Abstract
Small RNAs (sRNAs) are widespread in bacteria and play critical roles in regulating physiological processes. They are best characterized in Escherichia coli K-12 MG1655, where 83 sRNAs constitute nearly 2% of the gene complement. Most sRNAs act by base pairing with a target mRNA, modulating its translation and/or stability; many of these RNAs share only limited complementarity to their mRNA target, and require the chaperone Hfq to facilitate base pairing. Little is known about the evolutionary dynamics of bacterial sRNAs. Here, we apply phylogenetic and network analyses to investigate the evolutionary processes and principles that govern sRNA gene distribution in 27 E. coli and Shigella genomes. We identify core (encoded in all 27 genomes) and variable sRNAs; more than two-thirds of the E. coli K-12 MG1655 sRNAs are core, whereas the others show patterns of presence and absence that are principally due to genetic loss, not duplication or lateral genetic transfer. We present evidence that variable sRNAs are less tightly integrated into cellular genetic regulatory networks than are the core sRNAs, and that Hfq facilitates posttranscriptional cross talk between the E. coli–Shigella core and variable genomes. Finally, we present evidence that more than 80% of genes targeted by Hfq-associated core sRNAs have been transferred within the E. coli–Shigella clade, and that most of these genes have been transferred intact. These results suggest that Hfq and sRNAs help integrate laterally acquired genes into established regulatory networks.
Keywords: lateral genetic transfer, horizontal genetic transfer, sRNAs, regulatory networks, genome evolution, genetic loss
Introduction
A substantial fraction of the bacterial transcriptome is composed of small RNAs (sRNAs). In Escherichia coli, 83 sRNA transcripts represent nearly 2% of the genes verified in the species. sRNAs are widespread and are critical regulators of disparate physiological processes (Waters and Storz 2009; Gottesman and Storz 2010). Examples of sRNA-mediated regulation broadly emphasize the diverse roles that sRNAs play in posttranscriptional regulation of the interactions that underpin cellular adaptation, including transposition (Simons and Kleckner 1983), quorum sensing (Lenz et al. 2004), toxin–antitoxin systems (Fozo et al. 2008), plasmid replication (MacLellan et al. 2005), bacterial virulence (Lenz et al. 2004), and responses to environment (Wassarman 2002).
Although some characterized sRNAs bind to proteins and change their function (Babitzke and Romeo 2007), most sRNAs act through base pairing with target mRNAs, thereby modulating their translation and/or stability. mRNA-targeting sRNAs can be grouped into two broad classes, cis- and trans-encoded. The cis-encoded sRNAs are encoded at the same chromosomal locus as their regulated mRNA but on the opposite strand and thus necessarily share extensive complementarity with the corresponding transcript. In contrast, trans-encoded sRNAs are positioned at a distinct genetic location and generally share more limited complementarity with their targets (Storz et al. 2005). In many instances of trans-encoded sRNA regulation, the sRNA chaperone Hfq is required to facilitate base pairing between discontinuous stretches of limited complementarity between the sRNA and its target (Brennan and Link 2007).
Identification of sRNA targets and the regulators that control sRNA expression is a continuing challenge; however, recent efforts toward delineating complete maps of the interactions that exist among sRNAs, their targets, and associated transcriptional regulators have yielded preliminary sets of interaction partners. From these, sRNA-mediated regulation can be generalized as a network in which molecules (genes, proteins, and RNAs) are represented as nodes and regulatory interactions as edges. Within global regulatory networks, sRNAs participate in specific regulatory circuits—positive and negative feedback loops, feed-forward loops, single-input modules, and dense overlapping regulons—that together comprise networks of global regulation (Beisel and Storz 2010).
Despite growing appreciation of the functions and regulatory processes in which sRNAs participate, little is known of how sRNAs evolve within bacterial lineages (Waters and Storz 2009; Gottesman and Storz 2010). In many bacteria, strains within a species share a set of core genes but can differ substantially in their repertoire of variable genes, the presence or the absence of which is predominantly due to gene genesis, lateral genetic transfer (LGT), and genetic loss (Lerat et al. 2003). To what extent have these evolutionary processes shaped the sRNA content of extant genomes? The experimental validation of numerous sRNAs in E. coli K-12 MG1655 and the availability of complete genome sequences from multiple diverse strains of the E. coli–Shigella clade have now opened these issues to comparative analysis, although estimates of the variable sRNA content will necessarily be limited by which strains have been sequenced and the lack of characterization of sRNAs in strains other than E. coli K-12. Here, we address sRNA evolution within a framework that considers the evolutionary constraints potentially imposed by the molecular interactions that govern the participation of sRNAs in cellular regulatory networks. For the first time, we assign 83 sRNAs in E. coli K-12 MG1655 to the core or variable genomes of the E. coli–Shigella clade and address 1) patterns of conservation of sRNAs in this clade; 2) the relative contribution of LGT and genetic loss to sRNA phyletic distribution; 3) network properties of core and variable sRNAs; 4) the roles of Hfq and sRNAs in mediating posttranscriptional cross talk between the core and variable genomes; and 5) the role of Hfq-associated sRNAs in the regulation of genes that have been laterally transferred within the E. coli–Shigella lineage.
Materials and Methods
E. coli K-12 sRNAs
About 80 sRNA transcripts have been experimentally validated in E. coli. Applying a deep sequencing approach, Raghavan et al. (2011) recently detected and quantified the vast majority of these previously validated sRNAs. To obtain an up-to-date list of E. coli K-12 sRNAs, we merged the list of 80 known E. coli sRNAs provided by Raghavan et al. (2011) with a list of 79 E. coli K-12 sRNAs extracted from the sRNA database sRNAmap (Huang et al. 2009). Overall, 73 sRNAs were present in both the list provided by Raghavan et al. and sRNAmap, seven were unique to the Raghavan et al. (2011) list and six were unique to sRNAmap. We decided to include the 13 sRNAs that were not present in both the Raghavan et al. list and sRNAmap in our final analysis on the basis that they were each present in at least one of three additional E. coli databases: EcoCyc (Keseler et al. 2011), EcoGene (Rudd 2000), and RegulonDB (Gama-Castro et al. 2011). We excluded two mRNAs (ryfB and isrB) from our final list because, although they were initially identified as sRNAs, they have since been found to encode small proteins. We also removed psrN, which was initially reported to be a small RNA but has since been found to be the 5′ untranslated region of the alx gene (Nechooshtan et al. 2009). Thus, 83 known E. coli K-12 sRNAs went forward to phyletic distribution analysis.
Ongoing characterization of direct sRNA targets has started to reveal the mechanisms by which sRNAs enact regulation. Targets for numerous E. coli sRNAs have so far been identified and subsequently collected by RegulonDB and sRNAmap. Using these data, we separated the E. coli K-12 sRNAs with known targets into three main groups: Hfq-associated trans-encoded sRNAs, cis-encoded sRNAs, and protein-binding sRNAs (supplementary table S1, Supplementary Material online). We note that all characterized E. coli trans-encoded sRNAs require Hfq for regulation of their targets (Waters and Storz 2009).
In addition to detecting previously experimentally verified E. coli K-12 sRNAs, Raghavan et al. (2011) also detected 53 previously predicted sRNAs as well as 10 new sRNAs within the strain using RNA-seq. These 63 sRNAs also went forward to phyletic distribution analysis but were examined separately from the 83 known sRNAs as they are of unknown function and potential targets have not been confirmed. We used coordinates for these sRNAs as provided by Raghavan et al. (2011).
Presence and Absence of sRNAs across 27 E. coli and Shigella Strains
We investigated both syntenic and sequence-similarity based approaches to determine the presence or the absence of the E. coli K-12 MG1655 known sRNAs across 27 strains of E. coli and Shigella. Previous analyses have used sequence similarity alone to examine phyletic distributions of sRNAs (Hershberg et al. 2003). For this reason, we first investigated an approach based solely on sequence similarity. The complete genome sequences of 27 E. coli and Shigella genomes were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/) (supplementary table S2, Supplementary Material online) and the E. coli K-12 sRNA nucleotide sequences were extracted from the NC_000913.2 genome assembly. Each of the 83 known sRNAs was then compared using BLAST+ and legacy BLAST (Altschul et al. 1990, 1997) to the 27 complete genome sequences. A given sRNA was determined to be present in an E. coli or Shigella target genome if BLAST searches yielded one or more alignments satisfying both E value < 0.001 and target length ≥ 0.7 × query length. BLAST+ and legacy BLAST gave the same phyletic distributions across the 27 E. coli and Shigella strains for 71 of the 83 sRNAs examined. Given that BLAST+ has performance improvements over legacy BLAST in particular, yielding significant sRNA-genome alignments that were not found using legacy BLAST we decided to use the sRNA phyletic distributions determined using BLAST+ for the next stage of our analysis.
There are ways to investigate sRNA conservation across genomes other than by sequence similarity alone, for example, using synteny information. Mauve (Darling et al. 2004, 2010) is a sequence-similarity–based alignment tool, which incorporates synteny information to construct whole-genome alignments. Within a Mauve genome alignment, positional homology is implied among aligned regions of the genomes (Darling et al. 2010). The alignment can therefore be used to identify regions homologous to E. coli K-12 sRNAs in the other 26 E. coli and Shigella genomes. Whole-genome alignment of the 27 E. coli and Shigella strains was produced using the progressiveMauve program included in MAUVE version 2.3.0, with default parameter values (Darling et al. 2004). A custom python script was then used to identify regions aligned to the query sRNAs in the E. coli and Shigella genomes and thereby extract their phyletic distributions across the E. coli–Shigella clade.
For 52 (63%) of the 83 query sRNAs, both Mauve and BLAST+ yield the same phyletic distributions. This provides high confidence in the patterns of presence and absence that we have determined for these sRNAs. We examined the BLAST+ alignments for the 31 sRNAs for which BLAST+ and Mauve yield different phyletic distributions and found that 16 (52%) of these sRNAs yielded more than one significant BLAST alignment in a minimum of one of the target genomes. There are sRNAs included within this subset that represent highly repetitive elements.
Mauve uses an anchor-based strategy for genome alignment. Problems are known to arise if a particular subsequence occurs numerous times in each genome because it becomes unclear which combination of regions to align (Darling et al. 2004). This can result in unaligned repetitive regions or anomalous alignments. Comparing the disagreeing phyletic distributions, we find cases where Mauve implies an sRNA is absent from a particular genome where BLAST+ has found a perfect match. For these reasons, all sRNA phyletic distributions reported in the results were determined using BLAST+. The only exceptions are the phyletic distributions reported for OmrA and OmrB, for which we report phyletic distributions as determined using legacy BLAST. These sRNAs are known to have arisen through an ancestral duplication and share high sequence similarity (Argaman et al. 2001). At the thresholds specified, BLAST+ yields spurious alignments of OmrB to genomic regions encoding OmrA in a subset of genomes.
To further increase confidence in the sRNA phyletic distributions as determined by BLAST+, we examined all genomic regions yielding significant BLAST alignments to query sRNAs to determine if they overlap with protein-coding sequence, since we know that many of the query sRNAs are encoded in the intergenic regions of genomes. No unexpected alignments between protein-coding regions and query sRNAs were identified. In the case of cis-acting sRNAs, query sequences were expected to yield significant BLAST alignments to protein-coding regions. For example, IstR-1 and IstR-2 are encoded at the same locus as a toxic polypeptide (Vogel et al. 2004).
Finally, we investigated the effect of using different BLAST+ thresholds for determining the presence or the absence of an sRNA in a given genome. We repeated the analysis using different values for the minimum BLAST+ target length required to predict presence in a given genome and found that changing this threshold to values between 0.6 × query length and 0.8 × query length did not significantly affect our results. At 0.8 × query length, we find the pattern of presence and absence across the 27 genomes changes for only two (C0343 and ISI128) of the 83 sRNAs examined, and at a threshold of 0.6 × query length, the pattern changes for only one sRNA (sokB). Therefore, changing thresholds for the minimum target length required to predict the presence of an sRNA to values between 0.6 × query length and 0.8 × query length does not affect the main conclusions of the paper.
Using the sequence-similarity–based BLAST approach described above, we also determined the presence or the absence of the 83 known E. coli K-12 sRNAs in the recently determined genome of E. coli O104:H4 strain TY-2482 (E value < 0.001 and target length ≥ 0.7 × query length). The genome sequence of E. coli O104:H4 strain TY-2482 was obtained from the FTP site: ftp://ftp.genomics.org.cn/pub/Ecoli_TY-2482/ (11/11/2011).
In addition, we inferred phyletic distributions for the 53 previously predicted sRNAs and 10 new sRNAs reported by Raghavan et al. (2011). The phyletic distributions for these sRNAs were determined using BLAST+ (E value < 0.001 and target length ≥ 0.7 × query length).
Inference of Gene Families
Next, we used Mauve (Darling et al. 2004, 2010) to infer sets of putatively orthologous genes among the 27 E. coli and Shigella strains. A whole-genome alignment of 31 E. coli and Shigella strains, including the 27 complete E. coli and Shigella genomes and four additional draft genomes (E. coli 101-1, E. coli F11, E. coli O157H7 str EC440, and Shigella sp D9), was performed using the progressiveMauve program included in MAUVE version 2.3.0, with default parameter values (Darling et al. 2004). A list of all positionally homologous protein-coding gene sets for the 27 complete genome sequences was then derived from the alignment using the MAUVE “export orthologs” function. Draft genomes were included to increase the diversity of strains included in the alignment; however, we did not include sequences from these genomes in our sets of putative orthologs as annotation of these genomes was incomplete, and we wanted to examine only annotated protein-coding genes.
The amino acid sequences corresponding to the resulting 5,282 gene sets of size N ≥ 4 were extracted from GenBank and aligned using ProbCons (Do et al. 2005). Ambiguously aligned regions of the alignments were removed using GBLOCKS version 0.91b (Castresana 2000) with parameter settings as follows: minimum number of sequences for a conserved position: (n/2) + 1; minimum number of sequences for a flank position: (n/2) + 1; maximum number of contiguous nonconserved positions: 50; minimum length of a block: five; and allowed gap positions (n is the total number of sequences in the aligned data set). The resulting amino acid alignments were computationally reverse translated to nucleotide alignments using the equivalent nucleotide sequences from GenBank.
Construction of E. coli–Shigella Reference Tree
We inferred a Bayesian phylogenetic tree (Huelsenbeck and Ronquist 2001) for each of the 5,282 E. coli and Shigella gene families (N > 4) using the nucleotide alignments, then aggregated all strongly supported bipartitions (PP ≥ 0.95) using matrix representation with parsimony (MRP) (Ragan 1992) to generate a E. coli–Shigella supertree. We manually rooted this tree according to Touchon et al. (2009). Tree views were produced using Interactive Tree Of Life (Letunic and Bork 2007).
Bayesian inference of individual phylogenetic gene trees was performed using the software MrBayes version 3.1.2 (Huelsenbeck and Ronquist 2001; Huelsenbeck et al. 2001). All analyses were carried out using four Markov chains, three of which were “heated.” The heating parameter was fixed at 0.5. Given that the sequences within individual gene families were highly similar and, in many cases, contained only a small number of substitutions, the evolutionary model was set to the Hawegawa, Kishino and Yano model model (Hasegawa et al. 1985) with gamma-distributed rate variation across sites. Gene sets with <14 sequences were run for 1 million generations, while gene sets with ≥14 sequences were run for 5 million generations. All analyses used a burn-in of 50,000 generations and sampled tree space every 100 generations.
Parsimony Analysis of sRNA Gene Acquisition and Loss
Based on the differential presence or absence of the 83 known E. coli K-12 MG1655 sRNAs across 27 strains of E. coli and Shigella, we reconstructed the most parsimonious scenarios for genetic gain and loss of sRNAs on the rooted E. coli–Shigella reference phylogeny using generalized parsimony as implemented in PAUP* version 4.0. All putative genetic gains and losses reported in the results were estimated using the DELTRAN (delayed transformation) algorithm and relative penalties for gain and loss of 2:1 (i.e., gain/loss penalty = 2). A gain/loss penalty of 2 has been shown to be suitable for biologically reasonable estimation of genetic gain and loss events (Snel et al. 2002). Estimation of gains and losses under the ACCTRAN (accelerated transformation) algorithm (supplementary table S3, Supplementary Material online) and using a gain/loss penalty of 1 (supplementary table S4, Supplementary Material online) gave very similar results for the 83 known sRNAs.
We also estimated the most parsimonious scenarios for genetic gain and loss for the 53 previously predicted sRNAs and 10 new sRNAs reported by Raghavan et al. (2011). For these sRNAs, we again compared gains and losses inferred using the ACCTRAN and DELTRAN algorithms at gain/loss penalty = 2 and gain/loss penalty = 1 (supplementary tables S5 and S6, Supplementary Material online). We found that using a gain/loss penalty of one increased the number of inferred LGT events for this subset of sRNAs; however, across all sRNAs we examined, the preponderance of paraphyletic distribution remains for all algorithms and LGT/gene loss penalties.
Network Analyses
The sRNA interaction network of E. coli K-12 MG1655 was reconstructed by downloading the targets and regulators of the E. coli K-12 MG1655 sRNAs from the sRNAmap database (Huang et al. 2009) and RegulonDB (Gama-Castro et al. 2011). In addition, interactions from recent literature (Papenfort and Vogel 2009; Mandin and Gottesman 2010) were included. The final interaction network has 59 regulator-sRNA interactions and 157 sRNA-target interactions. The network view was generated using Cytoscape (Shannon et al. 2003).
Functional Analysis of sRNA Targets
For functional analysis of sRNA targets, all E. coli K-12 protein-coding sRNA target genes were assigned to a J Craig Venter Institute (JCVI) functional category. The role identifiers (Mainrole) for sRNA targets were retrieved from the JCVI Comprehensive Microbial Resource website http://cmr.jcvi.org/ (Peterson et al. 2001).
Evolutionary Analysis of Hfq-Associated sRNA Targets
For evolutionary analysis of the targets of Hfq-associated core sRNAs, each of the corresponding Bayesian nucleotide trees was compared to the E. coli–Shigella reference topology using the EEEP program (Beiko and Hamilton 2006) with a bootstrap collapse threshold of 95% and strict reference tree ratchet (-rR). EEEP identifies instances of discordance between a test trees and a reference tree. Discordance between the test tree and the E. coli–Shigella reference tree was interpreted as putative evidence of lateral transfer of the target within the E. coli–Shigella clade.
For detection of recombination in nucleotide sequences, we implemented the two-phase strategy described by Chan et al. (2007). First, three statistical measures (Maynard Smith 1992; Jakobsen and Easteal 1996; Bruen et al. 2006) were used to detect preliminary evidence of phylogenetic discrepancies within gene families. Where a minimum of two of these three statistical tests revealed high significance of phylogenetic discrepancy across sites in an alignment (P value < 0.1), recombination was inferred. Second, a rigorous phylogenetic approach as implemented in the software package DualBrothers (Minin et al. 2005) was used to infer tree topologies and evolutionary rates across sites within an alignment that, in the first phase, showed evidence of recombination. We used the DualBrothers parameter value settings and the classification system described by Chan, Darling, et al. (2009) to identify sequence sets presenting clear evidence of recombination breakpoints within the gene boundaries. Inference of one or more recombination breakpoints has been interpreted as evidence of within-gene genetic transfer of one or more genes in the corresponding gene set (Chan, Darling, et al. 2009).
Results
sRNA Genes Are Broadly Conserved in E. coli and Shigella
We selected E. coli for our analysis because 20 complete genome sequences, as well as several unfinished draft genomes, are available and sRNAs have been relatively well-characterized in one strain, E. coli K-12 MG1655. We included seven Shigella genomes because the Shigella phenotype has evolved multiple times from different E. coli clones (Pupo et al. 2000). The 27 complete genomes included in this study span diverse commensal and pathogenic lineages, and include representatives from the major E. coli phylogenetic groups (i.e., A, B1, B2, D, and E) described by multi-locus enzyme electrophoresis (Herzer et al. 1990) and genetic markers (Desjardins et al. 1995; Gordon et al. 2008).
For sRNA phyletic distribution analysis, the sequences of 83 known sRNAs characterized in E. coli K-12 MG1655 were compared by BLAST (Altschul et al. 1990, 1997) to the 27 E. coli and Shigella genome sequences. A sRNA-encoding gene was considered to be present or conserved in a given genome if BLAST searches yielded one or more alignments satisfying the similarity criteria outlined in Methods. sRNA alignments against the target genomes are highly conserved (supplementary fig. S1, Supplementary Material online) and show higher conservation than alignments of E. coli–Shigella protein-coding genes (supplementary fig. S2, Supplementary Material online). Among 2,023 most significant BLAST hits against the target genomes, 1,158 (57.2%) show 100% sequence identity. We find that 61 (73%) of the 83 query sRNAs yield a maximum of one significant BLAST alignment per target genome, while each of the remaining 22 yields more than one significant BLAST alignment in at least one target genome. We refer to these sRNA families as single-copy and multi-copy respectively. The latter include bona fide instances of within-genome sRNA gene duplication, as well as matches to genomic regions that are not considered to encode genuine sRNA synologs. For example, experimental studies have shown that E. coli sRNA rttR is encoded by one of the repeated sequence units of the tyrT operon (Bösl and Kersten 1991). Patterns of presence and absence for each of the 83 sRNAs across the 27 genomes are shown in supplementary table S1 (Supplementary Material online).
In many taxa of bacteria including E. coli–Shigella, all strains share a set of core genes but can differ substantially in their repertoire of variable genes. This variability arises predominantly from gene duplication, LGT and genetic loss (Lerat et al. 2003). Among these 83 sRNAs, 60 are core in the sense of having been identified in all 27 genomes. Knowledge of the size and composition of this core necessarily depends on which genomes happen to have been sequenced; but as these 27 strains are physiologically diverse and core-size estimates appear to be stabilizing as further genomes are sequenced (fig. 1), these 60 almost certainly provide a representative picture of the true E. coli–Shigella core sRNA set. With the exception of E. coli K-12 W3110, which has the same sRNA content as K-12 MG1655, only subsets of the query K-12 MG1655 sRNAs are conserved in each of the other genomes. Of all 83 sRNAs, 23 are variable. The phyletic distributions of these sRNAs are summarized in figure 2. Only one sRNA (C0293) is known to occur only in the two E. coli K-12 strains. As-yet uncharacterized sRNAs other than these 83 may occur in one or more genomes other than the K-12 strains, but necessarily contribute only to the variable, not the core, sRNA set.
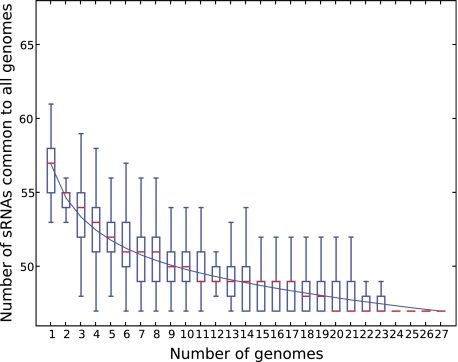
Fig. 1.—
Number of Escherichia coli single-copy core sRNAs estimated according to the number of genomes. This figure shows the number of sRNAs in common for a given number of genomes, analyzed for the different strains of E. coli and Shigella. The upper and lower edges of the boxes indicate the first quartile and third quartile, respectively, of all possible different input orders of the genomes. The central red horizontal line indicates the sample median. The central vertical lines extend from each box as far as the data extend, to a distance of at most 1.5 times the interquartile range. Data points that fall beyond this range are not shown. The blue line passing through all boxes passes through the sample mean of all possible different input orders of the genomes. At 27 sequenced genomes, there are 45 single-copy core sRNAs.
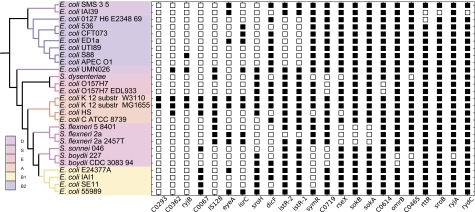
Fig. 2.—
Phyletic distributions of 23 variable sRNA genes across 27 Escherichia coli and Shigella genomes. A black box indicates that BLAST searches of the corresponding query E. coli K-12 MG1655 sRNA against the respective genome yielded one or more significant alignments (E value < 0.001 and target length ≥ 0.7 × query length), while a white box indicates that no significant alignments were found. Strains are ordered based on their placement in the E. coli–Shigella species tree shown on the left. The phylogeny was constructed using MRP from 5,282 Bayesian nucleotide trees. Colors on the MRP tree indicate membership in E. coli phylogenetic groups.
The different classes of sRNAs act differently and may therefore be subject to different evolutionary constraints. For example, many Hfq-binding sRNAs contain a highly conserved core region which is frequently involved in base pairing with targets (Sharma et al. 2007), suggesting that pairing constrains evolution (Gottesman and Storz 2010). Among the 23 sRNAs identified as variable within E. coli–Shigella clade most are of unknown function, three are cis-acting (IstR-1, IstR-2 and SokB) and four are Hfq-associated trans-acting (DicF, RseX, RydC and OmrB). Given these small numbers, we do not yet know if different classes of sRNA regulators are subject to different evolutionary constraints within the E. coli–Shigella clade.
OmrB represents a particularly intriguing example of a variable sRNA. It is one of two redundant sRNAs, OmrA and OmrB (formerly RygA and RygB), that arise from an ancestral gene duplication and (except as noted below) are encoded by adjacent regions of the intergenic space between aas and galR (Argaman et al. 2001). OmrA and OmrB almost invariably regulate the same targets (Papenfort and Vogel 2009). In particular, each mediates the repression of the same subset of outer membrane proteins (Guillier and Gottesman 2008). What advantage might this provide? Paperfort and Vogel (Papenfort and Vogel 2009) suggest that redundant sRNAs may guarantee tight control of regulator levels and target mRNA regulation. While OmrA is conserved in all 27 genomes in our study, and the intergenic region encoding OmrA and OmrB is conserved across many enterobacterial genomes, OmrB is absent from both strains of Shigella boydii in our data set. Whatever the advantage of this redundancy, it appears to be dispensable for at least two of these 27 strains.
Given that in some bacterial pathogens sRNAs are key mediators of virulence gene expression (Lenz et al. 2004), we also used BLAST to investigate the occurrence of the 83 known E. coli K-12 sRNAs in the newly available genome sequence of the rare enterohemorrhagic E. coli 0104:H4 strain TY-2482 (Beijing Genomics Institute, China) responsible for a severe outbreak in Europe in 2011. Of these 83 sRNAs, 78 yielded significant BLAST hits against the TY-2482 genome; sRNAs C0614, C0293, C0362, RyjB, and IS128 were not found. These five sRNAs are variable within our 27-genome data set and as they are of unknown function, their absence from TY-2482 is not directly informative about the virulence. Interestingly, among the 27 genomes in our main analysis, this particular subset of sRNAs is absent only from the two other enterohemorrhagic strains O157H7 and EDL933. Experimental characterization of novel sRNAs in E. coli 0104:H4 strain TY-2482 is needed to gain further insights into virulence of this particular strain.
Although sRNAs have been relatively well-characterized in E. coli, it is not yet clear if the majority of sRNAs present in the species have been identified (Waters and Storz 2009). A recent analysis using a deep sequencing approach to detect putative sRNA transcripts in E. coli K-12 (Raghavan et al. 2011) detected 53 previously predicted sRNAs, and identified a further 10 new putative sRNA transcripts. These 63 putative sRNAs are of unknown function. We wanted to determine if the high proportion of core genes observed for the 83 known sRNAs also occurred in the set of 63 sRNAs of unknown function. Using the same approach as for the 83 known sRNAs, we carried out sRNA phyletic distribution analysis on the additional set of 63 sRNAs. Among these 63 sRNAs 31 (49%) are core, a lower proportion than for the 83 known sRNAs, but still high enough to suggest that sRNAs of both known and unknown function are broadly conserved among strains of Shigella and E. coli. Phyletic distributions for the 53 previously predicted sRNAs and 10 new sRNAs are shown in supplementary tables S7 and S8 (Supplementary Material online), respectively.
Secondary Loss, Not Lateral Transfer, Is the Primary Determinant of Variable sRNA Distribution in E. coli and Shigella
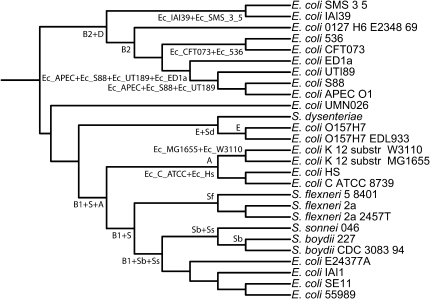
Patterns of presence and absence on phylogenetic trees can be used to infer evolutionary events that have shaped extant genomes (Snel et al. 2002). For example, the presence of a gene in all members of the clade is most parsimoniously explained by its presence in the most-recent common ancestor, whereas anomalous or sporadic absence is likely due to gene loss, and irregular or fragmented phyletic distributions can be indicative of LGT (Ragan 2001; Ragan and Charlebois 2002). To examine the evolutionary processes that have shaped the sRNA content of extant E. coli and Shigella genomes, we first constructed an E. coli–Shigella species tree. Bayesian phylogenetic trees were independently inferred for 5,282 gene families (see Materials and Methods), and all well-supported bipartitions (posterior probability [PP] ≥ 0.95) were aggregated using the method of MRP (Ragan 1992) to construct a robust reference tree (fig. 3), onto which we mapped observed distributions of the variable sRNAs to estimate sRNA gain and loss along each internal edge. Touchon et al. (2009) reconstructed the phylogenetic history of 20 E. coli and Shigella strains using a maximum likelihood approach from 1,878 concatenated E. coli and Shigella core gene sequences. Our MRP tree is remarkably congruent with this phylogeny: of the 52 bipartitions in the MRP tree, 49 are concordant with those recovered by Touchon et al.
Fig. 3.—
Escherichia coli and Shigella reference tree with internal branches labeled. The phylogeny was constructed using MRP from 5,282 Bayesian nucleotide trees.
From the estimates of sRNA gain and loss, we classified each of 23 variable sRNAs as monophyletic (present in the most-recent common ancestor and all its descendants), paraphyletic (present in the most-recent common ancestor and some of its descendants), or polyphyletic (present in some genomes but not in their most-recent common ancestor). For variable genes, monophyly indicates a scenario of gene appearance and descent; paraphyly, one or more instances of gene loss; and polyphyly, multiple LGT events. More complex and hybrid scenarios of gene loss and LGT are also possible (table 1). We find that most of the 23 variable sRNAs are paraphyletic on the MRP tree of these 27 genomes; one sRNA (C0293) is monophyletic, while RyjB is polyphyletic and has apparently been transferred into the E. coli lineage as many as three times. The instance of monophyly likewise reflects descent from a separate LGT event. Nonetheless, the preponderance of paraphyletic distributions clearly indicates that sRNAs are much more frequently lost from than recruited stably into E. coli and Shigella genomic lineages. With the exception of C0067, all paraphyletic sRNAs were inferred to be present in the common ancestor of E. coli–Shigella. In contrast, the phyletic distribution of C0067 supports paraphyly within a more restricted clade, and suggests that this sRNA was gained from an external lineage along the ancestral branch leading to the Shigella, B1 and A phylogenetic groups but was subsequently lost up to three times. We similarly carried out an analysis of sRNA gains and losses using the 32 variable sRNAs that were identified among the 63 putative sRNAs detected by Raghavan et al. (2011). Among these sRNAs, we likewise found a preponderance of paraphyletic (25/32, 78%) distributions (supplementary table S9, Supplementary Material online).
Table 1.
Gains and Losses of Variable sRNAs in the Escherichia coli–Shigella Lineage
| sRNA Name | Branches on Which Inferred sRNA Gains Occur | Branches on Which Inferred sRNA Losses Occur | Category |
| C0719 | E + Sd, Sb + Ss | Paraphyletic | |
| C0293 | Ec_MG1655 + ECW3110 | Monophyletic | |
| ryjB | Ec_S88, Ec_UMN026, Ec_MG1655 + ECW3110 | Polyphyletic | |
| istR-2 | B2, Sf_2a | Paraphyletic | |
| ryjA | Sb_277 | Paraphyletic | |
| sroB | Ec_IA139 | Paraphyletic | |
| istR-1 | B2, Sf_2a | Paraphyletic | |
| C0614 | E | Paraphyletic | |
| C0362 | B2 + D, E+Sd, Ec_C_ATCC, B1 + S | Paraphyletic | |
| C0465 | Sd, Sb_227 | Paraphyletic | |
| C0067 | B1+S+A | Ec_C_ATCC, Sf, Sb | Paraphyletic |
| dicF | Ec_IAI39, Ec_536, Sd, Ec_HS, Sf, Ss_046, Sb_227, Ec_SE11 | Paraphyletic | |
| sokB | Sf | Paraphyletic | |
| sokA | Sf | Paraphyletic | |
| eyeA | Ec_E2348, Ec_536 + Ec_CFT073, Ec_APEC + Ec_S88 + Ec_UT189, Ec_UMN026, E + Sd, Ec_C_ATCC + Ec_HS, Sf_5_8401, Sb + Ss, Ec_IAI1, Ec_SE11 | Paraphyletic | |
| rseX | Sd, Ec_HS, Sf | Paraphyletic | |
| sroH | B2 + D, Ec_C_ATCC, Sf, Ss_046 | Paraphyletic | |
| rydC | Ec_IA139 | Paraphyletic | |
| isrC | Ec_55989 | Ec_IAI39 + Ec_SMS_3, Ec_E2348, Ec_APEC + Ec_S88 + Ec_UT189, E + Sd, Ec_C_ATCC + Ec_HS, Sf_5_8401, B1 + Sb + Ss | Paraphyletic |
| rttR | Ec_IAI39, Ec_E2348 | Paraphyletic | |
| symR | Ec_55989 | Sd, Ec_HS, B1 + Sb + Ss | Paraphyletic |
| IS128 | B2 + D, E, B1 + Sb + Ss | Paraphyletic | |
| omrB | Sb | Paraphyletic |
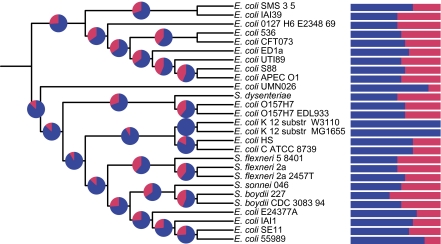
The numbers of inferred gains and losses vary only modestly among lineages implied by the MRP tree, with gene loss inferred considerably more frequently than gene gain (fig. 4). Almost all of the inferred gains occur along the lineage leading to E. coli K-12; this is a consequence of using only K-12 sRNAs as queries for the presence/absence analysis (in turn occasioned by the state of knowledge). We infer sRNA losses to have been particularly frequent in the most-recent common ancestor of the Shigella flexneri strains, and in the ancestor of Shigella dysenteriae. Although the numbers are small, this is in broad agreement with studies that show that Shigella genomes have lost more genomic material than have E. coli genomes (Yang et al. 2005). We also infer the ancestral lineage of the restricted–host-range pathogen E. coli E2348 to have incurred relatively many sRNA losses. Among these 27 genomes, S. boydii Sb227 shares the fewest sRNAs in common with E. coli K-12 MG1655.
Fig. 4.—
Reconstruction of gains and losses of variable sRNAs in the evolution of the Escherichia coli–Shigella lineage. The cladogram shows the phylogenetic relationships among the 27 E. coli and Shigella genomes, as in figure 2. Pie charts on internal nodes of the tree show the proportion of the 23 variable sRNAs that were inferred to be present in the ancestral strain corresponding to the node. Ancestral sRNA content as represented in the pie charts is based on the inferences of sRNA gains and losses reported in table 1. Dark blue indicates the presence and red indicates the absence. Similarly, the bars on the right show the proportion of the 23 variable sRNAs that were inferred to be present and the absence in the corresponding extant E. coli and Shigella genomes.
Core sRNAs Are More Tightly Integrated into Genetic Regulatory Networks Than Are Variable sRNAs
We extracted interactions between E. coli K-12 MG1655 sRNAs and their targets and transcriptional regulators from sRNAmap (Huang et al. 2009), RegulonDB (Gama-Castro et al. 2011) and recent literature (Papenfort and Vogel 2009; Mandin and Gottesman 2010) and used this knowledge to construct an sRNA interaction network (fig. 5) that encompasses 59 regulator-sRNA interactions and 157 sRNA-target interactions. Overall, 34 unique protein-coding regulators of sRNAs, 114 targets and 48 sRNAs are represented in the network.
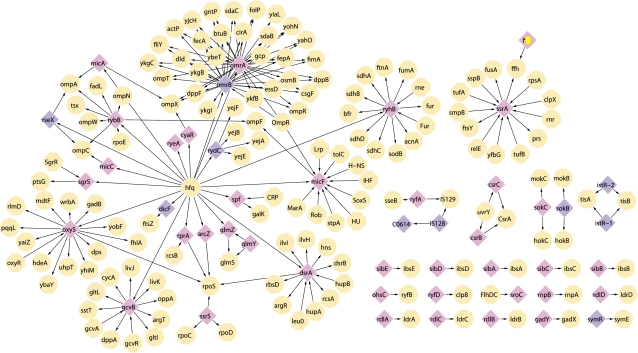
Fig. 5.—
Escherichia coli K-12 MG1655 sRNA interaction network. The network shows functional interactions between sRNAs and protein-related biomacromolecules (proteins, mRNAs, and genomic DNAs) in E. coli K-12 MG1655. This network was reconstructed using interactions obtained from sRNAmap (Huang et al. 2009) and recent literature (Papenfort and Vogel 2009; Mandin and Gottesman 2010). sRNAs are represented as diamonds, sRNA targets as ellipses, and sRNA regulators as rounded rectangles. sRNA nodes are colored accordingly: core sRNAs are pink and variable sRNAs are purple.
In a recent review, Beisel and Storz (2010) described the specific regulatory circuits that incorporate base-pairing sRNAs. These include single-input modules, dense overlapping regulons, negative feedback loops and feed-forward loops (Beisel and Storz 2010). Here we focus on the participation of sRNAs in single-input modules, as these regulatory circuits appear recurrently in the reconstructed sRNA interaction network. As defined by Beisel and Storz, in single-input modules a solitary regulator coordinates the expression of more than one target molecule. Thus, single sRNAs regulators that alone activate or repress the expression of more than one target are members of such modules. Most Hfq-binding sRNAs have been found to interact with multiple target mRNAs in response to particular environmental signals, and thereby form single-input modules (Beisel and Storz 2010). We find that all Hfq-associated sRNAs forming part of single-input modules in the E. coli K-12 sRNA interaction network are core (GcvB, RyhB, DsrA, OxyS, SsrS, RybB), suggesting that interaction with multiple targets constrains the evolution of these sRNAs.
Another type of regulatory circuit is the dense overlapping regulon. As defined by Beisel and Storz (2010), these circuits combine multiple overlapping single-input modules to coordinate responses to multiple biological signals. A well-established example of an sRNA-based dense overlapping regulon converging on one target in E. coli, is the co-ordinated regulatory control of the master regulator of the general stress response RpoS by ArcZ (originally named RyhA), RprA, DsrA and OxyS (Mandin and Gottesman 2010). Gottesman and Storz (2010) speculate that sRNAs with one or more shared targets may also share a common ancestor. We extended this to hypothesize that sRNAs that control the expression of a common set of genes will share the same phyletic distribution. Our results show that sRNAs that enact regulation of RpoS are all encoded by E. coli–Shigella core genes and thus share the same phyletic distribution within the E. coli–Shigella clade.
Variability implies that interactions need to be formed (after gene gain, e.g., by LGT) and dissolved (after gene loss). We find that, as a rule, variable sRNAs have fewer interaction partners than core sRNAs (fig. 4). In particular, with the exception of OmrB (see below), each variable sRNA interacts with at most two target molecules and therefore occupies a more peripheral position in the network. It is striking that all of the hub sRNA nodes (SsrA, RyhB, MicF, GcvB, RybB, DsrA, OxyS, OmrA), except OmrB, are core. All of the variable sRNAs represented in the network have been lost at least once since E. coli diverged from Salmonella, while only one (SymR) has been putatively transferred into the lineage. We thus hypothesize that variable sRNAs are less well-integrated into the E. coli regulatory circuits and are therefore more susceptible to genetic loss. The loss of such sRNAs is less disruptive to the posttranscriptional regulatory network, and therefore less likely to be detrimental to the population.
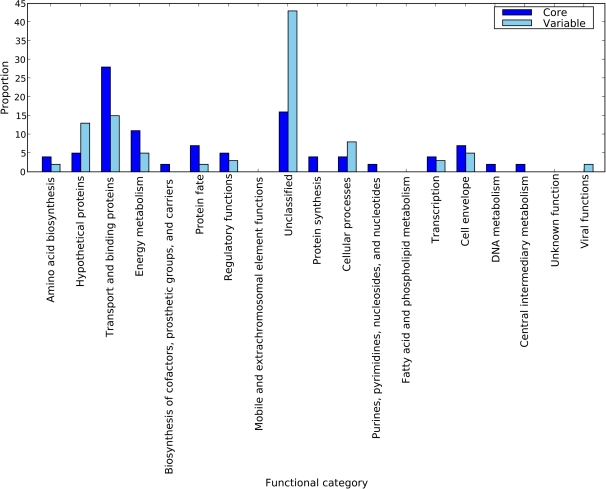
There are 114 sRNA targets represented in the reconstructed sRNA interaction network, 112 of which are protein-coding. Of these 112 protein-coding targets, 54 are core and 58 are variable within the E. coli–Shigella clade. To examine the functions controlled by E. coli K-12 sRNAs, we used annotations from The JCVI Comprehensive Microbial Resource (Peterson et al. 2001) to assign functional categories to the 112 sRNA targets. Figure 6 shows, for each functional category, the proportion of core and variable sRNA targets.
Fig. 6.—
Distribution of Escherichia coli K-12 sRNA target genes among JCVI functional categories. Shown for each functional category are proportions of core and variable sRNA target genes. Of 112 E. coli K-12 sRNA target genes, 54 are core and 58 are variable.
The distribution of sRNA targets among the JCVI functional categories broadly emphasizes the diverse range of functions controlled by E. coli K-12 sRNAs. The functional categories represented among the 112 sRNA targets include: Amino acid biosynthesis; Hypothetical proteins; Transport and binding proteins; Energy metabolism; Biosynthesis of cofactors, prosthetic groups, and carriers; Protein fate; Regulatory functions; Unclassified; Protein synthesis, Cellular processes; Purines, pyrimidines, nucleosides, and nucleotides; Transcription; Cell envelope; DNA metabolism; Central intermediary metabolism; and Viral functions. The majority of variable sRNA targets are of unknown function, while the majority of core sRNA targets fall within the “Transport and binding proteins” category.
The sRNA phyletic distribution analysis shows sRNA losses to have been particularly frequent in the ancestral lineage of the restricted–host-range pathogen E. coli E2348. Among the 58 variable sRNA target genes present in E. coli K-12, 20 (34.5%) are absent from E. coli E2348. Although the majority of these targets are of unknown function, genes among these 20 sRNA targets have been assigned JCVI functional categories Transport and binding proteins; Cellular processes; Transcription; Cell envelope; and Viral functions. Thus genes associated with a diverse range of sRNA-mediated processes have been lost in the lineage leading to E. coli E2348.
The Core and Variable Genomes Cross Talk Posttranscriptionally via Hfq and sRNAs
If laterally acquired genetic material is to persist in its new host, it must recruit interaction partners to regulate its expression (Navarre et al. 2007; Lercher and Pál 2008), as inappropriate expression could impose a substantial upfront cost on fitness of the host. Enteric bacteria are able to limit these costs at the level of transcriptional control by silencing the expression of foreign genes via the DNA-binding protein H-NS (Navarre et al. 2007). Do similar mechanisms of control exist at the level of posttranscriptional regulation? In E. coli, all characterized trans-encoded sRNAs are Hfq dependent (Waters and Storz 2009). Hfq is believed to facilitate posttranscriptional cross talk between core and variable genome regions and in some cases, potentially aids the integration of laterally acquired genes into existing posttranscriptional control networks (Papenfort and Vogel 2010; Chao and Vogel 2010). Here, we examine evidence for Hfq-mediated cross talk between E. coli–Shigella core and variable genes, and for selective targeting of genes, which have been subject to intraspecies LGT by Hfq-associated sRNAs.
Among the E. coli Hfq-associated sRNAs, we identified four which are variable within the E. coli–Shigella species: DicF, OmrB, RseX, and RydC. Each of these sRNAs has mRNA targets that are encoded by E. coli–Shigella core genes. DicF is a well-established example of sRNA lateral transfer; it is encoded within a cryptic prophage and inhibits translation of the host cell division gene ftsZ (Bouché F and Bouché JP 1989), which is core in E. coli–Shigella. The observed pattern of presence/absence indicates that DicF was present in the common ancestor of E. coli and Shigella but has subsequently been lost, potentially up to eight times. OmrB down regulates its own transcriptional activator, which is coded by E. coli core gene ompR (Guillier and Gottesman 2008). This potentially represents a unique case of a variable sRNA regulating a core gene, as the partially homologous sRNA OmrA, which unlike OmrB is core in E. coli and Shigella, almost invariably regulates the same targets as OmrB. Therefore, control of regulator levels and target mRNA regulation, including the repression of OmpR, by OmrA-OmrB is likely preserved in the Shigella boydii strains in which OmrB is no longer present.
Conversely, Hfq-associated core sRNAs might target the mRNAs of recently acquired lateral genes, thereby facilitating the integration of these genes into host-cell networks. From sRNAmap, we extracted 85 interactions between Hfq-associated core sRNAs and their targets, involving 15 unique sRNAs and 77 unique targets. Of these 77 target genes, 39 are core and 38 are variable in the E. coli–Shigella lineage, but almost all of the latter are widely conserved among these strains, that is, variable phyletic distributions are likely the result of occasional gene loss rather than LGT from an external clade. Moreover, 13 of the 15 Hfq-associated core sRNAs investigated have at least one core target. We thus focused our investigation on potential instances of posttranscriptional cross talk between core Hfq-associated sRNAs and target genes that have been subject to LGT within the E. coli–Shigella lineage.
Hfq-Associated sRNAs Regulate Genes Transferred Intact within the E. coli–Shigella Lineage
Conflicting (topologically incongruent) phylogenetic signals can be interpreted as putative evidence of LGT. We compared the inferred Bayesian phylogenetic tree for each of the 74 target genes sets of size N ≥ 4 with our E. coli–Shigella reference tree. Incongruence between one or more well-supported (PP ≥ 0.95) bipartitions of the gene tree and the E. coli–Shigella reference tree was interpreted as evidence of the within-species transfer of the corresponding gene. Remarkably, 61 (82%) of the 74 target genes yielded gene trees incongruent with the MRP reference (table 2).
Table 2.
Evidence for within-Species LGT of Genes Targeted by Core Hfq-Dependent sRNAs in the E. coli–Shigella Lineage
| Target Gene Name | bNumber | sRNA Core Regulator(s) | Number of E. coli and Shigella Strains in which Gene Is Present (N) | Core Or Variable | Nucleotide Tree Congruent Or Incongruent with E. coli–Shigella Reference Tree? | Evidence for Internal Recombination Breakpoint(s)? |
| rpoS | b2741 | oxyS, dsrA, arcZ, rprA | 22 | Variable | Congruent | No |
| fhlA | b2731 | oxyS | 24 | Variable | Incongruent | No |
| yobF | b1824 | oxyS | 18 | Variable | Congruent | No |
| yhiM | b3491 | oxyS | 19 | Variable | Incongruent | No |
| gadB | b1493 | oxyS | 27 | Core | Incongruent | No |
| uhpT | b3666 | oxyS | 27 | Core | Incongruent | No |
| wrbA | b1004 | oxyS | 21 | Variable | Incongruent | No |
| ybaY | b0453 | oxyS | 25 | Variable | Incongruent | Yes |
| mdtF | b3514 | oxyS | 25 | Variable | Incongruent | No |
| hdeA | b3510 | oxyS | 27 | Core | Congruent | No |
| rlmD | b2785 | oxyS | 27 | Core | Incongruent | No |
| dps | b0812 | oxyS | 27 | Core | Incongruent | No |
| yaiZ | b0380 | oxyS | 6 | Variable | Congruent | No |
| pqqL | b1494 | oxyS | 17 | Variable | Incongruent | No |
| rcsA | b1951 | dsrA | 26 | Variable | Incongruent | No |
| argR | b3237 | dsrA | 27 | Core | Incongruent | No |
| dsrB | b1952 | dsrA | 27 | Core | Incongruent | No |
| rbsD | b3748 | dsrA | 26 | Variable | Incongruent | No |
| hns | b1237 | dsrA | 27 | Core | Congruent | No |
| ilvI | b0077 | dsrA | 26 | Variable | Incongruent | No |
| ilvH | b0078 | dsrA | 27 | Core | Incongruent | No |
| fur | b0683 | ryhB | 27 | Core | Incongruent | No |
| bfr | b3336 | ryhB | 4 | Variable | Congruent | No |
| fumA | b1612 | ryhB | 27 | Core | Incongruent | Yes |
| acnA | b1276 | ryhB | 23 | Variable | Incongruent | No |
| ftnA | b1905 | ryhB | 27 | Core | Incongruent | No |
| sodB | b1656 | ryhB | 27 | Core | Incongruent | No |
| sdhA | b0723 | ryhB | 27 | Core | Incongruent | No |
| sdhC | b0721 | ryhB | 27 | Core | Iongruent | No |
| sdhB | b0724 | ryhB | 27 | Core | Congruent | No |
| sdhD | b0722 | ryhB | 27 | Core | Incongruent | No |
| ompW | b1256 | rybB | 27 | Core | Incongruent | No |
| ompF | b0929 | rybB, micF | 27 | Core | Incongruent | No |
| tsx | b0411 | rybB | 27 | Core | Incongruent | No |
| ompC | b2215 | rybB, micC | 27 | Core | Incongruent | No |
| ompN | b1377 | rybB | 21 | Variable | Incongruent | No |
| fadL | b2344 | rybB | 27 | Core | Incongruent | No |
| glmS | b3729 | glmZ | 27 | Core | Incongruent | No |
| ptsG | b1101 | sgrS | 27 | Core | Incongruent | No |
| actP | b4067 | omrA | 26 | Variable | Incongruent | Yes |
| btuB | b3966 | omrA | 25 | Variable | Incongruent | No |
| cirA | b2155 | omrA | 21 | Variable | Incongruent | No |
| fliY | b1920 | omrA | 10 | Variable | Incongruent | No |
| fepA | b0584 | omrA | 26 | Variable | Incongruent | No |
| ompT | b0565 | omrA | 3 | Variable | N < 4 | N < 4 |
| folP | b3177 | omrA | 27 | Core | Incongruent | No |
| yohN | b2107 | omrA | 10 | Variable | Incongruent | No |
| dld | b2133 | omrA | 27 | Core | Incongruent | No |
| ybeT | b0647 | omrA | 8 | Variable | Congruent | No |
| ompX | b0814 | omrA, micA, cyaR | 27 | Core | Congruent | No |
| gcp | b3064 | omrA | 27 | Core | Incongruent | Yes |
| ykfB | b0250 | omrA | 2 | Variable | N < 4 | N < 4 |
| fecA | b4291 | omrA | 5 | Variable | Congruent | No |
| gntP | b4321 | omrA | 17 | Variable | Incongruent | Yes |
| osmB | b1283 | omrA | 25 | Variable | Incongruent | No |
| sdaB | b2797 | omrA | 26 | Variable | Incongruent | No |
| sdaC | b2796 | omrA | 27 | Core | Incongruent | No |
| ompR | b3405 | omrA | 27 | Core | Incongruent | No |
| yahO | b0329 | omrA | 20 | Variable | Incongruent | No |
| dppB | b3543 | omrA | 27 | Core | Incongruent | No |
| yjcH | b4068 | omrA | 24 | Variable | Congruent | No |
| dppF | b3540 | omrA | 27 | Core | Incongruent | No |
| fimA | b4314 | omrA | 2 | Variable | N < 4 | N < 4 |
| ykgC | b0304 | omrA | 21 | Variable | Incongruent | No |
| csgF | b1038 | omrA | 23 | Variable | Incongruent | No |
| yiaL | b3576 | omrA | 22 | Variable | Incongruent | No |
| ompA | b0957 | micA, rybB | 27 | Core | Incongruent | No |
| gltI | b0655 | gcvB | 26 | Variable | Incongruent | Yes |
| dppA | b3544 | gcvB | 25 | Variable | Incongruent | No |
| argT | b2310 | gcvB | 25 | Variable | Incongruent | No |
| sstT | b3089 | gcvB | 27 | Core | Incongruent | Yes |
| oppA | b1243 | gcvB | 27 | Core | Incongruent | No |
| gltL | b0652 | gcvB | 26 | Variable | Incongruent | No |
| livK | b3458 | gcvB | 27 | Core | Congruent | No |
| cycA | b4208 | gcvB | 27 | Core | Incongruent | Yes |
| livJ | b3460 | gcvB | 27 | Core | Incongruent | Yes |
| galK | b0757 | spf | 27 | Core | Incongruent | No |
Units of genetic transfer and recombination do not necessarily correspond to intact genes (Chan, Beiko, et al. 2009; Chan, Darling, et al. 2009). We recently reported (Skippington and Ragan 2011) that among 5,282 E. coli–Shigella orthologous protein sets, 2,440 (42.2%) show evidence of topological incongruence with the reference tree. This is a much lower proportion of topological incongruence than observed for the 74 sRNA target gene sets. Among these 2,440 protein sets, 463 (19.0%) were independently inferred to contain at least one recombination breakpoint within the boundaries of the open reading frame. Applying a two-phase strategy for detecting recombination breakpoints (Chan et al. 2007) to the 74 target gene sets, we found evidence for internal recombination breakpoints in only nine (12%) gene sets (they also yielded incongruent gene trees). Thus, surprisingly, almost every gene transferred within this clade and targeted by a core Hfq-dependent sRNA is inferred to have been transferred intact. Because in E. coli, trans-acting sRNAs interact with their targets via discontinuous stretches of limited complementarity with the help of Hfq, transfer of the entire target could be crucial to establishment of appropriate spatial interactions between Hfq, the sRNA, and the mRNA target.
Discussion
We have integrated distributional, phylogenetic, and network analyses to explore the evolutionary dynamics of sRNAs in the E. coli–Shigella clade. sRNAs are broadly conserved among strains of Shigella and E. coli. More than two-thirds of E. coli sRNAs are core, a considerably higher proportion than for protein-coding genes. Twenty sequenced strains share a common core of about 1,976 orthologous genes, that is, about 46% of the 4,306 protein-coding genes individually encoded by E. coli K-12 MG1655 (Touchon et al. 2009). A potential explanation for the extensive within-species conservation of RNA regulators is the relative lesser cost to the cell of the maintenance of sRNA-coding genes compared with protein-coding genes, that is, in speed of synthesis and absence of need for translation.
All of the sRNAs examined in this study have been retained in E. coli K-12 MG1655, which has been maintained as a laboratory strain under relatively narrow growth conditions with minimal genetic manipulation (Blattner et al. 1997). Variable sRNAs that mediate responses to special environmental conditions would not necessarily have been retained in this genome. This potentially explains the high proportion of core sRNAs found among the 83 sRNAs examined. Before we can fully understand the contribution of core sRNAs to the overall sRNA content of the E. coli–Shigella clade, sRNAs must be identified and validated thoroughly in additional strains of E. coli and Shigella.
Despite the high number of core sRNAs identified, we also found a significant subset of sRNAs belonging to the variable genome: sRNAs are evolutionarily dynamic even within species. Secondary loss, not lateral transfer, is the primary determinant of variable sRNA phyletic distribution. Losses have been most frequent in the lineages leading to Shigella and to the restricted–host-range pathogen E. coli E2348. Given the central roles played by sRNA regulators in fine-tuning bacterial responses to changes in the environment (Beisel and Storz 2010), we hypothesize that the fact that other pathogenic strains of E. coli have not incurred such significant sRNA losses is a reflection of their broader host ranges. This could be tested by sequencing of further host-restricted pathogenic strains.
By examining the regulatory interactions of sRNAs in E. coli K-12, we identified general network properties that may place constraints on, or contribute to, patterns of sRNA conservation. In particular, core sRNAs are more tightly integrated into genetic regulatory networks than are variable sRNAs. This result provides a basis for predicting how readily integrated sRNAs may be lost: sRNA hubs in the sRNA interaction network are less susceptible to gene loss than are sRNAs that occupy more peripheral positions of the network. It is less clear how readily laterally acquired sRNAs might join and interact with the existing network. Waters and Storz (2009) suggest that the expression of a spurious transcript, whether cis-encoded or trans-encoded with limited complementarity to an mRNA target, could easily become fixed in a population should it provide a selective advantage to the host. We find evidence of LGT from an external clade for only two sRNAs in the interaction network, SgrS and SymR, each of which has only one known target.
Newly acquired sequences can decrease host fitness unless integrated into existing genetic regulatory networks. Regulation of transferred DNA by Hfq-associated sRNAs appears to represent an important posttranscriptional mechanism by which bacteria limit the cost on competitive fitness of inappropriate expression of transferred DNA (Chao and Vogel 2010). Here, we have presented clear evidence that Hfq mediates posttranscriptional cross talk between the core and variable E. coli genome and, moreover, that Hfq-associated core sRNAs interact with targets that have been transferred intact within the E. coli–Shigella lineage. More than 80% of the mRNAs targeted by Hfq-mediated sRNAs show evidence of LGT within the E. coli–Shigella clade.
An integrated understanding of the evolution of sRNAs is still in its infancy, but continued efforts to determine the prevalence of sRNAs and their interacting partners in bacteria will enable further insight into the evolutionary dynamics that have shaped the extant sRNA content of bacterial species and hence their ability to fine-tune responses to the environment.
Supplementary Material
Supplementary tables S1–S9 and figures S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Aaron Darling for the Escherichia coli–Shigella Mauve alignment and for advice, and Cheong Xin Chan for breakpoint-detection scripts. All phylogenetic analyses were performed using high-performance computing resources of the National Computational Infrastructure National Facility at the Australian National University. This work was supported by the Australian Research Council grant CE0348221. E.S. is supported by an Australian Postgraduate Award and a Queensland Government Smart State PhD Scholarship.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argaman L, et al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Beiko RG, Hamilton N. Phylogenetic identification of lateral genetic transfer events. BMC Evol Biol. 2006;6:15. doi: 10.1186/1471-2148-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev. 2010;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bösl M, Kersten H. A novel RNA product of the tyrT operon of Escherichia coli. Nucleic Acids Res. 1991;19:5863–5870. doi: 10.1093/nar/19.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché F, Bouché JP. Genetic evidence that DicF, a second division inhibitor encoded by the Escherichia coli dicB operon, is probably RNA. Mol Microbiol. 1989;3:991–994. doi: 10.1111/j.1365-2958.1989.tb00249.x. [DOI] [PubMed] [Google Scholar]
- Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chan CX, Beiko RG, Darling AE, Ragan MA. Lateral transfer of genes and gene fragments in prokaryotes. Genome Biol Evol. 2009;1:429–438. doi: 10.1093/gbe/evp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CX, Beiko RG, Ragan MA. A two-phase strategy for detecting recombination in nucleotide sequences. S Afr Comput J. 2007;38:20–27. [Google Scholar]
- Chan CX, Darling AE, Beiko RG, Ragan MA. Are protein domains modules of lateral genetic transfer? PLoS One. 2009;4:e4524. doi: 10.1371/journal.pone.0004524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao YJ, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins P, Picard B, Kaltenböck B, Elion J, Denamur E. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment-length polymorphism. J Mol Evol. 1995;41:440–448. doi: 10.1007/BF00160315. [DOI] [PubMed] [Google Scholar]
- Do CB, Mahabhashyam MSP, Brudno M, Batzoglou S. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol Rev. 2008;72:579–589. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Castro S, et al. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units) Nucleic Acids Res. 2011;39:D98–D105. doi: 10.1093/nar/gkq1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DM, Clermont O, Tolley H, Denamur E. Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environ Microbiol. 2008;10:2484–2496. doi: 10.1111/j.1462-2920.2008.01669.x. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2010 doi: 10.1101/cshperspect.a003798. 3:a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res. 2008;36:6781–6794. doi: 10.1093/nar/gkn742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano TA. Dating of the human ape splitting by a molecular clock of mitochondrial-DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Hershberg R, Altuvia S, Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzer PJ, Inouye S, Inouye M, Whittam TS. Phylogenetic distribution of branched RNA-linked multicopy single-stranded-DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, et al. sRNAMap: genomic maps for small non-coding RNAs, their regulators and their targets in microbial genomes. Nucleic Acids Res. 2009;37:D150–D154. doi: 10.1093/nar/gkn852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Evolution—Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Jakobsen IB, Easteal S. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput Appl Biosci. 1996;12:291–295. doi: 10.1093/bioinformatics/12.4.291. [DOI] [PubMed] [Google Scholar]
- Keseler IM, et al. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39:D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, et al. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Moran NA. From gene trees to organismal phylogeny in prokaryotes: the case of the γ-Proteobacteria. PLoS Biol. 2003;1:101–109. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Pál C. Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Mol Biol Evol. 2008;25:559–567. doi: 10.1093/molbev/msm283. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- MacLellan SR, Smallbone LA, Sibley CD, Finan TM. The expression of a novel antisense gene mediates incompatibility within the large repABC family of α-proteobacterial plasmids. Mol Microbiol. 2005;55:611–623. doi: 10.1111/j.1365-2958.2004.04412.x. [DOI] [PubMed] [Google Scholar]
- Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- Minin VN, Dorman KS, Fang F, Suchard MA. Dual multiple change-point model leads to more accurate recombination detection. Bioinformatics. 2005;21:3034–3042. doi: 10.1093/bioinformatics/bti459. [DOI] [PubMed] [Google Scholar]
- Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS—facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. A pH-responsive riboregulator. Genes Dev. 2009;23:2650–2662. doi: 10.1101/gad.552209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res Microbiol. 2009;160:278–287. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell Host Microbe. 2010;8:116–127. doi: 10.1016/j.chom.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. The comprehensive microbial resource. Nucleic Acids Res. 2001;29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo GM, Lan RT, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci U S A. 2000;97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan MA. Phylogenetic inference based on matrix representation of trees. Mol Phylogenet Evol. 1992;1:53–58. doi: 10.1016/1055-7903(92)90035-f. [DOI] [PubMed] [Google Scholar]
- Ragan MA. Detection of lateral gene transfer among microbial genomes. Curr Opin Genet Dev. 2001;11:620–626. doi: 10.1016/s0959-437x(00)00244-6. [DOI] [PubMed] [Google Scholar]
- Ragan MA, Charlebois RL. Distributional profiles of homologous open reading frames among bacterial phyla: implications for vertical and lateral transmission. Int J Syst Evol Microbiol. 2002;52:777–787. doi: 10.1099/00207713-52-3-777. [DOI] [PubMed] [Google Scholar]
- Raghavan R, Groisman EA, Ochman H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 2011;21:1487–1497. doi: 10.1101/gr.119370.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd KE. EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 2000;28:60–64. doi: 10.1093/nar/28.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons RW, Kleckner N. Translational control of Is10 transposition. Cell. 1983;34:683–691. doi: 10.1016/0092-8674(83)90401-4. [DOI] [PubMed] [Google Scholar]
- Skippington E, Ragan MA. Within-species lateral genetic transfer and the evolution of transcriptional regulation in Escherichia coli and Shigella. BMC Genomics. 2011;12:532. doi: 10.1186/1471-2164-12-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 2002;12:17–25. doi: 10.1101/gr.176501. [DOI] [PubMed] [Google Scholar]
- Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu Rev Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- Touchon M, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Argaman L, Wagner EGH, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Wassarman KM. Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell. 2002;109:141–144. doi: 10.1016/s0092-8674(02)00717-1. [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, et al. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33:6445–6458. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]