Abstract
Sex-biased gene expression (i.e., the differential expression of genes between males and females) is common among sexually reproducing species. However, genes often differ in their sex-bias classification or degree of sex bias between species. There is also an unequal distribution of sex-biased genes (especially male-biased genes) between the X chromosome and the autosomes. We used whole-genome expression data and evolutionary rate estimates for two different Drosophilid lineages, melanogaster and obscura, spanning an evolutionary time scale of around 50 Myr to investigate the influence of sex-biased gene expression and chromosomal location on the rate of molecular evolution. In both lineages, the rate of protein evolution correlated positively with the male/female expression ratio. Genes with highly male-biased expression, genes expressed specifically in male reproductive tissues, and genes with conserved male-biased expression over long evolutionary time scales showed the fastest rates of evolution. An analysis of sex-biased gene evolution in both lineages revealed evidence for a “fast-X” effect in which the rate of evolution was greater for X-linked than for autosomal genes. This pattern was particularly pronounced for male-biased genes. Genes located on the obscura “neo-X” chromosome, which originated from a recent X-autosome fusion, showed rates of evolution that were intermediate between genes located on the ancestral X-chromosome and the autosomes. This suggests that the shift to X-linkage led to an increase in the rate of molecular evolution.
Keywords: gene expression, dN/dS, sex-biased genes, transcriptomics
Introduction
Sexual dimorphism, that is, the physical differentiation of males and females of a species, is widespread across the animal kingdom. Most of these differences can be attributed to the evolution of differential gene expression in the two sexes (reviewed by Ellegren and Parsch 2007). Genes that differ in expression level between males and females are commonly referred to as sex-biased genes and can be further divided into male- and female-biased genes, depending on which sex shows higher expression, while genes with similar expression levels in the two sexes are referred to as unbiased. Previous population genetic and comparative genomic studies of Drosophila melanogaster and its sister species D. simulans revealed that male-biased genes have increased levels of amino acid divergence between species (Zhang et al. 2004; Gnad and Parsch 2006) and particularly high rates of adaptive evolution (Pröschel et al. 2006; Sawyer et al. 2007; Baines et al. 2008). For species outside the well-studied melanogaster subgroup, the situation is less clear. Initial studies of D. ananassae and D. pseudoobscura that used a limited number of genes could not confirm the pattern of faster evolution for male-biased genes (Metta et al. 2006; Grath et al. 2009). However, a whole-genome study reported accelerated rates of protein evolution for male-biased genes between two closely related species in the obscura group (Jiang and Machado 2009).
Many factors contribute to variation in rates of evolution between different proteins in Drosophila by either influencing the rate of evolution itself or imposing evolutionary constraints. Larracuente et al. (2008) identified expression level, intron and protein length, intron number, number of protein–protein interactions, recombination rate, and translational selection as possible affectors. In one recent study, protein secondary structure was found to influence rates of positive selection in Drosophila (Ridout et al. 2010). These authors found that amino acids forming disordered regions, for example, random coils, are more likely to experience positive selection than amino acids situated in helices and β-structures. For D. melanogaster and Mus musculus, it has been shown that sex-biased genes showing tissue-specific gene expression in reproductive tissues, and thus having a narrow expression profile within the organism, show faster rates of evolution than unbiased genes with tissue-specific expression or those with broader expression profiles (Meisel 2011).
The chromosomal location of genes (i.e., whether they are sex-linked or autosomal) may also influence their rate of evolution. Theory predicts that X-linked genes (or Z-linked genes in female heterogametic taxa) should exhibit a “fast-X” effect if adaptation occurs primarily through new beneficial mutations that are, on average, recessive (Charlesworth et al. 1987). This is because recessive X-linked mutations are immediately subject to selection in hemizygous males. However, a fast-X effect is not expected when adaptation occurs from standing genetic variation (Orr and Betancourt 2001), as may be the case when there is rapid environmental change (Karasov et al. 2010). In addition, factors such as the overall effective population size and the relative effective population sizes of males and females, may also affect rates of evolution of the X chromosome and autosomes differently (Vicoso and Charlesworth 2009). These factors may explain why a clear fast-X (or fast-Z) effect has been observed in mammals and birds, but not in Drosophila (Mank et al. 2009). In Drosophila, the fast-X effect is expected to be small (Mank et al. 2009) and is typically not observed in whole genome studies that examine ratios of nonsynonymous-to-synonymous divergence (e.g., Thornton et al. 2006). However, if selection on sex-biased genes occurs mainly in the sex that shows enriched expression, as appears to be the case in Drosophila (Connallon and Clarke 2011), then male-biased genes should show the strongest fast-X effect because they are most often subject to selection in a hemizygous background (Baines et al. 2008). Consistent with this, male-biased genes have been observed to exhibit a large fast-X effect in the D. melanogaster subgroup (Baines et al. 2008).
In this report, we examine the influence of the degree and conservation of sex-biased gene expression, as well as chromosomal location, on the rate of molecular evolution in two independent Drosophilid lineages, melanogaster and obscura. These lineages diverged from each other around 50 Ma (Bergman et al. 2002; Tamura et al. 2004). An interesting difference between the two lineages is the presence of a “neo-X” chromosome in the obscura lineage, which is the result of a fusion of an autosome and the X chromosome that occurred about 8–12 Ma (Tamura et al. 2004; Richards et al. 2005; Gurbich and Bachtrog 2008; Bachtrog et al. 2009). We find that there is a positive correlation between the degree of male-biased expression and the rate of protein evolution in both lineages. Genes with conserved male-biased expression between the lineages show the fastest rates of evolution. For both lineages, we observe a fast-X effect that is especially strong for male-biased genes. This effect is greater for the ancestral X chromosome but is also present on the neo-X chromosome of the obscura lineage.
Materials and Methods
Data Sets and Species
For both the melanogaster and the obscura lineage, we extracted male/female (M/F) expression ratios and data on the rate of protein evolution measured by the ratio of nonsynonymous to synonymous substitutions (dN/dS) from the Sebida database (v. 2.0; Gnad and Parsch 2006; http://www.sebida.de). The melanogaster data originate from a meta-analysis of D. melanogaster sex-biased gene expression over several studies (Parisi et al. 2003; Ranz et al. 2003; Gibson et al. 2004; Parisi et al. 2004; McIntyre et al. 2006; Ayroles et al. 2009), while data for the obscura lineage come from a microarray analysis of D. pseudoobscura (Jiang and Machado 2009). All M/F expression ratios were log2 transformed. We excluded genes for which no FlyBase identifier (FBgn) could be associated, as well as genes lacking information on chromosomal location, dN/dS, or expression state. For the melanogaster lineage, dN/dS values come from a comparison of D. melanogaster and D. simulans. For the obscura lineage, dN/dS values come from a comparison of D. pseudoobscura and D. persimilis. In both cases, we excluded genes with dN/dS ≥ 9, as they tend to be unreliable estimates where dS is equal to (or very close to) zero. The final data set contained 12,419 genes (10,437 autosomal and 1,982 X linked) for the melanogaster lineage and 10,118 genes (6,641 autosomal, 1,657 located on the left arm of the X chromosome [XL], and 1,820 located on the right arm of the X chromosome [XR]) for the obscura lineage. Orthologs between D. melanogaster and D. pseudoobscura could be identified for 8,439 genes. In addition, we used expression data from D. ananassae (Zhang et al. 2007) to infer M/F expression ratios of genes of this species. Drosophila ananassae is phylogenetically situated at the split of the melanogaster subgroup from the melanogaster group (Drosophila 12 Genomes Consortium et al. 2007; Larracuente et al. 2008) and was used to infer gains or losses of sex-biased expression within the melanogaster group. Orthologs among all three species could be identified for 5,336 genes.
Analysis of Rates of Evolution with Respect to Degree of Sex-Bias
Correlations between log2(M/F) and the rate of protein evolution (dN/dS) were assessed using the nonparametric Spearman's rank correlation for each lineage. In addition, correlations were determined separately for male-, female-, and unbiased genes. For each lineage, we also ranked all genes by their M/F expression ratio and separated them into five equally sized groups (highly male biased, weakly male biased, unbiased, weakly female biased, and highly female biased). Comparisons among groups were performed using Kruskal–Wallis tests. Significant differences were further investigated using pairwise Mann–Whitney U tests between groups with Bonferroni correction.
Analysis of Rates of Evolution with Respect to Tissue-Specific Expression
We used FlyAtlas (Chintapalli et al. 2007) and the approach of Meisel (2011) to determine the expression breadth of all genes in our D. melanogaster data set. We considered 10 somatic tissues shared by males and females (brain, eye, thoracioabdominal ganglion, salivary gland, crop, midgut, Malpighian tubule, hindgut, heart, and fat body), two male-specific tissues (accessory gland and testis), and two female-specific tissues (spermatheca and ovary). A gene was considered as expressed in a given tissue if its mean microarray signal intensity was ≥100 (Meisel 2011). Since tissue-specific expression data were not available for D. pseudoobscura, we made the assumption that D. pseudoobscura genes shared the expression pattern of their D. melanogaster orthologs. In the end, we were able to assign tissue expression patterns to 11,082 D. melanogaster genes and 7,757 D. pseudoobscura orthologs. For comparisons of evolutionary rates, we separated sex-biased genes (male and female) into two groups: 1) those with expression limited to male (or female) reproductive tissues and 2) those expressed in one or more nonreproductive tissue. To control for the possible accelerated evolution of genes with tissue-specific expression (Haerty et al. 2007; Meisel 2011), we also compared the above genes to a set of unbiased genes that showed expression in only a single somatic tissue.
Inferring the Conservation of Sex-Biased Gene Expression
To examine the conservation of sex-biased gene expression, we investigated the M/F ratios of orthologous genes between the two lineages, melanogaster and obscura. First, we compared the rate of protein evolution of genes with conserved sex-biased expression between D. melanogaster and D. pseudoobscura to that of genes that differed in their sex-bias classification between species. We divided the set of genes into nine categories according to their expression conservation (see supplementary table 1, Supplementary Material online). Second, we analyzed the conservation of degree of sex bias between these orthologs for male- and female-biased genes. We ranked the sex-biased genes according to their degree of sex-bias within each species and compared the overlap of genes between the species for the top 10% and top 25% of genes. Subsequently, data from D. ananassae were used to infer changes in sex-biased gene expression along the phylogeny. Each category from the above analysis (supplementary table 1, Supplementary Material online) can be subdivided into three groups (male-, female-, or unbiased) according to the expression state in D. ananassae. Thus, the expression pattern in the three species can be separated into 27 groups (see supplementary table 2, Supplementary Material online). A gain or loss of sex-biased gene expression along the phylogeny was inferred using parsimony. First, we compared the groups with gene expression states conserved over all three species. Second, we performed comparisons between groups of genes with conserved sex-biased gene expression and those that differed in sex-bias classification among species. All comparisons were performed using pairwise Mann–Whitney U tests between groups with Bonferroni correction and were carried out on the complete data sets and on autosomal and X-linked genes separately. Spearman rank correlations were used to assess the correlation between rates of protein evolution of orthologs on the two lineages, as well as the correlation between M/F expression ratios on the two lineages.
Influence of Chromosomal Location and Gene Conservation
To test for a possible “fast-X” effect, we compared the rates of evolution (measured as dN/dS) between autosomal and X-linked genes. For the obscura lineage, X-linked genes were further divided into genes situated on the XR and genes situated on the XL. Genes on XR originate from a recent (∼8–12 Ma) X-autosome fusion in the obscura clade and are generally located on the autosomal arm 3L in D. melanogaster. Differences between groups were determined using Kruskal–Wallis tests and subsequent pairwise Mann–Whitney U tests with Bonferroni correction. To test for lineage-specific differences and the influence of homologous gene conservation between lineages, we also performed the above analyses on the set of orthologous genes between the melanogaster and obscura lineages. All statistical analyses were performed using R 2.10.1 (R Development Core Team 2010).
Results
Correlation between Degree of Male-Bias and Rate of Protein Evolution
For both the melanogaster and the obscura lineage, there was a significant positive correlation between the rate of protein evolution (dN/dS) and the ratio of male-to-female (M/F) expression (Spearman's rank correlation, melanogaster: ρ = 0.16, P < 0.001; obscura: ρ = 0.17, P < 0.001). This was true for the complete set of genes, as well as the autosomal and X-linked genes considered separately (melanogaster autosomes: ρ = 0.17, P < 0.001; obscura autosomes: ρ = 0.17, P < 0.001; melanogaster X: ρ = 0.18, P < 0.001; obscura X: ρ = 0.25, P = 0.03). Particularly for the melanogaster lineage, the above correlations were much stronger for male-biased genes than for female- and unbiased genes (table 1).
Table 1.
Correlations between dN/dS and M/F Expression Ratio
| Genesa | Chromosome |
Melanogaster Lineage |
Obscura Lineage |
||||
| Nb | ρc | Pd | Nb | ρc | Pd | ||
| All | All | 12,419 | 0.16 | <0.001 | 10,118 | 0.17 | <0.001 |
| Auto | 10,437 | 0.17 | <0.001 | 6,641 | 0.17 | <0.001 | |
| X (or XL) | 1,982 | 0.18 | <0.001 | 1,657 | 0.25 | <0.001 | |
| XR | — | — | — | 1,820 | 0.17 | <0.001 | |
| M | All | 3,381 | 0.34 | <0.001 | 3,295 | 0.07 | <0.001 |
| Auto | 2,350 | 0.30 | <0.001 | 2,350 | 0.07 | <0.001 | |
| X (or XL) | 395 | 0.38 | <0.001 | 470 | 0.15 | <0.001 | |
| XR | — | — | — | 475 | 0.07 | 0.11 | |
| F | All | 4,983 | −0.01 | 0.31 | 4,761 | 0.07 | <0.001 |
| Auto | 4,053 | −0.01 | 0.52 | 2,954 | 0.07 | <0.001 | |
| X (or XL) | 929 | −0.03 | 0.42 | 840 | 0.06 | 0.09 | |
| XR | — | — | — | 967 | 0.08 | 0.01 | |
| U | All | 4,055 | −0.01 | 0.50 | 2,062 | 0.01 | 0.52 |
| Auto | 3,397 | −0.01 | 0.79 | 1,337 | 0.02 | 0.55 | |
| X (or XL) | 658 | −0.03 | 0.39 | 347 | 0.01 | 0.81 | |
| XR | — | — | — | 378 | 0.02 | 0.75 | |
“M” indicates male-biased genes, “F” indicates female-biased genes, and “U” indicates unbiased genes.
Number of genes.
Spearman's ρ.
As determined by Spearman's rank correlation.
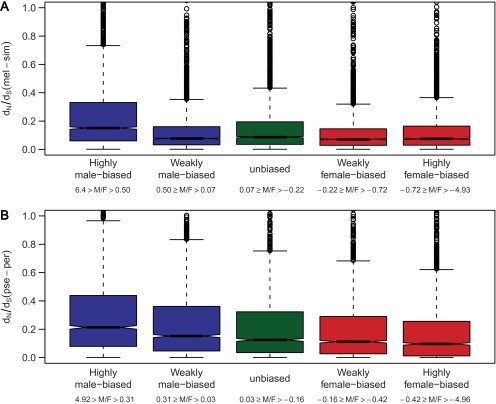
When the genes were placed into categories according to their type and degree of sex-biased expression, a general pattern of faster evolution for highly male-biased genes was evident (fig. 1). This pattern held for both autosomal and X-linked genes (supplementary table 3, Supplementary Material online).
FIG. 1.—
Evolutionary rates of male-, female-, and unbiased genes. Genes were ranked according to M/F expression ratios and divided into five equally sized groups for each lineage. The M/F cutoffs (on a log2 scale) used to create these groups are given in parentheses. Panel (A) shows comparisons for the melanogaster lineage. Each group contains 2,483 or 2,484 genes. Panel (B) shows comparisons for the obscura lineage. Each group contains 2,023 or 2,024 genes. The heavy horizontal line in each box indicates the median, with notches at the side indicating its 95% confidence interval. The edges of each box represent the bounds of the upper and lower quartiles, that is, the box shows the interquartile range. The dotted lines (“whiskers”) on either side of the box indicate the adjacent values. The upper (lower) adjacent value is the value of the largest (smallest) observation that is less (greater) than or equal to the upper (lower) quartile plus 1.5 times the length of the interquartile range. For both lineages, highly male-biased genes had significantly higher dN/dS than genes of all other categories (Mann–Whitney U test, P < 0.001 in all cases). Significance levels for all comparisons are given in supplementary table 3 (Supplementary Material online).
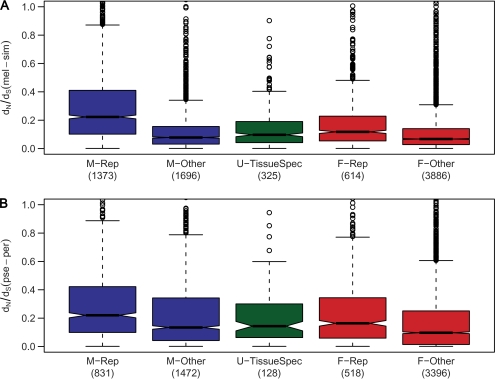
Previous studies have shown that the vast majority of sex-biased genes differ in expression between male and female reproductive tissues (Parisi et al. 2003) and that reproductive tissue-expressed genes show an accelerated rate of evolution relative to sex-biased genes expressed in nonreproductive tissues (Meisel 2011). We could confirm this pattern in our data set (fig. 2). Although tissue-specific genes are known to evolve faster than those with broad expression patterns (Haerty et al. 2007; Meisel 2011), this cannot completely explain our observations, as genes expressed in male or female reproductive tissues had significantly faster rates of evolution than unbiased genes that were expressed in a single somatic tissue (fig. 2A). Because tissue-specific expression has not been systematically analyzed in D. pseudoobscura, we could not directly test for its effects on evolutionary rate. However, under the assumption that D. pseudoobscura genes share the expression pattern of their D. melanogaster orthologs, we find that the above results also hold for the obscura lineage (fig. 2B). Note that this approach may exclude a disproportionate number of male-biased or male reproductive tissue-expressed genes, as they tend to show high turnover between species (Zhang et al. 2004; Levine et al. 2006; Baines et al. 2008). Despite this limitation, we see remarkable similarity between the two lineages, especially for male reproductive genes (fig. 2).
FIG. 2.—
Evolutionary rates of reproductive and nonreproductive tissue-specific genes. Genes expressed specifically in male or female reproductive tissues are labeled as M-Rep and F-Rep, respectively. Genes with male- or female-biased expression that is not limited to reproductive tissues are labeled M-Other and F-Other, respectively. Genes with unbiased expression that is limited to a single somatic tissue are labeled U-TissueSpec. Panel (A) displays results for the melanogaster lineage. Panel (B) displays results for the obscura lineage. Genes with expression limited to male-reproductive tissues show significantly faster rates of evolution than all other groups (Mann–Whitney U test, P < 0.001 in all cases). Genes with expression limited to female-reproductive tissues show significantly faster rates of evolution than other female-biased genes for both lineages (P < 0.001). Furthermore, these genes show significantly faster rates of evolution than unbiased genes with tissue-specific expression for the melanogaster lineage (P = 0.009). For the obscura lineage, the same tendency can be observed but is not significant (P = 0.30).
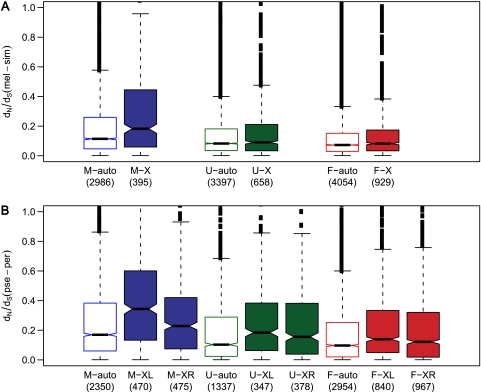
We also compared dN/dS of the X-linked and autosomal genes within each expression class. In all cases, X-linked genes had faster rates of protein evolution than autosomal genes (fig. 3). For male-biased genes, the difference was significant in both lineages (table 2). For the obscura lineage, genes located on the “neo-X” chromosome (XL) consistently showed faster rates of evolution than genes located on XR (fig. 3 and table 2).
FIG. 3.—
Comparison of autosomal and X-linked genes. Panel (A) displays results for the melanogaster lineage. Panel (B) displays results for the obscura lineage. Numbers of genes per group are given in parentheses. “M” indicates male-biased genes, “F” indicates female-biased genes, and “U” indicates unbiased genes. Significance levels for relevant comparisons are given in table 2.
Table 2.
Significance Levels for Comparisons of dN/dS between Autosomal and X-Linked Genes
| Lineage | Expressiona | Comparison | Pb |
| Melanogaster | M | Auto versus X | <0.001* |
| F | Auto versus X | 0.1741 | |
| U | Auto versus X | 0.0245 | |
| Obscura | M | Auto versus XL | <0.001* |
| Auto versus XR | 0.0047* | ||
| XL versus XR | <0.001* | ||
| U | Auto versus XL | <0.001* | |
| Auto versus XR | 0.0013* | ||
| XL versus XR | 0.1812 | ||
| F | Auto versus XL | <0.001* | |
| Auto versus XR | 0.0052* | ||
| XL versus XR | 0.0077 |
“M” indicates male-biased genes, “F” indicates female-biased genes, and “U” indicates unbiased genes.
As determined by Mann–Whitney U tests. Values marked with asterisks are significant after Bonferroni correction.
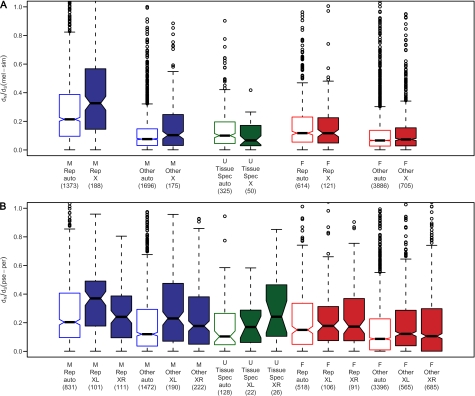
When both sex- and tissue-specific expression patterns were considered, there was a highly significant fast-X effect for male-biased reproductive tissue-specific genes in the melanogaster lineage (fig. 4 and table 3). A weaker fast-X effect was observed for male-biased genes with expression outside of reproductive tissues, but was not significant after correcting for multiple tests (table 3). There was no evidence for faster evolution of X-linked genes with expression limited to female reproductive tissues. In the obscura lineage, male-biased genes located on chromosome arm XL showed a highly significant fast-X effect, regardless of whether their expression was limited to reproductive tissues or not (fig. 4 and table 3). Female-biased genes expressed outside of reproductive tissues located on XL showed significantly faster evolution than autosomal genes (table 3).
FIG. 4.
Comparison of autosomal and X-linked reproductive tissue-specific and nonreproductive tissue-specific genes. Panel (A) displays results for the melanogaster lineage. Panel (B) displays results for the obscura lineage. Numbers of genes per group are given in parentheses. “M” indicates male-biased genes, “F” indicates female-biased genes, and “U” indicates unbiased genes. “Rep” indicates genes with expression limited to male (or female) reproductive tissues, “Other” indicates the remaining male-biased (or female-biased) genes, and “TissueSpec” indicates genes with expression in only a single somatic tissue. Significance levels for relevant comparisons are given in table 3.
Table 3.
Significance Levels for Comparisons of dN/dS between Autosomal and X-Linked Reproductive Tissue-Specific and Nonreproductive Tissue-Specific Genes
| Lineage | Expressiona | Comparison | Pb |
| Melanogaster | M-Rep | Auto versus X | <0.0001* |
| F-Rep | Auto versus X | 0.9904 | |
| U-TissueSpec | Auto versus X | 0.0380 | |
| M-Other | Auto versus X | 0.0201 | |
| F-Other | Auto versus X | 0.0527 | |
| Obscura | M-Rep | Auto versus XL | 0.0001* |
| Auto versus XR | 0.8055 | ||
| XL versus XR | 0.0028* | ||
| M-Other | Auto versus XL | <0.0001* | |
| Auto versus XR | 0.0256 | ||
| XL versus XR | 0.0304 | ||
| U-TissueSpec | Auto versus XL | 0.4508 | |
| Auto versus XR | 0.0106 | ||
| XL versus XR | 0.1501 | ||
| F-Rep | Auto versus XL | 0.5156 | |
| Auto versus XR | 0.2353 | ||
| XL versus XR | 0.6717 | ||
| F-Other | Auto versus XL | <0.0001* | |
| Auto versus XR | 0.0495 | ||
| XL versus XR | 0.0275 |
“M” indicates male-biased genes, “F” indicates female-biased genes, and “U” indicates unbiased genes. M-Rep/F-Rep = reproductive tissue-specific male- or female-biased genes, M-Other/F-Other = all other male- or female-biased genes, U-TissueSpec = tissue-specific genes, but only expressed in nonreproductive tissues.
As determined by Mann–Whitney U tests. Values marked with asterisks are significant after Bonferroni correction.
Influence of Gain/Loss and Conservation of Sex-Biased Expression
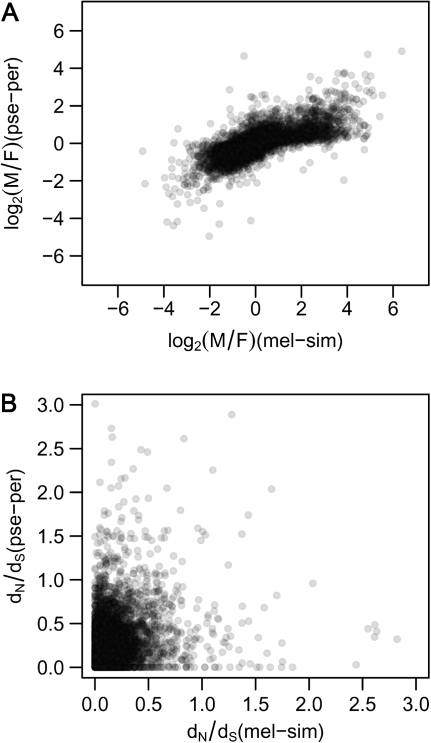
For the set of orthologous genes between D. melanogaster and D. obscura, we determined the conservation of sex-biased gene expression and separated the genes into nine groups (supplementary table 1, Supplementary Material online). There were significant differences in the rate of protein evolution among the nine groups (Kruskal–Wallis test, P < 0.001). We also performed pairwise comparisons of groups using Mann–Whitney U tests with Bonferroni correction. Genes with conserved male-biased expression had significantly higher dN/dS values than genes of all other groups, whereas there was typically no significant difference between genes with male-biased expression in only one species (supplementary table 4, Supplementary Material online). Furthermore, conserved female-biased genes did not show differences in the rate of evolution compared with genes with female-biased gene expression in only one species (supplementary table 4, Supplementary Material online). There was a significant positive correlation between the M/F ratios for the orthologous genes of the two lineages (Spearman's ρ = 0.76, P < 0.001; fig. 5A). However, we observed only moderate conservation of the degree of sex bias for both male- and female-biased genes. For each lineage, genes with sex-biased expression (1,681 male-biased genes and 3,390 female-biased genes) were ranked according to their degree of sex bias and the overlap between the top 10% and top 25% of genes in each species was determined. The proportion of genes that were in the top 10% for the melanogaster lineage and also in the top 10% for the obscura lineage was 41% for male-biased genes and 45% for female-biased genes. For the top 25% of genes, the overlap was 54% for the male-biased genes and 50% for the female-biased genes.
FIG. 5.
Correlation of M/F expression ratios and evolutionary rates between lineages. Orthologous genes between the melanogaster and the obscura lineage were compared. Spearman rank correlations were used to determine correlations between M/F expression ratios and evolutionary rates for both lineages. Panel (A) displays the relationship between M/F expression ratios (ρ = 0.76, P < 0.001). Panel (B) displays the relationship between evolutionary rates measured by dN/dS (ρ = 0.37, P < 0.001). For clarity, 22 points lying outside the boundaries of the x and y axes (dN/dS > 3.0) are not shown.
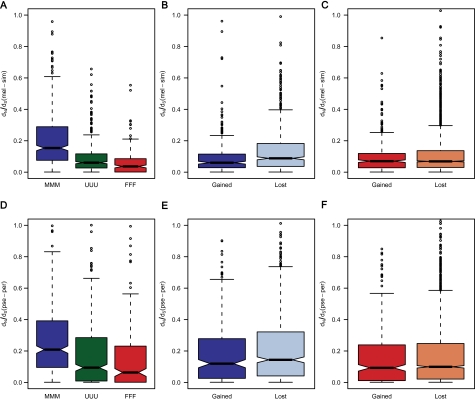
In addition, we investigated whether or not the rate of evolution of sex-biased genes was affected by gain or loss of sex-biased gene expression along the Drosophilid phylogeny. Estimates of sex-biased gene expression in D. ananassae were used to infer gains or losses of sex-biased expression within the melanogaster group. This allowed the genes to be separated into 27 groups (supplementary table 2, Supplementary Material online) according to sex-bias conservation. There were significant differences among the groups in both lineages (Kruskal–Wallis test, P < 0.001). Consistent with previous studies (Metta et al. 2006; Baines et al. 2008), genes with conserved male-biased expression showed significantly higher rates of evolution than female-biased or unbiased genes (fig. 6A and D). However, some groups only consisted of very few genes and showed high variation in dN/dS.
FIG. 6.—
Comparison of orthologous genes among D. melanogaster, D. ananassae, and D. pseudoobscura. Panels (A–C) display results using dN/dS between D. melanogaster and D. simulans. Panels (D–F) display results using dN/dS values between D. pseudoobscura and D. persimilis. “M” indicates male-biased genes, “F” indicates female-biased genes, and “U” indicates unbiased genes. Panels (A) and (D) show a comparison of the genes with conserved expression states. The first letter indicates the expression state in D. melanogaster, the second letter indicates the expression state in D. ananassae, and the third letter indicates the expression state in D. pseudoobscura. There were significant differences among the groups in both lineages (Kruskal–Wallis test, P < 0.001) and all pairwise comparisons between groups revealed significant differences (Mann–Whitney U test, P < 0.001 in all cases). Panels (B) and (E) show results for genes that have recently gained male-biased expression (UMU, MUU) compared with genes that have recently lost male-biased expression (UMM, MUM). Genes that recently lost male-biased expression showed higher rates of evolution in both comparisons (P < 0.001 for melanogaster; P = 0.06 for obscura). Panels (C) and (F) display the equivalent results for female-biased genes. There are no significant differences between the groups (P = 0.34 for melanogaster; P = 0.30 for obscura).
Because some of the above groups contained few genes, we focused on the comparison of genes that recently gained sex-biased expression to those that recently lost sex-biased expression. For both male- and female-biased genes, we compared genes that gained sex-biased expression in either D. melanogaster or D. ananassae (UMU, MUU and UFU, FUU) with genes that lost sex-biased expression in one of the two species (UMM, MUM and UFF, FUF). For male-biased genes, genes that recently lost male-biased expression showed higher rates of evolution in both lineages (Mann–Whitney U test, P < 0.001 for melanogaster; P = 0.06 for obscura, fig. 6B and E). However, there are no significant differences between the two groups for genes with female-biased expression (Mann–Whitney U test, P = 0.34 for melanogaster; P = 0.30 for obscura, fig. 6C and F).
The Influence of Sex-Linkage on the Rate of Protein Evolution
Of the 8,439 orthologous genes shared between the melanogaster and the obscura lineages, 5,509 were autosomal in both lineages and 1,227 genes were X-linked in both lineages (i.e., located on the X chromosome in melanogaster and on XL in obscura). A total of 1,527 genes were located on the “neo-X” chromosome in D. pseudoobscura (XR) and on the homologous autosomal arm 3L in D. melanogaster. In addition to these, 88 genes were autosomal in D. melanogaster and located on XL in D. pseudoobscura, 56 genes were X-linked in D. melanogaster and autosomal in D. pseudoobscura, 11 genes were X-linked in D. melanogaster and located on XR in D. pseudoobscura, and 21 genes were autosomal in D. melanogaster (but not on 3L) and located on XR in D. pseudoobscura.
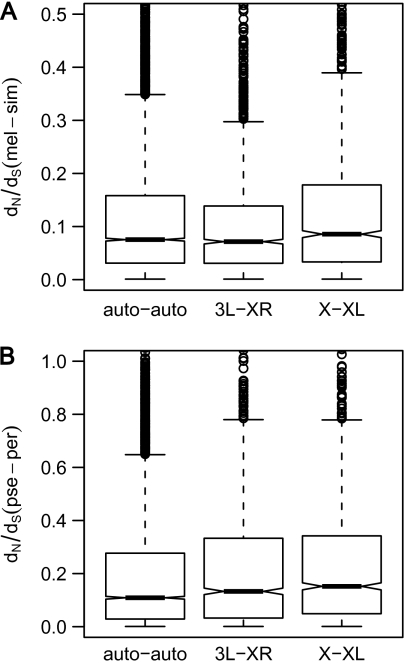
We compared rates of evolution among three groups of orthologous genes: 1) genes that were autosomal in both lineages (auto-auto), 2) genes that were X-linked in both lineages (X-XL), and 3) genes that were located on 3L in melanogaster and on XR in obscura (3L-XR). Kruskal–Wallis tests revealed significant differences among these three groups in both lineages (P < 0.001 in both cases). For the melanogaster lineage, X-linked genes showed significantly higher dN/dS values than 3L-XR genes (Mann–Whitney U test, P < 0.001). After Bonferroni correction, there were no significant differences between either autosomal and X-linked or autosomal and 3L-XR genes (Mann–Whitney U test, P = 0.02 in both cases, fig. 7A). However, when we pooled autosomal and 3L-XR genes into one group, the X-linked genes again had significantly faster rates of protein evolution (Mann–Whitney test, P < 0.001). For the obscura lineage, autosomal genes showed the slowest rates of evolution, followed by genes located on 3L-XR and X-linked genes (fig. 7B). The differences between groups were significant for the two comparisons involving autosomal genes (Mann–Whitney U test, auto-auto vs. 3L-XR: P = 0.0013; auto-auto vs. X-linked: P < 0.001). When we considered genes that were autosomal in the melanogaster lineage and located on the “neo-X” in the obscura lineage (3L-XR), rates of molecular evolution were significantly higher for the obscura lineage, again indicating faster-X evolution (Mann–Whitney U test, P < 0.001). In accordance with previous findings (Sturgill et al. 2007; Jiang and Machado 2009), there is a general paucity of male-biased genes on XR relative to the autosomes (χ2 test, P < 0.001).
FIG. 7.—
Comparison of genes with conserved chromosomal location and genes located on 3L-XR. Panel (A) displays results using dN/dS between D. melanogaster and D. simulans. Panel (B) displays results using dN/dS values between D. pseudoobscura and D. persimilis. “Auto-auto” indicates genes with conserved autosomal location, “3L-XR” indicates genes located on 3L and XR, respectively, and “X-XL” indicates genes with conserved X-linkage. There were significant differences among the groups in both lineages (Kruskal–Wallis test, P < 0.001). For all pairwise tests, Bonferroni correction has been applied. For the melanogaster lineage, genes with conserved X-linkage show significantly higher dN/dS than genes located on 3L-XR (Mann–Whitney U test, P < 0.001). There are no significant differences between auto-auto and 3L-XR or between auto-auto and X-XL (P = 0.02 in both cases) (Panel A). For the obscura lineage, the differences were significant for the two comparisons involving conserved autosomal genes (auto-auto vs. 3L-XR: P = 0.0013; auto-auto vs. X-XL: P < 0.001). There is no significant difference between genes with conserved X-linkage and genes located on 3L-XR (P = 0.02) (Panel B).
Comparison of Orthologous Genes
Because our previous assessment of the correlation between dN/dS and the M/F expression ratio treated each lineage independently, the number and identity of genes were not the same in the two lineages. This is because some genes did not have clear one-to-one orthologs between lineages or expression data were lacking for some genes in one of the lineages. In order to utilize a common set of genes in the two lineages, we repeated the analysis using the set of orthologous genes for which expression data were available for both lineages. Overall, there was a positive correlation between the M/F ratios of orthologs in the two lineages (Spearman's ρ = 0.76, P < 0.001; fig. 5A). There was also a positive correlation between the rates of protein evolution of orthologous genes in the two lineages (Spearman's ρ = 0.37, P < 0.001; fig. 5B), which was robust to different dS and dN/dS cutoffs (supplementary table 5, Supplementary Material online). The correlation between M/F and dN/dS was significant for both lineages (melanogaster: ρ = 0.12, P < 0.001; obscura: ρ = 0.14, P < 0.001, see also supplementary table 6, Supplementary Material online).
In addition, we looked at orthologous genes that showed conservation of chromosomal location and those that showed conserved expression state between the two lineages but not necessarily conservation of chromosomal location. Again, the positive correlations were significant for both lineages and were particularly pronounced for male-biased genes in the melanogaster lineage (supplementary table 6http://www.gbe.oxfordjournals.org/lookup/suppl/doi:10.1093/gbe/evs012/-/DC1, Supplementary Material online). This indicates that there are differences in sex-biased gene evolution between lineages. We performed group-based comparisons for both the sets of all orthologs between the two lineages and for all orthologs with conserved expression state (MM, FF, and UU, respectively). We ordered the genes according to their M/F expression ratios and divided the sets into five equally sized groups. For both data sets, the observed pattern is consistent with what we found before for all genes in the two lineages (supplementary table 7, Supplementary Material online). The rate of evolution (dN/dS) differed significantly among the expression groups (Kruskal–Wallis test, P < 0.001 in all cases) and there was a general pattern of faster evolution for genes with a higher degree of male-biased expression but not for genes with a higher degree of female-biased expression (supplementary table 7, Supplementary Material online).
Discussion
By investigating the molecular evolution of sex-biased genes in two independent distantly related Drosophilid lineages, we have been able to uncover some common features of sex-biased gene evolution, as well as features that differ between lineages. Consistent with a previous study of the melanogaster lineage (Meisel 2011), we find that the rate of protein evolution (dN/dS) is positively correlated with the ratio of male-to-female gene expression (M/F). This correlation is mainly driven by male-biased genes, which show the strongest correlation between dN/dS and M/F in both lineages (table 1). Furthermore, highly male-biased genes and those expressed specifically in reproductive tissues show significantly higher dN/dS than all other categories of genes in both lineages (figs. 1 and 2). We also find that orthologous genes that show conserved male-biased expression in both lineages show the fastest rates of protein evolution (fig. 6).
For female-biased genes there is a slightly negative, but nonsignificant, correlation between dN/dS and M/F in the melanogaster lineage (table 1). In other words, genes with highly female-biased expression tend to evolve faster than those with weakly female-biased expression. This agrees qualitatively with Meisel (2011) and our categorical analysis of sex-biased genes (fig. 1A). The negative correlation between dN/dS and M/F is much stronger when we consider only genes that are expressed exclusively in female reproductive tissues (ρ = −0.11, P = 0.008) and these genes show a stronger female-bias in expression than those that are expressed in other tissues (median log2(M/F) values of −1.25 and −0.66, respectively; Mann–Whitney U test, P < 0.001). However, in the obscura lineage, there is a positive correlation between dN/dS and M/F for female-biased genes (table 1), suggesting that highly female-biased genes evolve more slowly than weakly female-biased genes in this lineage.
Our findings can explain some previous observations about sex-biased gene evolution in the obscura and ananassae groups. For example, Grath et al. (2009) found that a sample of genes with male-biased expression in D. ananassae did not show accelerated rates of protein evolution in the ananassae lineage, although those that showed conserved male-biased expression between D. ananassae and D. melanogaster did. The lack of an observed accelerated rate of evolution for D. ananassae male-biased genes may be attributable to the fact that the examined genes were not chosen because they showed strong male-biased expression in D. ananassae, but instead were chosen because they were highly male-biased in D. melanogaster (Pröschel et al. 2006; Baines et al. 2008). Because the degree of sex-biased expression is not strongly correlated between lineages, the male-biased genes that were examined were not those with very high levels of male-biased expression D. ananassae (Grath et al. 2009). Since it is the highly male-biased genes that show the fastest rate of evolution, their absence from the D. ananassae data set may explain why an increased rate of molecular evolution was not detected on the ananassae lineage.
An early study of sex-biased gene expression in D. pseudoobscura found that genes with conserved male-biased expression between D. pseudoobscura and D. melanogaster showed accelerated rates of protein evolution, but those with male-biased expression only in D. pseudoobscura did not (Metta et al. 2006). In this case, the expression was determined by SAGE (serial analysis of gene expression) in D. pseudoobscura and, given the limited depth of SAGE sequencing, would be expected to identify genes with highly male-biased expression. However, because the correlation between M/F and dN/dS for male-biased genes is weaker in the obscura lineage than the melanogaster lineage (table 1), it may be that the limited sample size of Metta et al. (2006) prevented the detection of a difference in molecular evolutionary rate among groups of genes. Consistent with this, a recent study using whole-genome microarrays and comparative genomics of D. pseudoobscura and D. persimilis detected a significantly elevated rate of protein evolution for male-biased genes (Jiang and Machado 2009).
It is likely that estimates of dN/dS between the closely related D. pseudoobscura and D. persimilis are inflated by the presence of shared ancestral polymorphism (Machado et al. 2002; Machado et al. 2007; Noor et al. 2007; Kulathinal et al. 2009). However, even if it inflates overall divergence, the presence of ancestral polymorphism is unlikely to affect our conclusions. This is because our analyses were performed on groups of genes (male-, female-, and unbiased) compared between the same two species. For the elevated dN/dS of male-biased genes to be explained by shared ancestral polymorphism, there would have to be more shared polymorphism in male-biased genes than in the other groups of genes. This would require either an overall elevation in nonsynonymous polymorphism in male-biased genes, or balancing selection maintaining more ancestral nonsynonymous polymorphism in male-biased genes. Neither of these possibilities is supported by observations in the melanogaster or ananassae lineages, where divergence has been calculated between more distantly related species (Pröschel et al. 2006; Baines et al. 2008; Grath et al. 2009), although they cannot be ruled out in the obscura lineage with the data presently at hand.
Another common pattern that we observed is that X-linked genes showed elevated dN/dS relative to autosomal genes (fig. 3 and table 2). This is consistent with previous findings in the melanogaster lineage (Baines et al. 2008) and extends the observation to the obscura lineage. A previous study did not find evidence for faster-X evolution in these two lineages (Thornton et al. 2006). However, that study had several limitations: 1) only sex-biased expression data from D. melanogaster were used, 2) a complete genome sequence from a second species of the obscura lineage was not available, and 3) only genes that could be identified as conserved orthologs between D. melanogaster and D. pseudoobscura were analyzed. All of these factors, but especially the last one, contribute to the inability to detect a significant fast-X effect (Baines et al. 2008).
In principle, faster-X evolution could result from genetic drift having a greater impact on the X chromosome than on the autosomes. This would be expected if the effective population size of the X chromosome was smaller than that of the autosomes, allowing a higher proportion of neutral (or slightly deleterious) mutations to become fixed on the X chromosome (Vicoso and Charlesworth 2009; Mank et al. 2010). However, in Drosophila there appears to be little difference in the effective population sizes of the X chromosome and the autosomes, except in recently derived populations (Andolfatto 2001; Kauer et al. 2002; Hutter et al. 2007; Parsch et al. 2009). Furthermore, there is abundant evidence across multiple Drosophila lineages that positive selection is the predominant force driving protein divergence between species (Sella et al. 2009; Wilson et al. 2011) and the influence of positive selection appears to be greater on the X chromosome than on the autosomes (Baines et al. 2008; Müller et al. 2012).
In both the melanogaster and obscura lineages, the fast-X effect was strongest for male-biased genes (fig. 3 and table 2). If faster-X evolution is mainly driven by an increased rate of adaptive substitution on the X chromosome due to the efficient selection of recessive mutations in hemizygous males (Charlesworth et al. 1987), then this pattern should be expected for two reasons. First, male-biased genes tend to show the highest rates of adaptive evolution between species (Pröschel et al. 2006), indicating that a larger fraction of amino acid replacements in male-biased genes could be potential targets of positive selection. Second, male-biased genes are expected to experience selection mainly in a male (hemizygous) genetic background where recessive X-linked mutations are immediately exposed to selection. Given the strong correlation between M/F and dN/dS observed for male-biased genes, a fast-X effect could also be caused by X-linked genes having stronger male-biased expression than autosomal genes. However, this is not the case, as autosomal male-biased genes show higher M/F than X-linked male-biased genes on both lineages (Mann–Whitney U test, P < 0.05 in both lineages; see also supplementary table 8, Supplementary Material online).
Unbiased genes typically will be subject to selection in both sexes and thus, will sometimes encounter selection in a hemizygous background. This may explain why unbiased genes show a fast-X effect that is smaller than that of male-biased genes, but still significant, in both the melanogaster and obscura lineages (table 2). Female-biased genes, in contrast, should mainly encounter selection in the female genetic background, where X-linked recessive mutations have no fixation advantage over autosomal recessive mutations. This may explain why a fast-X effect is not observed for female-biased genes on the melanogaster lineage (table 2). However, female-biased genes do show a significant fast-X effect in the obscura lineage (table 2). We investigated if this could be a result of X-linked or female-biased genes having lower rates of synonymous divergence (dS). Whereas genes located on XL have higher dS than genes located on XR or on the autosomes (see below), there is indeed lower synonymous divergence for female-biased genes in comparison to both male- and unbiased (Mann–Whitney U test, P value < 0.001 in both cases). This relative reduction in dS may result from there being greater constraint on synonymous codon usage for female-biased genes (Hambuch and Parsch 2005; Singh et al. 2005), possibly because they show greater expression breadth than male-biased or unbiased genes (Meisel 2011). However, differences in dS cannot explain the observed fast-X effect, as X-linked female-biased genes have higher dS than autosomal female-biased genes (Mann–Whitney U test, P < 0.001).
In the obscura lineage, the fast-X effect is strongest for comparisons between the autosomes and XL (the ancestral X chromosome) but is also evident in comparisons between the autosomes and XR (the neo-X chromosome; table 2). This cannot be explained by a reduction in dS on chromosome arm XL, as dS is significantly higher for XL than for XR and all autosomes (Kruskal–Wallis test, P < 0.001, pairwise Mann–Whitney U test, P < 0.001 in all cases). Instead, the large fast-X effect observed for XL is caused by elevated nonsynonymous divergence. This is consistent with there being a long-term pattern of faster-X evolution and not a brief burst of accelerated evolution following the shift from autosomal to X-linkage.
In summary, the combination of transcriptomic and comparative genomic data has allowed us to investigate patterns of sex-biased gene evolution across Drosophila lineages that diverged up to 50 Ma. In both the melanogaster and obscura lineages, we observed an accelerated rate of protein evolution for male-biased genes, especially those expressed in reproductive tissues, and a positive correlation between the degree of male-biased expression and dN/dS. For male-biased genes, this correlation is stronger in the melanogaster lineage than in the obscura lineage. The fastest evolving genes are those that show conserved male-biased expression between lineages. These findings can explain some differences between lineages observed in studies that used smaller data sets. The separation of genes into sex-biased expression groups also reveals a fast-X effect that is particularly pronounced for male-biased genes, as would be expected if positive selection acts on recessive X-linked mutations in hemizygous males. In the pseudoobscura lineage, a fast-X effect is also observed for male-biased genes on the neo-X chromosome, although the effect is smaller than that for genes on the ancestral X chromosome. This suggests that long-term X-linkage promotes rapid adaptive evolution of genes expressed predominantly in males.
Supplementary Material
Supplementary tables S1–S8 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank C. A. Machado and Y. Zhang for providing data and two anonymous reviewers for their constructive comments on the manuscript. This work was supported by Deutsche Forschungsgemeinschaft grants PA 903/4-1 and PA 903/6-1.
References
- Andolfatto P. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol Biol Evol. 2001;18:279–290. doi: 10.1093/oxfordjournals.molbev.a003804. [DOI] [PubMed] [Google Scholar]
- Ayroles JF, et al. System genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Jensen JJ, Zhang Z. Accelerated adaptive evolution on a newly formed X chromosome. PLoS Biol. 2009;7:e82. doi: 10.1371/journal.pbio.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines JF, Sawyer SA, Hartl DL, Parsch J. Effects of X-linkage and sex-biased gene expression on the rate of adaptive protein evolution in Drosophila. Mol Biol Evol. 2008;25:1639–1650. doi: 10.1093/molbev/msn111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman CM, et al. Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol. 2002;3:research0086. doi: 10.1186/gb-2002-3-12-research0086. 1–0086.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 1987;130:113–146. [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Connallon T, Clarke AG. Association between sex-biased gene expression and mutations with sex-specific phenotypic consequences in Drosophila. Genome Biol Evol. 2011;3:151–155. doi: 10.1093/gbe/evr004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- Gibson G, et al. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics. 2004;167:1791–1799. doi: 10.1534/genetics.104.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Parsch J. Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics. 2006;22:2577–2579. doi: 10.1093/bioinformatics/btl422. [DOI] [PubMed] [Google Scholar]
- Grath S, Baines JF, Parsch J. Molecular evolution of sex-biased genes in the Drosophila ananassae subgroup. BMC Evol Biol. 2009;9:219. doi: 10.1186/1471-2148-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurbich TA, Bachtrog D. Gene content evolution on the X chromosome. Curr Opin Genet Dev. 2008;18:493–498. doi: 10.1016/j.gde.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty E, et al. Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics. 2007;177:1321–1335. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambuch TM, Parsch J. Patterns of synonymous codon usage in Drosophila melanogaster genes with sex-biased expression. Genetics. 2005;170:1691–1700. doi: 10.1534/genetics.104.038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter S, Li H, Beisswanger S, De Lorenzo D, Stephan W. Distinctly different sex ratios in African and European populations of Drosophila melanogaster inferred from chromosomewide single nucleotide polymorphism data. Genetics. 2007;177:469–480. doi: 10.1534/genetics.107.074922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z-F, Machado CA. Evolution of sex-dependent gene expression in three recently diverged species of Drosophila. Genetics. 2009;183:1175–1185. doi: 10.1534/genetics.109.105775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov T, Messer PW, Petrov DA. Evidence that adaptation in Drosophila is not limited by mutation at single sites. PLoS Genet. 2010;6:e1000924. doi: 10.1371/journal.pgen.1000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer M, Zangerl B, Dieringer D, Schlötterer C. Chromosomal patterns of microsatellite variability contrast sharply in African and non-African populations of Drosophila melanogaster. Genetics. 2002;160:247–256. doi: 10.1093/genetics/160.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MA. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 2009;5:e1000550. doi: 10.1371/journal.pgen.1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente AM, et al. Evolution of protein-coding genes in Drosophila. Trends Genet. 2008;24:114–123. doi: 10.1016/j.tig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci U S A. 2006;103:9935–9939. doi: 10.1073/pnas.0509809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Haselkorn TS, Noor MA. Evaluation of the genomic extent of effects of fixed inversion differences on intraspecific variation and interspecific gene flow in Drosophila pseudoobscura and D. persimilis. Genetics. 2007;175:1289–1306. doi: 10.1534/genetics.106.064758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Kliman RM, Markert JA, Hey J. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol Biol Evol. 2002;19:472–488. doi: 10.1093/oxfordjournals.molbev.a004103. [DOI] [PubMed] [Google Scholar]
- Mank JE, Nam K, Ellegren H. Faster-Z evolution is predominantly due to genetic drift. Mol Biol Evol. 2010;27:661–670. doi: 10.1093/molbev/msp282. [DOI] [PubMed] [Google Scholar]
- Mank JE, Vicoso B, Berlin S, Charlesworth B. Effective population size and the faster-X effect: empirical results and their interpretation. Evolution. 2009;64:663–674. doi: 10.1111/j.1558-5646.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- McIntyre LM, et al. Sex-specific expression of alternative transcripts in Drosophila. Genome Biol. 2006;7:R79. doi: 10.1186/gb-2006-7-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP. Towards a more nuanced understanding of the relationship between sex-biased gene expression and rates of protein coding sequence evolution. Mol Biol Evol. 2011;28:1893–1900. doi: 10.1093/molbev/msr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metta M, Gudavalli R, Gibert J-M, Schlötterer C. No accelerated rate of protein evolution in male-biased Drosophila pseudoobscura genes. Genetics. 2006;174:411–420. doi: 10.1534/genetics.106.057414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller L, Grath S, von Heckel K, Parsch J. Inter- and intraspecific variation in Drosophila genes with sex-biased expression. Int J Evol Biol. 2012;2012:963976. doi: 10.1155/2012/963976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Garfield DA, Schaeffer SW, Machado CA. Divergence between the Drosophila pseudoobscura and D. persimilis genome sequences in relation to chromosomal inversions. Genetics. 2007;177:1417–1428. doi: 10.1534/genetics.107.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, Betancourt AJ. Haldane's sieve and adaptation from the standing genetic variation. Genetics. 2001;157:875–884. doi: 10.1093/genetics/157.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, et al. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, et al. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 2004;5:R40. doi: 10.1186/gb-2004-5-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J, Zhang Z, Baines JF. The influence of demography and weak selection on the McDonald-Kreitman test: an empirical study in Drosophila. Mol Biol Evol. 2009;26:691–698. doi: 10.1093/molbev/msn297. [DOI] [PubMed] [Google Scholar]
- Pröschel M, Zhang Z, Parsch J. Widespread adaptive evolution of Drosophila genes with sex-biased expression. Genetics. 2006;174:893–900. doi: 10.1534/genetics.106.058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing [Internet]. Vienna (Austria): R Foundation for Statistical Computing. 2010. [cited 2012 Feb 16]. Available from: http://www.R-project.org/. [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Mol Biol Evol. 2003;13:735–748. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- Richards S, et al. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridout KE, Dixon CJ, Filatov DA. Positive selection differs between protein secondary structure elements in Drosophila. Genome Biol Evol. 2010;2010:166–179. doi: 10.1093/gbe/evq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SA, Parsch J, Zhang Z, Hartl DL. Prevalence of positive selection among nearly neutral amino acid replacements in Drosophila. Proc Natl Acad Sci U S A. 2007;104:6504–6510. doi: 10.1073/pnas.0701572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G, Petrov DA, Przeworski M, Andolfatto P. Pervasive natural selection in the Drosophila genome? PloS Genet. 2009;5:e1000495. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh ND, Davis JC, Petrov DA. X-linked genes evolve higher codon bias in Drosophila and Caenorhabditis. Genetics. 2005;171:145–155. doi: 10.1534/genetics.105.043497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill D, Zhang Y, Parisi M, Oliver B. Demasculinization of X chromosomes in the Drosophila genus. Nature. 2007;450:238–241. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Thornton K, Bachtrog D, Andolfatto P. X chromosomes and autosomes evolve at similar rates in Drosophila: no evidence for faster-X protein evolution. Genome Res. 2006;16:498–504. doi: 10.1101/gr.4447906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. Effective population size and the faster-X effect: an extended model. Evolution. 2009;63:2413–2426. doi: 10.1111/j.1558-5646.2009.00719.x. [DOI] [PubMed] [Google Scholar]
- Wilson DJ, Hernandez RD, Andolfatto P, Przeworski M. A population genetics-phylogenetics approach to inferring natural selection in coding sequences. PLoS Genet. 2011;7:e1002395. doi: 10.1371/journal.pgen.1002395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature. 2007;450:233–237. doi: 10.1038/nature06323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hambuch TM, Parsch J. Molecular evolution of sex-biased genes in Drosophila. Mol Biol Evol. 2004;21:2130–2139. doi: 10.1093/molbev/msh223. [DOI] [PubMed] [Google Scholar]