon and Oxelman 2008a; Sloan et al. 2009). Our analyses based on data from complete plastid genomes produced similar ambiguities (supplementary fig. S3, Supplementary Material online). Likewise, an analysis of a small number of nuclear genes across this genus found that some loci support monophyly between the S. noctiflora and the S. conica lineages, whereas others do not (Rautenberg et al. forthcoming). Therefore, the question of single versus multiple origins of accelerated organelle genome evolution in Silene remains unresolved. Efforts are underway to produce deep transcriptome sequencing coverage of multiple Silene species. The resulting data set should help disentangle the phylogenetic relationships within Silene as well as elucidate the cytonuclear interactions that have shaped the extreme patterns of organelle genome evolution in this genus.
Supplementary Material
Supplementary figures S1–S3 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Dave McCauley for providing S. vulgaris tissue for DNA extraction and John Chuckalovcak for his 454 sequencing efforts. This work was supported by the National Science Foundation (DEB-0808452, MCB-1022128) and the Jefferson Scholars Foundation.
References
- Abdelnoor RV, et al. Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc Natl Acad Sci U S A. 2003;100:5968–5973. doi: 10.1073/pnas.1037651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams KL, Palmer JD. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol. 2003;29:380–395. doi: 10.1016/s1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- Alverson AJ, Zhuo S, Rice DW, Sloan DB, Palmer JD. The mitochondrial genome of the legume Vigna radiata and the analysis of recombination across short mitochondrial repeats. PLoS One. 2011;6:e16404. doi: 10.1371/journal.pone.0016404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- Arrieta-Montiel MP, Shedge V, Davila J, Christensen AC, Mackenzie SA. Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics. 2009;183:1261–1268. doi: 10.1534/genetics.109.108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker FT, Breman F, Merckx V. DNA sequence evolution in fast evolving mitochondrial DNA nad1 exons in Geraniaceae and Plantaginaceae. Taxon. 2006;55:887–896. [Google Scholar]
- Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006;11:101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Benning C, Xu C, Awai K. Non-vesicular and vesicular lipid trafficking involving plastids. Curr Opin Plant Biol. 2006;9:241–247. doi: 10.1016/j.pbi.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Blazier JC, Guisinger MM, Jansen RK. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae) Plant Mol Biol. 2011;76:263–272. doi: 10.1007/s11103-011-9753-5. [DOI] [PubMed] [Google Scholar]
- Bubunenko MG, Schmidt J, Subramanian AR. Protein substitution in chloroplast ribosome evolution. A eukaryotic cytosolic protein has replaced its organelle homologue (L23) in spinach. J Mol Biol. 1994;240:28–41. doi: 10.1006/jmbi.1994.1415. [DOI] [PubMed] [Google Scholar]
- Campagna ML, Downie SR. The intron in chloroplast gene rpl16 is missing from the flowering plant families Geraniaceae, Goodeniaceae and Plumbaginaceae. Trans Illin Acad Sci. 1998;91:1–11. [Google Scholar]
- Cappadocia L, et al. Crystal structures of DNA-whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell. 2010;22:1849–1867. doi: 10.1105/tpc.109.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Giraud E, Whelan J. Protein transport in organelles: dual targeting of proteins to mitochondria and chloroplasts. FEBS J. 2009;276:1187–1195. doi: 10.1111/j.1742-4658.2009.06876.x. [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Small I. A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res. 2007;35:e114. doi: 10.1093/nar/gkm640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Mower JP, Qiu YL, Palmer JD. Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc Natl Acad Sci U S A. 2004;101:17741–17746. doi: 10.1073/pnas.0408302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley TW, et al. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 2006;23:2175. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A, Madesis P. DNA replication, recombination, and repair in plastids. In: Bock R, editor. Cell and molecular biology of plastids. Berlin (Germany): Springer; 2007. pp. 65–119. [Google Scholar]
- Delannoy E, Fujii S, des Francs CC, Brundrett M, Small I. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol. 2011;28:2077–2086. doi: 10.1093/molbev/msr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie SR, Llanas E, Katz-Downie DS. Multiple independent losses of the rpoC1 intron in angiosperm chloroplast DNA's. Syst Bot. 1996;21:135–151. [Google Scholar]
- Downie SR, et al. Six independent losses of the chloroplast DNA rpl2 intron in dicotyledons: molecular and phylogenetic implications. Evolution. 1991;45:1245–1259. doi: 10.1111/j.1558-5646.1991.tb04390.x. [DOI] [PubMed] [Google Scholar]
- Drescher A, Ruf S, Calsa T, Jr, Carrer H, Bock R. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 2000;22:97–104. doi: 10.1046/j.1365-313x.2000.00722.x. [DOI] [PubMed] [Google Scholar]
- Drouin G, Daoud H, Xia J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol Phylogenet Evol. 2008;49:827–831. doi: 10.1016/j.ympev.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erixon P, Oxelman B. Reticulate or tree-like chloroplast DNA evolution in Sileneae (Caryophyllaceae)? Mol Phylogenet Evol. 2008a;48:313–325. doi: 10.1016/j.ympev.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Erixon P, Oxelman B. Whole-gene positive selection, elevated synonymous substitution rates, duplication, and indel evolution of the chloroplast clpP1 gene. PLoS One. 2008b;3:e1386. doi: 10.1371/journal.pone.0001386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk HT, Berg S, Krupinska K, Maier UG, Krause K. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 2007;7:45. doi: 10.1186/1471-2229-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Goulding SE, Olmstead RG, Morden CW, Wolfe KH. Ebb and flow of the chloroplast inverted repeat. Mol Gen Genet. 1996;252:195–206. doi: 10.1007/BF02173220. [DOI] [PubMed] [Google Scholar]
- Greiner S, et al. The complete nucleotide sequences of the 5 genetically distinct plastid genomes of Oenothera, subsection Oenothera: II. A microevolutionary view using bioinformatics and formal genetic data. Mol Biol Evol. 2008;25:2019–2030. doi: 10.1093/molbev/msn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Genome-wide analyses of geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc Natl Acad Sci U S A. 2008;105:18424–18429. doi: 10.1073/pnas.0806759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 2011;28:583–600. doi: 10.1093/molbev/msq229. [DOI] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol. 2008;66:350–361. doi: 10.1007/s00239-008-9086-4. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, Daniell H. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 2007;104:19369. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 2005;395:348–384. doi: 10.1016/S0076-6879(05)95020-9. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Wojciechowski MF, Sanniyasi E, Lee SB, Daniell H. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae) Mol Phylogenet Evol. 2008;48:1204–1217. doi: 10.1016/j.ympev.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H, Ogihara Y. Phylogenetic affinities of the grasses to other monocots as revealed by molecular analysis of chloroplast DNA. Curr Genet. 1996;29:572–581. doi: 10.1007/BF02426962. [DOI] [PubMed] [Google Scholar]
- Kim E, Archibald J. Diversity and evolution of plastids and their genomes. In: Aronsson H, Sandelius AS, editors. The chloroplast—interactions with the environment. Berlin (Germany): Springer-Verlag; 2009. pp. 1–39. [Google Scholar]
- Knoop V. When you can’t trust the DNA: RNA editing changes transcript sequences. Cell Mol Life Sci. 2011;68:567–586. doi: 10.1007/s00018-010-0538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox EB, Downie SR, Palmer JD. Chloroplast genome rearrangements and the evolution of giant lobelias from herbaceous ancestors. Mol Biol Evol. 1993;10:414–430. [Google Scholar]
- Knox EB, Palmer JD. The chloroplast genome arrangement of Lobelia thuliniana (Lobeliaceae): expansion of the inverted repeat in an ancestor of the Campanulales. Plant Syst Evol. 1999;214:49–64. [Google Scholar]
- Kode V, Mudd EA, Iamtham S, Day A. The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 2005;44:237–244. doi: 10.1111/j.1365-313X.2005.02533.x. [DOI] [PubMed] [Google Scholar]
- Koller B, Fromm H, Galun E, Edelman M. Evidence for in vivo trans splicing of pre-mRNAs in tobacco chloroplasts. Cell. 1987;48:111–119. doi: 10.1016/0092-8674(87)90361-8. [DOI] [PubMed] [Google Scholar]
- Kuroda H, Maliga P. The plastid clpP1 protease gene is essential for plant development. Nature. 2003;425:86–89. doi: 10.1038/nature01909. [DOI] [PubMed] [Google Scholar]
- Li R, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Logacheva MD, Penin AA, Valiejo-Roman CM, Antonov AS. Structure and evolution of junctions between inverted repeat and small single copy regions of chloroplast genome in non-core Caryophyllales. Mol Biol. 2009;43:757–765. [PubMed] [Google Scholar]
- Logacheva MD, Samigullin TH, Dhingra A, Penin AA. Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. ancestrale–a wild ancestor of cultivated buckwheat. BMC Plant Biol. 2008;8:59. doi: 10.1186/1471-2229-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- Magee AM, et al. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 2010;20:1700–1710. doi: 10.1101/gr.111955.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Zapater JM, Gil P, Capel J, Somerville CR. Mutations at the Arabidopsis CHM locus promote rearrangements of the mitochondrial genome. Plant Cell. 1992;4:889–899. doi: 10.1105/tpc.4.8.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy SR, Kuehl JV, Boore JL, Raubeson LA. The complete plastid genome sequence of Welwitschia mirabilis: an unusually compact plastome with accelerated divergence rates. BMC Evol Biol. 2008;8:130. doi: 10.1186/1471-2148-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen RS, et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13:645–658. doi: 10.1105/tpc.13.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Wernegreen JJ. Lifestyle evolution in symbiotic bacteria: insights from genomics. Trends Ecol Evol. 2000;15:321–326. doi: 10.1016/s0169-5347(00)01902-9. [DOI] [PubMed] [Google Scholar]
- Mower JP, Sloan DB, Alverson AJ. Plant mitochondrial diversity—the genomics revolution. In: Wendel JF, editor. Plant genome diversity. Berlin (Germany): Springer-Verlag; Forthcoming. [Google Scholar]
- Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD. Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol. 2007;7:135. doi: 10.1186/1471-2148-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol Syst. 1992;23:263–286. [Google Scholar]
- Palmer JD. Isolation and structural analysis of chloroplast DNA. Meth Enzymol. 1986;118:167–186. [Google Scholar]
- Palmer JD, Herbon LA. Plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence. J Mol Evol. 1988;28:87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- Parkinson CL, et al. Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae. BMC Evol Biol. 2005;5:73. doi: 10.1186/1471-2148-5-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, et al. Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J Biol Chem. 2004;279:4768–4781. doi: 10.1074/jbc.M309212200. [DOI] [PubMed] [Google Scholar]
- Picardi E, Regina TM, Brennicke A, Quagliariello C. REDIdb: the RNA editing database. Nucleic Acids Res. 2007;35:D173–D177. doi: 10.1093/nar/gkl793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran JH, Gao H, Wang XQ. Fast evolution of the retroprocessed mitochondrial rps3 gene in conifer II and further evidence for the phylogeny of gymnosperms. Mol Phylogenet Evol. 2010;54:136–149. doi: 10.1016/j.ympev.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Raubeson LA, Jansen RK. Chloroplast genomes of plants. In: Henry RJ, editor. Plant diversity and evolution: genotypic and phenotypic variation in higher plants. Wallingford (UK): CABI; 2005. pp. 45–68. [Google Scholar]
- Rautenberg A, Sloan DB, Aldén V, Oxelman B. Phylogenetic relationships of Silene multinervia (Caryophyllaceae) and Silene section Conoimorpha. Syst Bot. Forthcoming [Google Scholar]
- Redei GP. Extra-chromosomal mutability determined by a nuclear gene locus in Arabidopsis. Mutat Res. 1973;18:149–162. [Google Scholar]
- Roussell DL, Thompson DL, Pallardy SG, Miles D, Newton KJ. Chloroplast structure and function is altered in the NCS2 maize mitochondrial mutant. Plant Physiol. 1991;96:232–238. doi: 10.1104/pp.96.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan BA, Oldenburg DJ, Bendich AJ. RecA maintains the integrity of chloroplast DNA molecules in Arabidopsis. J Exp Bot. 2010;61:2575–2588. doi: 10.1093/jxb/erq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, et al. The plastid chromosome of spinach (Spinacia oleracea): complete nucleotide sequence and gene organization. Plant Mol Biol. 2001;45:307–315. doi: 10.1023/a:1006478403810. [DOI] [PubMed] [Google Scholar]
- Shedge V, Arrieta-Montiel M, Christensen AC, Mackenzie SA. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell. 2007;19:1251–1264. doi: 10.1105/tpc.106.048355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T, et al. The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 2001;42:264–273. doi: 10.1093/pcp/pce031. [DOI] [PubMed] [Google Scholar]
- Sloan DB, et al. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10:e1001241. doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Alverson AJ, Storchova H, Palmer JD, Taylor DR. Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia. BMC Evol Biol. 2010;10:274. doi: 10.1186/1471-2148-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Barr CM, Olson MS, Keller SR, Taylor DR. Evolutionary rate variation at multiple levels of biological organization in plant mitochondrial DNA. Mol Biol Evol. 2008;25:243–246. doi: 10.1093/molbev/msm266. [DOI] [PubMed] [Google Scholar]
- Sloan DB, MacQueen AH, Alverson AJ, Palmer JD, Taylor DR. Extensive loss of RNA editing sites in rapidly evolving Silene mitochondrial genomes: selection vs. retroprocessing as the driving force. Genetics. 2010;185:1369–1380. doi: 10.1534/genetics.110.118000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Oxelman B, Rautenberg A, Taylor DR. Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae (Caryophyllaceae) BMC Evol Biol. 2009;9:260. doi: 10.1186/1471-2148-9-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Taylor DR. Evolutionary rate variation in organelle genomes: the role of mutational processes. In: Bullerwell CE, editor. Organelle genetics. Heidelberg (Germany): Springer-Verlag; 2012. pp. 123–146. [Google Scholar]
- Smith SA, Donoghue MJ. Rates of molecular evolution are linked to life history in flowering plants. Science. 2008;322:86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- Stanne TM, Sjogren LL, Koussevitzky S, Clarke AK. Identification of new protein substrates for the chloroplast ATP-dependent clp protease supports its constitutive role in Arabidopsis. Biochem J. 2009;417:257–268. doi: 10.1042/BJ20081146. [DOI] [PubMed] [Google Scholar]
- Tesler G. GRIMM: genome rearrangements web server. Bioinformatics. 2002;18:492–493. doi: 10.1093/bioinformatics/18.3.492. [DOI] [PubMed] [Google Scholar]
- Tillich M, et al. Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J. 2005;43:708–715. doi: 10.1111/j.1365-313X.2005.02484.x. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Whittle CA, Johnston MO. Male-driven evolution of mitochondrial and chloroplastidial DNA sequences in plants. Mol Biol Evol. 2002;19:938–949. doi: 10.1093/oxfordjournals.molbev.a004151. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Ems SC, Palmer JD. Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: loss or accelerated sequence evolution of tRNA and ribosomal protein genes. J Mol Evol. 1992;35:304–317. doi: 10.1007/BF00161168. [DOI] [PubMed] [Google Scholar]
- Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet. 2008;9:383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Lai YT, Lin CP, Wang YN, Chaw SM. Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: selection toward a lower-cost strategy. Mol Phylogenet Evol. 2009;52:115–124. doi: 10.1016/j.ympev.2008.12.026. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Xu YZ, et al. MutS HOMOLOG1 is a nucleoid protein that alters mitochondrial and plastid properties and plant response to high light. Plant Cell. 2011;23:3428–3441. doi: 10.1105/tpc.111.089136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Noguchi K. Interaction between chloroplasts and mitochondria: activity, function, and regulation of the mitochondrial respiratory system during photosynthesis. In: Kempken F, editor. Plant mitochondria. New York: Springer; 2011. pp. 383–409. [Google Scholar]
- Zaegel V, et al. The plant-specific ssDNA binding protein OSB1 is involved in the stoichiometric transmission of mitochondrial DNA in Arabidopsis. Plant Cell. 2006;18:3548–3563. doi: 10.1105/tpc.106.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G, Clegg MT. Evolution of higher-plant chloroplast DNA-encoded genes: implications for structure-function and phylogenetic studies. Annu Rev Plant Phys. 1987;38:391–418. [Google Scholar]