Abstract
Telomeres, which form the protective ends of eukaryotic chromosomes, are a ubiquitous and conserved structure of eukaryotic genomes but the basic structural unit of most telomeres, a repeated minisatellite motif with the general consensus sequence TnAmGo, may vary between eukaryotic groups. Previous studies on several species of green algae revealed that this group exhibits at least two types of telomeric sequences, a presumably ancestral type shared with land plants (Arabidopsis type, TTTAGGG) and conserved in, for example, Ostreococcus and Chlorella species, and a novel type (Chlamydomonas type, TTTTAGGG) identified in Chlamydomonas reinhardtii. We have employed several methodical approaches to survey the diversity of telomeric sequences in a phylogenetically wide array of green algal species, focusing on the order Chlamydomonadales. Our results support the view that the Arabidopsis-type telomeric sequence is ancestral for green algae and has been conserved in most lineages, including Mamiellophyceae, Chlorodendrophyceae, Trebouxiophyceae, Sphaeropleales, and most Chlamydomonadales. However, within the Chlamydomonadales, at least two independent evolutionary changes to the Chlamydomonas type occurred, specifically in a subgroup of the Reinhardtinia clade (including C. reinhardtii and Volvox carteri) and in the Chloromonadinia clade. Furthermore, a complex structure of telomeric repeats, including a mix of the ancestral Arabidopsis-type motifs and derived motifs identical to the human-type telomeric repeats (TTAGGG), was found in the chlamydomonadalean clades Dunaliellinia and Stephanosphaeria. Our results indicate that telomere evolution in green algae, particularly in the order Chlamydomonadales, is far more dynamic and complex than thought before. General implications of our findings for the mode of telomere evolution are discussed.
Keywords: TRAP, dot-blot hybridization, terminal restriction fragments (TRFs), 18S rDNA phylogeny, telomere evolution, green algae
Introduction
Telomeres are regarded as highly conserved features of eukaryotic genomes. These nucleoprotein structures protect the ends of linear chromosomes and distinguish them from double strand breaks (McClintock 1938). They are typically maintained by a special reverse transcriptase, telomerase, which adds telomeric repeats at chromosome ends to elongate telomeres. Telomerase consists of two subunits, a protein subunit (TERT) and an RNA subunit (TR). Telomeric DNA is formed by tandem repeats of very few variants of minisatellite sequence motifs TnAmGo that are conserved in individual groups of organisms, for example, TTAGGG in vertebrates and fungi (designed here as the human type), TTTAGGG in plants (Arabidopsis type), or TTAGG in insects (Richards and Ausubel 1988; Meyne et al. 1989; Okazaki et al. 1993). The type of minisatellite motif produced by telomerase is directed by a short region inside the TR subunit that serves as a template for telomeric DNA synthesis. Besides telomerase-based telomere maintenance, alternative mechanisms are known, for example, retrotransposons in telomeres of Drosophila or satellite repeats in Chironomus (for review, see Biessmann and Mason 2003). There are also groups of organisms where more types of telomere sequence were described and in some of these, the evolutionary switch points between the types of telomere sequences were identified in their phylogeny (fig. 1). In plants, two lineages were described with an evolutionary change or loss of the telomeric sequence. One is within the Solanaceae family, where the loss of the typical telomeric sequence was observed in a subgroup comprising the genera Cestrum, Vestia, and Sessea, whereas for example, the model Nicotiana and Solanum species retain the ancestral Arabidopsis-type telomeric sequence (Sykorova et al. 2003a). Two switch points were found in telomere evolution in the monocotyledonous plant order Asparagales, where the ancestral Arabidopsis type of the telomeric sequence was replaced by that of the human type in several families (Sykorova et al. 2003b) and later on, during evolution of the Alliaceae family, the human type was lost upon divergence of the genus Allium but remained in the other Alliaceae genera (Sykorova et al. 2006). In these cases, the changes in the telomere sequence variants exhibited a simple phylogenetic pattern, suggesting that the telomere type may be a useful synapomorphic character.
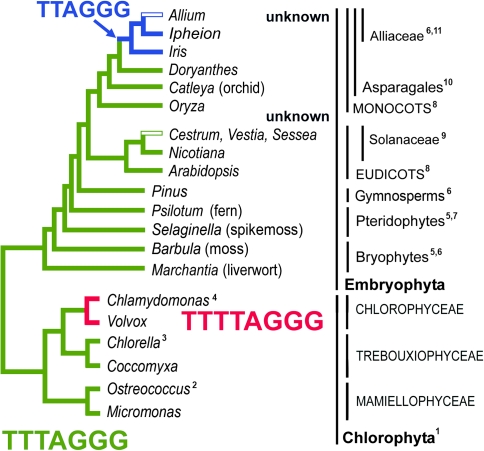
FIG. 1.—
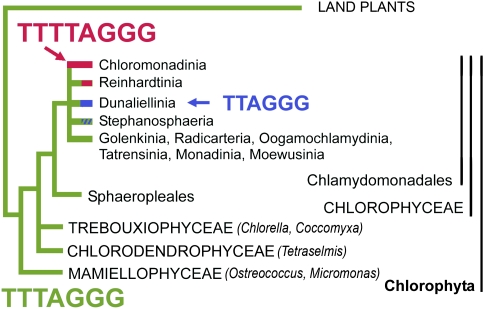
A summary of knowledge about different telomeric types in green algae and plants (Chloroplastida). The ancestral telomeric motif TTTAGGG is present in most green algae and land plants. Variation in this sequence is seen in the monocotyledonous plant order Asparagales (TTTAGGG–TTAGGG). The loss of minisatellite telomeric sequence and its replacement with an unknown sequence has been observed in Solanaceae and Alliaceae. In green algae, the switch to the telomere variant TTTTAGGG has been described in Chlamydomonas reinhardtii. Superscripts indicate references: 1) this work, 2) Derelle et al. (2006), 3) Higashiyama et al. (1995), 4) Petracek et al. (1990), 5) Suzuki (2004), 6) Fuchs et al. (1995), 7) Fuchs and Schubert (1996), 8) Richards and Ausubel (1988), 9) Sykorova et al. (2003a), 10) Sykorova et al. (2003b), 11) Sykorova et al. (2006).
Algae are a heterogeneous group of organisms broadly defined as photosynthetic autotrophic eukaryotes. Genome sequencing projects have enabled description of telomere sequences for diatoms and brown algae (TTAGGG; Armbrust et al. 2004; Bowler et al. 2008; Cock et al. 2010) and red algae (AATGGGGGG; Matsuzaki et al. 2004). Model species of green algae (fig. 1) possess two related telomeric types: a novel type (TTTTAGGG) found in Chlamydomonas reinhardtii, a member of the class Chlorophyceae (Petracek et al. 1990), and a type shared with most land plants (TTTAGGG, Arabidopsis type) documented from Chlorella vulgaris and Chlorella variabilis (class Trebouxiophyceae; Higashiyama et al. 1995; Blanc et al. 2010) and Ostreococcus tauri (class Mamiellophyceae; Derelle et al. 2006). The presence of a unique telomere sequence variant in C. reinhardtii raises the question about when and how in the green algal phylogeny the presumably ancestral Arabidopsis type was replaced by the novel Chlamydomonas type.
Algal telomeres and telomerase have not been subjects of any systematic study, despite their potential to reveal new features of the telomere maintenance system and despite the interesting questions of the evolutionary origin of telomere sequence diversity in different algal subgroups. As a part of a broader project aimed at filling in this substantial gap in our knowledge of eukaryotic genome biology, we employed a combination of approaches to investigate minisatellite telomeric repeats and telomerase activity in chlorophytes with a focus on Chlamydomonadales.
Materials and Methods
Algal Material, Control of Biological Contaminations, and DNA Extraction
The algal material used in this study originated from culture collections as specified in supplementary table S1 (Supplementary Material online). Algae were grown in the recommended liquid media BBM (Bold's Basal Medium) or MASM (Modified Artificial Seawater Medium) (www.ccap.ac.uk/media/pdfrecipes.htm) or on agar plates supplemented with suitable media. Only axenic algal cultures or cultures with only prokaryotic contaminants were accepted for this study. The absence/presence of contaminants was monitored microscopically and using algal cultures grown on LB and BBM agar plates. Genomic DNA for polymerase chain reaction (PCR) amplification of 18S rDNA sequences was isolated using the “modified IRRI” method (Collard et al. 2007) that produces raw DNA suitable, for example, for genotyping as well as for amplification of high copy number sequences. Genomic DNA from a control alga C. vulgaris (TEL01, supplementary table S1, Supplementary Material online) suitable for Southern hybridization and primer extension experiments was isolated by the standard protocol of Dellaporta et al. (1983) or by the CTAB method (Saghai-Maroof et al. 1984). For a wide range of algal samples, obtaining a higher yield of purified genomic DNA proved difficult using classical methods such as CTAB, the Dellaporta protocol, or commercially available DNA purification kits (not shown). Isolation of DNA from the majority of algal samples was thus performed according to a protocol similar to that for preparation of high-molecular-weight samples in agarose plugs (Sykorova et al. 2006). The algae were harvested from liquid cultures or agar plates, spun down, and the pellet was lyophilized. The samples were then incubated overnight at 55 °C in 2 ml tubes with slow rotation in lysis buffer (60 mM Tris–Cl, pH 8.0, 100 mM ethylenediaminetetraacetic acid, 0.5% sodium dodecyl sulfate [SDS]) supplemented with proteinase K (Sigma–Aldrich) to a final concentration of 500 μg/ml. DNA was purified using phenol:chlorophorm extraction and precipitated. Finally, the samples were gently dissolved in TE buffer and RNase A was added to a final concentration of 200 μg/ml. This protocol does not allow quantification of DNA concentration by spectrophotometry, so sample concentrations were estimated from agarose gels by a comparison to a series of standards of known concentration.
Dot-Blot Hybridization, Restriction Digestion, and Southern Hybridization
Genomic DNA samples (1–5 μg) were digested by restriction endonucleases RsaI, AluI, MboI, or TaqI (NEB) and run on a 0.9% agarose gel in TAE buffer. DNA fragments were alkali blotted onto Hybond-XL nylon membrane (Amersham) using a standard protocol (Sambrook et al. 1989) and hybridized with radioactively end-labeled oligonucleotide probes (ATSB, CHSB, HUSB, CASB, TTCAGGG-SB, CHTRTRev2, TTTAGGC-SB, T3AG2-SB, T3G3-SB, supplementary table S2, Supplementary Material online) as described in Sykorova et al. (2003b) with minor modifications according to Neplechova et al. (2005). Briefly, membranes were hybridized at 55 °C for 16 h and washed at 55 °C under low stringency conditions (2 × saline sodium citrate (SSC), 0.1% SDS); the final wash for the ATSB oligonucleotide was done using high stringency washing buffer (0.6 × SSC, 0.1% SDS) to avoid cross hybridization. Membranes for rehybridization with another probe were gently washed three times in 0.5% SDS at 80 °C. The same hybridization and washing conditions were used for dot-blot membranes. Dot-blot experiments used approximately 1 μg of genomic DNA per sample as described in Sykorova et al. (2003b). The control 18S ribosomal DNA (rDNA) probe (TEL220 Chlamydomonas debaryana) was prepared by PCR using specific primers (see above) and final rehybridization was done at 65 °C overnight and membranes were washed under high stringency conditions (0.2 × SSC, 0.1% SDS). Membranes were exposed to autoradiography screens and signals were visualized using a FLA5000 (FujiFilm) and evaluated by Multi Gauge software (FujiFilm).
Pulsed Field Gel Electrophoresis
Agarose plugs with high-molecular-mass DNA samples were prepared from lyophilized algal samples and BAL-31 and restriction enzyme digestion was performed as described in Sykorova et al. (2006). Briefly, agarose plugs with high-molecular-weight DNA (TEL168 Tetracystis excentrica, TEL157 Chloromonas perforata, TEL180 Neospongiococcum gelatinosum) were digested with BAL-31 nuclease (all NEB) for 15 or 45 min, and then by the restriction endonuclease ScaI-HF (TEL157, TEL180) or HindIII (TEL168) (all from NEB). The DNA was then analyzed by pulse-field gel electrophoresis using the CHEF Mapper (BioRad) under the following conditions: 1% agarose (Biorad) gel in 0.5 × TBE buffer, 6 V/cm, pulses 0.5–26 s for 20 h at 13 °C. Gels were alkali blotted and hybridized subsequently with the telomeric probes ATSB and HUSB.
Telomere Repeat Amplification Protocol
Telomerase activity was investigated using a protocol originally developed for plant telomerases (Fitzgerald et al. 1996; Sykorova et al. 2003b) and applied with modifications to dinoflagellates (Fojtova et al. 2010). Briefly, total proteins were extracted from 35 to 100 mg of lyophilized algal samples ground in liquid nitrogen and after centrifugation at 17,000 × g for 15 min, the telomerase-enriched fraction was purified from the supernatant by precipitation with 10% polyethylene glycol (PEG) 8,000 and the pellet was dissolved in telomerase extraction buffer. Alternatively, the samples of crude protein extracts (without PEG precipitation) were used as specified in Results. The amount of total protein in extracts was determined using the Bradford method (Bradford 1976). A control telomerase extract was prepared from 7 days-old Arabidopsis thaliana (Col-0) seedlings. The telomere repeat amplification protocol (TRAP) assay was performed in two phases (Sykorova et al. 2003b). In the extension step, 10 pmol of a substrate primer was elongated at 26 °C for 45 min in a reaction mix with the telomerase-enriched extract containing 0.01–1 μg of total protein. After the extension step, samples were heat inactivated and then a mixture containing 10 pmol of a reverse primer (TELPR30-3A) and 2 units of DyNazyme II Polymerase (Finnzymes) was added and PCR amplification of the TRAP product was performed (Sykorova et al. 2003b). Alternatively, the substrate primers 47F (Fojtova et al. 2002), CAMV (Fajkus et al. 1998), or GG(21) (Fitzgerald et al. 1996) and reverse primers representing different telomeric variants (TELPR, TELPR30-3A, CHTPR, HUTC, HUTPR, T3AG2-C, T3G3-C, supplementary table S2, Supplementary Material online) were used. Products were analyzed by polyacrylamide gel electrophoresis (PAGE), stained by GelStar(R) Nucleic Acid Gel Stain (LONZA) and visualized on a LAS3000 (FujiFilm). TRAP products from chosen algal species were cloned into the pCRIITOPO vector (Invitrogen) according to the manufacturer's recommendations and sequenced (Macrogen).
PCR and Sequencing of the 18S Ribosomal RNA Gene
To build a phylogeny for the strains investigated, we sequenced the 18S rDNA region of those strains (46 in total) for which it was not available from previous studies or for which the reported sequence seemed questionable (substandard). In a few cases, we relied on 18S rDNA sequences previously determined from presumably identical strains from a culture collection different from that used here for telomere investigations. To confirm that the previously sequenced strains were the same as our cultures, we checked the respective cultures by careful microscopical observation and in some cases, by sequencing the internal transcribed spacer (ITS) regions (data not shown). Genomic DNA isolated via the modified IRRI protocol (Collard et al. 2007) was used as a template for amplification of the 18S rDNA region using DyNazyme II Polymerase (Finnzymes) under PCR conditions described in Katana et al. (2001). For difficult DNA templates, we alternatively used Robust KAPA Polymerase (KAPA). The PCR products were gel purified using a Gel Extraction Kit (Qiagen) and subjected to direct sequencing using specific primers (Katana et al. 2001). PCR products that revealed unclear results from direct sequencing were cloned and sequenced (Macrogen). Newly determined sequences were deposited in GenBank with accession numbers JN903973–JN904007, JN968580–JN968586, JN968588, JN968589, and JN982286 (supplementary table S1, Supplementary Material online).
Phylogenetic Analyses
Initial analyses of newly obtained sequences and alignment construction were performed as described (Nemcova et al. 2011; Neustupa et al. 2011). A set of sequences for the final phylogenetic analysis was constructed to comprise representatives of all primary clades of Chlamydomonadales delineated by Nakada et al. (2008), other lineages of Chlorophyceae (Sphaeropleales, Oedogoniales, Chaetophorales, and Chaetopeltidales), and the remaining groups of the “core” chlorophytes (Trebouxiophyceae, Ulvophyceae, and Chlorodendrophyceae). 18S rDNA sequences from the trebouxiophytes C. variabilis NC64A and Coccomyxa sp. C-169 were extracted from the genomic scaffolds retrieved from the respective databases at the Joint Genome Institute (http://genome.jgi.doe.gov/ChlNC64A_1/ChlNC64A_1.home.html, scaffold_3, contig 410, 9,283–12,032 bp; and http://genome.jgi.doe.gov/Coc_C169_1/Coc_C169_1.home.html, scaffold_4, 2,976,163–2,978,399 bp). After removing unreliably aligned regions, the final alignment comprised 150 taxa and 1,647 positions (the alignment is available upon request). A maximum-likelihood (ML) tree was inferred using RAxML 7.2.8 available on the CIPRES Portal (Miller et al. 2010; http://www.phylo.org/sub_sections/portal/), employing a rapid bootstrapping algorithm followed by a thorough ML search on the original data set with the GTR + Γ substitution model (Stamatakis et al. 2008). A Bayesian inference was performed using the program MrBayes 3.1 (Huelsenbeck and Ronquist 2001). Two parallel Markov chain Monte Carlo runs were carried out for 3 million generations, each with one cold and three heated chains employing the GTR + Γ + I + COV evolutionary model. Trees were sampled every 100 generations. The initial 5,001 trees from each run were discarded as “burn-in” based on plotting log likelihood values. Posterior probabilities of tree bipartitions were calculated on the basis of the consensus of the remaining 50,000 trees.
Results
Sample Collection and Analyses
For this study, we tested a large range of algal strains from culture collections (see Materials and Methods) to sample broadly the phylogenetic diversity of the Chlorophyta and particularly the order Chlamydomonadales (class Chlorophyceae, fig. 1). All strains were examined for non-algal contaminants and 66 axenic algal strains or strains with bacterial contamination only (i.e., contaminants lacking telomeres and telomerase) were used for telomere/telomerase analyses. In attempt to determine what forms the ends of chromosomes (i.e., what is synthesized by telomerase), we investigated 62 algal strains by the TRAP assay for telomerase activity and cloned the TRAP products from 39 strains. In a subset of these strains (34 in total), the occurrence of variant minisatellite telomeric repeats was investigated by Southern hybridization (terminal restriction fragment [TRF] analysis and/or dot-blot hybridization) using telomeric oligonucleotide probes (table 1).
Table 1.
Hybridization Probes and Primers Used in This Study for Investigating Green Algal Minisatellite Telomeric Repeats
| Reference Organism Telomere Type | Minisatellite Repeat Unit | Hybridization Southern Blot | TRAP C-Rich Reverse Primer |
| Arabidopsis | TTTAGGG | ATSB | TELPR |
| Chlamydomonas | TTTTAGGG | CHSB | CHTPR |
| Human | TTAGGG | HUSB | HUTPR, HUTC |
| Chlorarachniophyte Nucleomorph | TCTAGGG | CASB | n.a. |
| Arabidopsis-derived | |||
| Exchange TTTAGGG | |||
| TTCAGGG | TTCAGGG | TTCAGGG-SB | n.a. |
| TTTAGGC | TTTAGGC | TTTAGGC-SB | n.a. |
| Deletion TTTAGGG | |||
| T3AG2 | TTTAGG | T3AG2-SB | T3AG2-C |
| T3G3 | TTTGGG | T3G3-SB | T3G3-C |
Note.— SB, Southern Blot, n.a., not analyzed.
A Phylogenetic Framework for the Telomere Sequence Evolution in Chlorophyceae
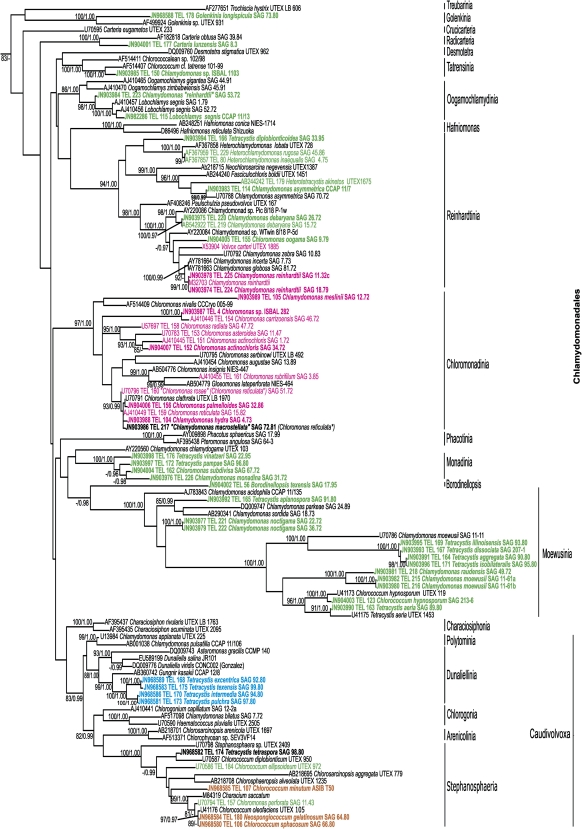
We used a standard and widely used marker for green algal phylogeny, the 18S ribosomal RNA gene, to infer the phylogenetic position of the algal strains investigated in this study for the type of telomeric sequences. The overall topology of the resulting tree (fig. 2; supplementary fig. S1, Supplementary Material online) is in accord with previous analyses and shows the basal split of the “core” chlorophytes into the four major lineages (Leliaert et al. 2012), Chlorodendrophyceae (represented by the genus Tetraselmis), Ulvophyceae, Trebouxiophyceae, and Chlorophyceae. The Chlorophyceae is further divided into two previously defined major clades (Turmel et al. 2008), OCC (Oedogoniales, Chaetophorales, Chaetopeltidales), and CS (Chlamydomonadales and Sphaeropleales). As usual in 18S rDNA-based phylogenies, most of these major groups are poorly supported by bootstrap or posterior probability values. Within Chlamydomonadales (=Volvocales), all clades defined by Nakada et al. (2008) were reconstructed, generally with high statistical support. Strains for which the telomeric sequences have been determined previously, or in this study, represent four major groups—Chlorodendrophyceae, Trebouxiophyceae, Sphaeropleales, and Chlamydomonadales. Their phylogenetic position fits the expectation based on previous taxonomic and phylogenetic studies, with the exception of several coccoid or capsal strains (attributed to the genera Chlorococcum, Tetracystis, and Neospongiococcum) that have not been previously studied by molecular means and that are very difficult to classify based on morphological features only; all these strains diverge within clades previously defined for the order Chlamydomonadales. As previously shown, some traditional genera of chlamydomonadalean algae (Chlamydomonas, Chlorococcum, and Tetracystis) are polyphyletic in the 18S rDNA tree. Borodinellopsis texensis SAG 17.95 belongs to the Chlamydomonadales, but does not fall into any of the major chlamydomonadalean clades. It may be sister to the Moewusinia clade, but the statistical support for this position is inconclusive, hence it potentially represents a new hitherto unrecognized chlamydomonadalean lineage.
FIG. 2.—
A maximum-likelihood phylogenetic tree based on 18S ribosomal RNA sequences from “core” chlorophytes with a denser sampling of the order Chlamydomonadales. Only the part of the tree corresponding to Chlamydomonadales is shown here; the full version of the tree is available as supplementary fig. S1 (Supplementary Material online). ML bootstrap percentage values/Bayesian posterior probabilities values higher than 80%/0.95 are shown above branches. Accession numbers of sequences from GenBank are given before the species name, and newly determined sequences are highlighted in bold. The “true” telomeric types are indicated in color (Arabidopsis type, green; Chlamydomonas type, magenta; human type, blue; not determined, brown). Major clades in the order Chlamydomonadales are named according to Nakada et al. (2008), *species names according Proschold et al. (2001).
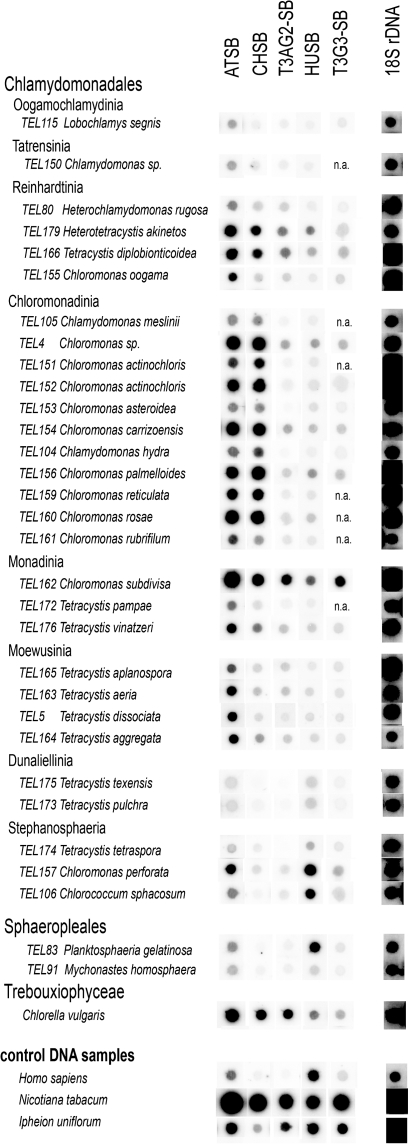
Dot-Blot Hybridization Screening of Chlamydomonadales and Sphaeropleales
We investigated 29 strains from Chlamydomonadales and two species from Sphaeropleales for the occurrence of variant minisatellite telomeric repeats by dot-blot hybridization (fig. 3 and table 1). Sonicated human genomic DNA and genomic DNA of the model plants Nicotiana tabacum (Solanaceae, Arabidopsis-type telomere) and Ipheion uniflorum (Asparagales, human-type telomere) were used as controls (fig. 1). Dot-blot hybridization results showed the presence of several types of telomeric minisatellite repeats in genomes of Chlamydomonadales and Sphaeropleales and an abundance of the Arabidopsis-type variant of telomeric sequence (ATSB), except in species from the clade Dunaliellinia (fig. 3). The Chlamydomonas-type variant (CHSB) revealed strong signals comparable to the Arabidopsis-type variant in all samples from the clade Chloromonadinia suggesting similar telomeric features in the species of this group. In several species, the abundance of the Arabidopsis-type variant was reflected in the abundance of the repeats that are most closely related to it; these variants occur in the following order of abundance: Arabidopsis type > Chlamydomonas type > T3AG2 type > human type. A human-type variant probe hybridized strongly to genomic DNA of the samples C. perforata (TEL157) and Chlorococcum sphacosum (TEL106) (Stephanosphaeria) and the sample Planktosphaeria gelatinosa (TEL83) (Sphaeropleales).
FIG. 3.—
Genomic DNA dot-blot hybridization data and their correlation with the green algal clades in the 18S rDNA-based phylogeny of the group (fig. 2; supplementary fig. S1, Supplementary Material online). Genomic DNA was hybridized with different types of minisatellite sequences representing typical telomeric types—the Arabidopsis type (ATSB), the Chlamydomonas type (CHSB), the human type (HUSB), and their variants T3AG2-SB, T3G3-SB, (see table 1, supplementary table S2, Supplementary Material online). All membranes were rehybridized with the control 18S rDNA probe. Control samples were from the green alga Chlorella vulgaris (Trebouxiophyceae), sonicated human DNA, and from plants with the typical Arabidopsis-type telomere sequence (Nicotiana tabacum, Solanaceae), and the human-type telomere sequence (Ipheion uniflorum, Asparagales). Accession numbers for all algal strains (identified by the TEL number) are listed in supplementary table S1 (Supplementary Material online). It should be noted that DNA concentration had to be estimated from agarose gels due to the purification method used, and the loaded amount of genomic DNAs might vary among samples.
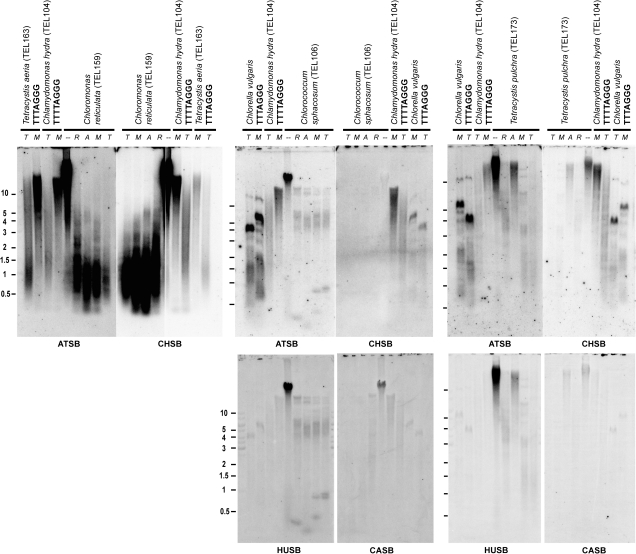
TRF Analysis
The typical telomere lacks recognition sites for restriction enzymes due to its minisatellite repeat sequence organized in tandem. Thus, most restriction enzymes will cut DNA at the closest site in the subtelomere, splitting a TRF from the rest of the chromosome. Digestion of genomic DNA, separation of fragments on agarose gel, and Southern hybridization with a telomeric probe enable estimation of the length of algal telomeres as TRF. The dot-blot results suggested a large abundance of the Arabidopsis- and the Chlamydomonas-type variants in species from the Chloromonadinia clade and of the human-type variant in the genomes of C. sphacosum (TEL106) and C. perforata (TEL157). To characterize the distribution and occurrence of potential telomere-like minisatellites, we investigated species from Chloromonadinia, Reinhardtinia, Stephanosphaeria, and Dunaliellinia clades for the presence of several types of minisatellite repeat sequences using an analysis of the TRF length (fig. 4; supplementary fig. S2, Supplementary Material online). Southern hybridization results showed a smear of TRF fragments between 0.3 and 2 kb in Chloromonas actinochloris (TEL151) and Chloromonas reticulata (TEL159) (Chloromonadinia) and revealed colocalization of the Arabidopsis-type and the Chlamydomonas-type variants in TRF of both genomes (fig. 4; supplementary fig. S2, Supplementary Material online). The TRFs from C. sphacosum (TEL106) (Stephanosphaeria) and Tetracystis pulchra (TEL173) (Dunaliellinia) were 4–8 kb and 1–2.5 kb long, respectively, with the same distribution visualized by the Arabidopsis-type (ATSB) and the human-type (HUSB) probe, suggesting a mixture of both sequences in the DNA fragments (fig. 4; supplementary fig. S2, Supplementary Material online). However, weak hybridization signals suggested that only a small portion of TRFs was formed by telomeric-type sequences, and these minisatellites were also scattered in the genomes. Therefore, restriction digestion split these signals into many fragments giving weak signals. Heterochlamydomonas inaequalis (TEL80) demonstrated short TRFs (300–750 bp) hybridizing predominantly with ATSB, as well as the presence of longer fragments probably from internal genome regions (supplementary fig. S2, Supplementary Material online). In addition, we rehybridized the membranes with minisatellite probes TTTAGGC-SB and TTCAGGG-SB, which represent sequences related to the Arabidopsis-type variant (TTTAGGG), and with CASB (TCTAGGG) representing a variant found in the chlorarachniophyte nucleomorph telomere (Gilson and McFadden 1995). None of these variant repeats showed a specific hybridization pattern.
FIG. 4.—
Results of terminal restriction fragment (TRF) analysis. Genomic DNA samples from the Chloromonadinia (TEL159), the Stephanosphaeria (TEL106), and the Dunaliellinia (TEL173) were digested by TaqI (T), MboI (M), AluI (A), or RsaI (R) restriction endonuclease (−, non-digested) and separated on an 0.9% agarose gel (marker lengths in kilo bases). Control algal DNA samples for the Arabidopsis type (TTTAGGG) and the Chlamydomonas type (TTTTAGGG) were included. The hybridization pattern of the probes specific for the telomeric types—the Arabidopsis type (ATSB), the Chlamydomonas type (CHSB), the human type (HUSB), and the chlorarachniophyte nucleomorph type (CASB) is shown (table 1). Only the part of minisatellite probes is shown here, full version of the TRF analysis is available as supplementary fig. S2 (Supplementary Material online)
Telomerase Activity Screening in Chlorophyceae Using TRAP Assay
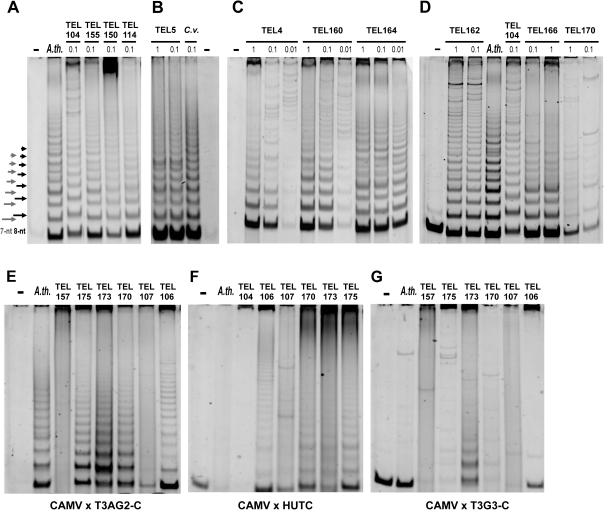
We investigated 60 algal strains from Chlorophyceae (table 2, supplementary table S1, Supplementary Material online) for the presence of a telomerase activity. Using the Arabidopsis-type sequence as a reverse primer, products were obtained for the majority of samples (examples at figs. 5A–D). A comparison of the TRAP pattern periodicity with control samples showed that the algal species could be assigned to three groups (table 2) according to the ladder of TRAP products, specifically with 1) a seven-nucleotide periodicity (typical for the Arabidopsis-type telomeric repeat), 2) an eight-nucleotide periodicity (suggesting the Chlamydomonas type), and 3) low or absent telomerase activity or smeared products. The group (3) represented a majority of species from the clades Stephanosphaeria and Dunaliellinia (fig. 2). The absence of a telomerase activity in the TRAP assay could be caused by technical problems with inefficient purification of telomerase extract (supplementary fig. S3A, Supplementary Material online), by a different substrate primer preference, and/or by an inadequate reverse primer sequence. To exclude technical problems in group (3) samples, we also used a crude telomerase extract in the TRAP assay (supplementary fig. S3A, Supplementary Material online) and alternative substrate primers (47F, GG(21), CAMV) were used to meet possible different substrate sequence requirements for the telomerase action. However, omitting the PEG purification step or alteration of the substrate primer did not increase the telomerase activity in samples (not shown). When we used reverse primers with variant minisatellite telomeric sequences—CHTPR (Chlamydomonas type, not shown), HUTPR, HUTC (human type), T3AG2-C (TTTAGG type), or T3G3-C type (TTTGGG type), ladders of TRAP products with a six-nucleotide periodicity were observed for reactions containing the reverse primers HUTC, HUTPR (not shown), and T3AG2-C in samples TEL106, 175, 173, 170 (fig. 5E–G).
Table 2.
Results of TRAP Assays
| Class/Order Clade | TEL | Species | TRAP Products Laddera (type) /CLONED | Number of Clones | Number of Repeats | T-SplippagebT(n)AGGG |
Variant Repeats (% of all) | G-Slippage | Mis-Incorporation | |||
| T(2) hu | T(3) at | T(4) ch | T(5,6) | |||||||||
| Chlorodendrophyceae | 211 | Tetraselmis chui | 7-nt (tttaggg) | n.a. | ||||||||
| Trebouxiophyceae | 01 | Chlorella vulgaris | TTTAGGG | 10 | 35 | 0 | 30 | 3 | 1 | 11.4c | 0 | 3 |
| Chlorophyceae/Sphaeropleales | 83 | Planktosphaeria gelatinosa | TTTAGGG | 5 | 27 | 0 | 27 | 0 | 0 | 0.0 | 0 | 0 |
| 87 | Coelastrella vacuolata | TTTAGGG | 2 | 17 | 0 | 17 | 0 | 0 | 0.0 | 0 | 0 | |
| 91 | Mychonastes homosphaera | TTTAGGG | 3 | 33 | 0 | 32 | 0 | 0 | 0.0 | 0 | 1 | |
| 138 | Pseudomuriella aurantiaca | 7-nt (tttaggg) | n.a. | |||||||||
| 188 | Bracteacoccus cohaerens | 7-nt (tttaggg) | n.a. | |||||||||
| Chlorophyceae/Chlamydomonadales Golenkinia | 178 | Golenkinia longispicula | TTTAGGG | 3 | 55 | 1 | 53 | 0 | 0 | 1.8 | 2 | 0 |
| Radicarteria | 177 | Carteria lunzensis | TTTAGGG | 3 | 80 | 0 | 74 | 6 | 0 | 7.5 | 2 | 0 |
| Tatrensinia | 150 | Chlamydomonas sp | 7-nt (tttaggg) | n.a. | ||||||||
| Oogamochlamydinia | 115 | Lobochlamys segnis | TTTAGGG | 5 | 65 | 5 | 53 | 5 | 1 | 16.9 | 3 | 2 |
| 223 | Chlamydomonas 'reinhardtii' | TTTAGGG | 4 | 86 | 1 | 57 | 25 | 3 | 33.7 | 1 | 0 | |
| Reinhardtinia | 80 | Heterochlamydomonas inaequalis | TTTAGGG | 8 | 230 | 0 | 153 | 63 | 6 | 30.0 | 8 | 10 |
| 166 | Tetracystis diplobionticoidea | TTTAGGG | 2 | 27 | 1 | 25 | 0 | 1 | 7.4 | 0 | 0 | |
| 229 | Heterochlamydomonas rugosa | TTTAGGG | 4 | 90 | 0 | 62 | 26 | 0 | 32.2 | 4 | 2 | |
| 114 | Chlamydomonas asymmetrica | TTTAGGG | 2 | 39 | 3 | 35 | 1 | 0 | 10.2 | 0 | 0 | |
| 179 | Heterotetracystis akinetos | TTTAGGG | 4 | 86 | 1 | 77 | 7 | 0 | 9.3 | 1 | 1 | |
| 155 | Chloromonas oogama | TTTAGGG | 3 | 66 | 1 | 56 | 8 | 0 | 13.6 | 1 | 1 | |
| 219 | Chlamydomonas debaryana | 7-nt (tttaggg) | n.a. | |||||||||
| 220 | Chlamydomonas debaryana | TTTAGGG | 5 | 71 | 1 | 64 | 6 | 0 | 9.8 | 4 | 1 | |
| 224 | Chlamydomonas reinhardtii | 8-nt (ttttaggg) | n.a. | |||||||||
| 225 | Chlamydomonas reinhardtii | +/− TTTTAGGG | 4 | 56 | 0 | 0 | 56 | 0 | 0.0 | 0 | 0 | |
| Chloromonadinia | 4 | Chloromonas sp. | TTTTAGGG | 7 | 99 | 0 | 7 | 89 | 2 | 9.0 | 0 | 1 |
| 104 | Chlamydomonas hydra | TTTTAGGG | 4 | 63 | 0 | 10 | 47 | 5 | 23.8 | 0 | 0 | |
| 105 | Chlamydomonas meslinii | TTTTAGGG | 4 | 77 | 0 | 10 | 64 | 0 | 12.9 | 1 | 2 | |
| 151 | Chloromonas actinochloris | TTTTAGGG | 5 | 104 | 1 | 15 | 88 | 0 | 14.4 | 1 | 2 | |
| 152 | Chloromonas actinochloris | TTTTAGGG | 5 | 47 | 0 | 12 | 35 | 0 | 25.5 | 0 | 1 | |
| 153 | Chloromonas asteroidea | +/− 8-nt (ttttaggg) | n.a. | |||||||||
| 154 | Chloromonas carrizoensis | TTTTAGGG | 5 | 72 | 1 | 10 | 57 | 3 | 18.0 | 1 | 1 | |
| 156 | Chloromonas palmelloides | TTTTAGGG | 3 | 58 | 2 | 8 | 48 | 0 | 13.8 | 5 | 0 | |
| 158 | Chloromonas radiata | TTTTAGGG | 3 | 48 | 0 | 6 | 40 | 2 | 16.7 | 1 | 0 | |
| 159 | Chloromonas reticulata | 8-nt (ttttaggg) | n.a. | |||||||||
| 160 | Chloromonas rosae | TTTTAGGG | 5 | 100 | 0 | 3 | 85 | 10 | 13.9 | 1 | 2 | |
| 161 | Chloromonas rubrifilum | TTTTAGGG | 5 | 171 | 0 | 82 | 85 | 2 | 49.1 | 5 | 2 | |
| 217 | Chlamydomonas macrostellata | Neg. | n.a. | |||||||||
| Monadinia | 162 | Chloromonas subdivisa | 7-nt (tttaggg) | n.a. | ||||||||
| 172 | Tetracystis pampae | 7-nt (tttaggg) | n.a. | |||||||||
| 176 | Tetracystis vinatzeri | 7-nt (tttaggg) | n.a. | |||||||||
| 226 | Chlamydomonas monadina | TTTAGGG | 3 | 65 | 1 | 60 | 4 | 0 | 7.7 | 0 | 0 | |
| Borodinelopsis | 56 | Borodinellopsis texensis | TTTAGGG | 4 | 76 | 27 | 40 | 7 | 1 | 46.0 | 0 | 1 |
| Moewusinia | 163 | Tetracystis aeria | TTTAGGG | 3 | 50 | 0 | 50 | 0 | 0 | 0.0 | 0 | 0 |
| 164 | Tetracystis aggregata | TTTAGGG | 7 | 109 | 0 | 106 | 3 | 0 | 2.7 | 1 | 0 | |
| 165 | Tetracystis aplanospora | 7-nt (tttaggg) | n.a. | |||||||||
| 5 | Tetracystis dissociata | TTTAGGG | 6 | 101 | 0 | 99 | 0 | 0 | 0.0 | 3 | 0 | |
| 171 | Tetracystis isobilateralis | 7-nt (tttaggg) | n.a. | |||||||||
| 169 | Tetracystis illinoisensis | 7-nt (tttaggg) | n.a. | |||||||||
| 215 | Chlamydomonas moewusii | TTTAGGG | 4 | 35 | 0 | 35 | 0 | 0 | 0.0 | 0 | 0 | |
| 216 | Chlamydomonas moewusii | 7-nt (tttaggg) | n.a. | |||||||||
| 218 | Chlamydomonas raudensis | TTTAGGG | 5 | 69 | 0 | 66 | 3 | 0 | 4.3 | 0 | 0 | |
| 221 | Chlamydomonas noctigama | TTTAGGG | 4 | 32 | 0 | 32 | 0 | 0 | 0.0 | 0 | 0 | |
| 222 | Chlamydomonas noctigama | TTTAGGG | 4 | 54 | 2 | 50 | 2 | 0 | 7.4 | 0 | 0 | |
| 123 | Chlorococcum hypnosporum | 7-nt (tttaggg) | n.a. | |||||||||
| Dunaliellinia | 168 | Tetracystis excentrica | TTAGGG | 4 | 114 | 74 | 30 | 4 | 1 | 36.8 | 4 | 2 |
| 170 | Tetracystis intermedia | +/− TTAGGG | 4 | 38 | 21 | 11 | 5 | 0 | 42.1 | 0 | 1 | |
| 173 | Tetracystis pulchra | +/− TTAGGG | 1 | 18 | 16 | 0 | 2 | 0 | 11.1 | 3 | 2 | |
| 175 | Tetracystis texensis | TTAGGG | 3 | 75 | 59 | 14 | 0 | 0 | 17.5 | 4 | 0 | |
| Stephanosphaeria | 106 | Chlorococcum sphacosum | +/− | n.a. | ||||||||
| 107 | Chlorococcum minutum | +/− | n.a. | |||||||||
| 157 | Chloromonas perforata | +/− TTTAGGG | 4 | 52 | 4 | 45 | 2 | 0 | 11.5 | 1 | 1 | |
| 180 | Neospongiococcum gelatinosum | +/− | n.a. | |||||||||
| 184 | Chlorococcum ellipsoideum | +/− 7-nt (tttaggg) | n.a. | |||||||||
NOTE.—nt, nucleotide, n.a., not analyzed. Major telomere type is underlined.
+/− low telomerase activity in TRAP, neg. no ladder, negative.
Various number of T residues categorizes repeats to human type (hu), Arabidopsis type (at), and Chlamydomonas type (ch).
TRAP products were cloned using the primer sets CAMV × TELPR (5 clones) and CAMV × HUTPR (5 clones), variant repeats were identified only in the TRAP products resulting from CAMV × HUTPR primer combination.
FIG. 5.—
TRAP assay of Chlorophytes. Telomerase activity of a representative set of algal samples (indicated by TEL numbers, see supplementary table S1, Supplementary Material online) is shown, using different sets of the substrate primers GG(21) (A,C,D), CAMV (B) and the Arabidopsis-type repeat reverse primers TELPR30-3A (A,C,D) or TELPR (B) (table 1, supplementary table S2, Supplementary Material online). Telomerase-enriched extracts from Arabidopsis thaliana (Ath) seedlings and from Chlamydomonas hydra (TEL104, Chloromonadinia clade) were used as a pattern control of a seven- and an eight- nucleotide (nt) periodicity ladder, respectively; arrows with numbers (A) denote the length increments of telomerase products. The 7-nt periodicity was observed in the TRAP products from control alga Chlorella vulgaris (B, TEL01) and from species in clades Tatrensinia (A, TEL150), Reinhardtinia (A, TEL155, TEL114; D, TEL166), Moewusinia (B, TEL5; C, TEL164), and Monadinia (D, TEL162). The 8-nt periodicity was seen in samples from the Chloromonadinia (C, TEL4, TEL160). An unclear TRAP pattern was revealed T. intermedia (D, TEL170) from Dunaliellinia. The amount of total protein is indicated in micrograms (A–D). The negative control (−) contained no extract. In addition, variant combinations of the substrate primer CAMV and the reverse primers T3AG2-C (E), HUTC (F, human-type repeat), T3G3-C (G) (table 1) were used for samples from the Dunaliellinia (TEL170, TEL173, TEL175), and the Stephanosphaeria (TEL106, TEL107, TEL157) clades.
Testing for Telomeric Localization of Minisatellite Repeats Using BAL-31 Digestion
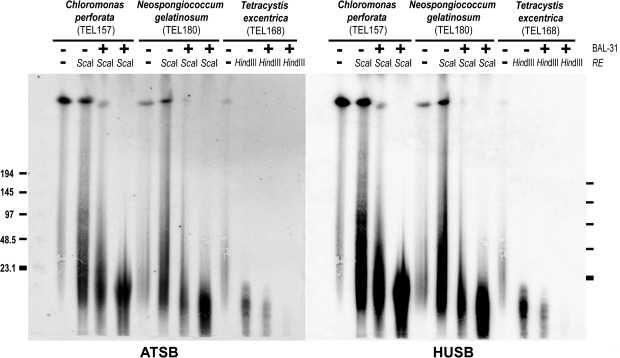
BAL-31 nuclease progressively shortens DNA fragments at both termini. Therefore, the hybridization signal for terminally positioned sequences diminishes and moves to lower molecular mass during digestion. Hence, BAL-31 digestion of high-molecular-weight genomic DNA can prove the terminal location of a sequence. Dot-blot hybridization and TRAP results suggested a synthesis of the human-type variant in several species from the Dunaliellinia clade and gave an unclear pattern of occurrence of minisatellite repeats in the Stephanosphaeria clade. We investigated the genomic position of these sequences in T. excentrica (TEL168), C. sphacosum (TEL157), and N. gelatinosum (TEL180). With an increasing duration of the BAL-31 nuclease digestion (15 and 45 min), there was a gradual decrease in the intensity of the HUSB and ATSB signal together with a shortening of TRFs in all three samples (fig. 6). This indicates that both Arabidopsis-type (ATSB) and human-type (HUSB) sequences are terminally positioned in these species. However, differences in signal intensities were observed between the probes, with the ATSB signal markedly weaker than HUSB. TRF patterns of TEL157 and TEL180 generated with ScaI-HF were smeared within the 20–200 kb range, with a mean length of ca. 50 kb. In the sample TEL168, the TRF pattern generated with HindIII showed several discrete bands between 15 and 40 kb. These numbers, however, do not reflect exact telomere lengths since the TRFs presumably also contain relatively long telomere-associated sequences.
FIG. 6.—
BAL-31 nuclease treatment of intact genomic DNA from the Dunaliellinia (TEL168) and the Stephanosphaeria (TEL157, TEL180). High-molecular weight DNA samples after BAL-31 nuclease treatment (15 and 45 min) and restriction endonuclease (RE) digestion (−, non digested) were analyzed by pulse-field gel electrophoresis (marker lengths in kilobase). The hybridization pattern of the probes specific for the human-type (HUSB) and the Arabidopsis-type (ATSB) telomeric sequence revealed their terminal position by a reduction of signal intensity with increasing duration of BAL-31 digestion.
Cloning the TRAP Products Reveals the “True” Telomere-Type Sequence Synthesized by Telomerase
Telomerase synthesizes telomeric repeats using a short template region of its RNA subunit (TR). The sequence type of the telomeric repeat then can be estimated from cloned and sequenced TRAP products. To confirm the observed TRAP pattern and to determine the sequence of the telomerase product, we sequenced TRAP products of 36 algal strains from the chlamydomonadalean clades Golenkinia, Radicarteria, Oogamochlamydinia, Reinhardtinia, Chloromonadinia, Monadinia, and Moewusinia, and of three species from the sister group Sphaeropleales (supplementary fig. S1, Supplementary Material online). For cloning, we chose reactions that expressed high enzyme processivity starting with the combination TS21 × TELPR30-3A (i.e., the reverse primer has the Arabidopsis type of sequence). To avoid artifacts, the TRAP products from another combination of a substrate primer (CAMV, 47F, GG(21)) and an alternative Arabidopsis-type reverse primer TELPR (differs from TELPR30-3A at the 3′-end) were cloned. Sequencing results confirmed the Arabidopsis-type (22 strains, table 2) and the Chlamydomonas-type variant (12 strains, table 2) synthesized by telomerase as deduced from the periodicity of TRAP products resolved on polyacrylamide gels (see above). The sequence analysis of TRAP products revealed different fidelity of telomerases independently of their phylogeny position, with T-slippage errors most common (table 2, examples of a detailed analysis are shown in supplementary fig. S4, Supplementary Material online). These errors are probably caused by inaccurate annealing of the telomere DNA strand to the anchor site of the TR template region that results in synthesis of a telomeric repeat shorter or longer by one “T” residue; however, the “errors” are incorporated randomly. Unusually high error rates were observed in products synthesized by telomerases of H. inaequalis (TEL80) and Chloromonas rubrifilum (TEL161), resulting in a mix of Arabidopsis- and Chlamydomonas-type telomeric repeats (table 2). The TRAP products of H. inaequalis contained predominantly the Arabidopsis-type variant and the products from C. rubrifilum contained a similar number of the Arabidopsis-type and the Chlamydomonas-type repeats in sequences of all clones. Considering the phylogenetic relationship between algal species, we also cloned TRAP products from Heterochlamydomonas rugosa (TEL229), which revealed results similar to H. inaequalis (TEL80) (table 2). In conclusion, the major Chlamydomonas-type variant was identified in all species investigated from the clade Chloromonadinia, making this clade uniform with respect to telomeres. Surprisingly, only two C. reinhardtii accessions (TEL224, 225) from the Reinhardtinia clade revealed synthesis of the Chlamydomonas-type telomeric sequence, whereas the telomerase of the remaining accession of C. reinhardtii (TEL223) synthesized predominantly Arabidopsis-type repeats (table 2; supplementary fig. S3B, Supplementary Material online).
Since Tetracystis intermedia (TEL170), T. pulchra (TEL173) and Tetracystis texensis (TEL175) from the Dunaliellinia clade did not show a clear TRAP pattern using the primer combinations mentioned above, TRAP products were cloned from reactions utilizing the primer set CAMV × T3AG2-C (fig. 5E). The TRAP products of T. excentrica (TEL168) were cloned using primer set GG(21) × TELPR30-3A (supplementary fig. S4, Supplementary Material online). All four species investigated from the Dunaliellinia clade (TEL168, 170, 173, 175) revealed telomerases producing human-type repeats with an error rate similar to telomerases from algal groups with a “noncanonical” telomeric sequence (table 2). The cloning of TRAP products from the Stephanosphaeria species showed synthesis of the Arabidopsis-type variant in C. sphacosum (TEL157), but the sequenced products from other species showed non-telomeric sequences resulting probably from PCR artifacts (not shown).
Discussion
For the purpose of interpreting the data reported in this study, it is important to distinguish between the telomeric sequences synthesized by telomerase, which form only the ends of chromosomes and are designated as “true” telomeric type in figures 1 and 7, and the telomere DNA, which consist of these distal sequences and of telomeric sequence variants present in more proximal telomeric regions constituting the telomere structure. Our data suggest that a change of the “true” telomere type happened independently in at least three green algal groups when the ancestral Arabidopsis type of telomere sequence (TTTAGGG) was repeatedly replaced by the Chlamydomonas type (TTTTAGGG) or by the human-type (TTAGGG) variant (fig. 7). Mapping the distribution of the telomere types onto a green algal phylogeny inferred from 18S rDNA sequences showed that the Arabidopsis type of telomeric sequence is ancestral not only for the whole Chlorophyceae group (figs. 1 and 2; supplementary fig. S1, Supplementary Material online) but also for the order Chlamydomonadales, whereas the Chlamydomonas type of telomeric sequence is present in only some chlamydomonadalean clades (Chloromonadinia and Reinhardtinia; fig. 3 and table 2). The human-type variant occurs in terminal positions in genomes of several species from the clades Dunaliellinia and Stephanosphaeria (fig. 6). These derived telomeric types are discussed separately in more detail below.
FIG. 7.—
Telomere variability in green algae. The predominant and apparently ancestral telomeric motif TTTAGGG is present in most green algae. A variation in this sequence is seen in the Chloromonadinia clade (TTTAGGG–TTTTAGGG) and in the subgroup of the Reinhardtinia (Chlamydomonas reinhardtii, Volvox carteri). The synthesis of the TTAGGG motif by telomerase was observed in the Dunaliellinia and the occurrence of this minisatellite variant in genomes of the Stephanosphaeria algal strains.
At Least Two Separate Origins of the Chlamydomonas Type of Telomeres in the Chlamydomonadales
Initial investigations of the telomeric sequences in the standard strain NO+ and a field isolate of C. reinhardtii (Petracek et al. 1990) revealed a novel telomeric sequence type, which has been confirmed by sequencing the whole genome of another C. reinhardtii accession (CC-503 cw92 mt+, GenBank accession number ABCN01000000; Merchant et al. 2007). To reevaluate the telomeric data for C. reinhardtii, we investigated three more accessions of this species from the culture collection. Furthermore, we tested several additional strains from the Reinhardtinia clade to determine the distribution of the Chlamydomonas-type telomeric sequences. Surprisingly, this derived telomeric type could be identified in only two C. reinhardtii accessions (table 2; supplementary table S1, Supplementary Material online), and based on the genome sequence reported by Prochnik et al. (2010), it is also present in a related alga Volvox carteri (GenBank accession number ACJH00000000). Two of the C. reinhardtii strains studied (TEL 224 and TEL 225) belong to standard strains of this species (Proschold et al. 2005). However, the C. reinhardtii accession (TEL223) that possesses the Arabidopsis-type variant in telomeres and a telomerase producing a high number of errors in the repeats synthesized in the in vitro testing system (table 2) has been apparently misidentified, as it occupies a phylogenetically remote position in the Oogamochlamydinia clade. This strain, a minus (−) mating type originally labeled as Chlamydomonas smithii, was found, unlike the plus (+) mating type strain of C. smithii, not to be able to interbreed (to mate) with standard C. reinhardtii strains (Harris 1989). It differs from standard C. reinhardtii also in morphology. A very different ITS sequence rather similar to C. culleus and homothallic zygote formation in clonal cultures, were also reported (Coleman and Mai 1997) and support our findings.
According to our 18S rDNA-based phylogeny, C. reinhardtii and V. carteri may represent a subclade within Reinhardtinia, including some other species (e.g., Chlamydomonas incerta, Chlamydomonas globosa, and Chlamydomonas zebra) that are yet to be studied with respect to their telomeres. A possible hypothesis is that the switch to the derived Chlamydomonas-type telomere sequence occurred in the stem lineage of this subclade, but a much denser sampling and a more robustly reconstructed phylogeny of the Reinhardtinia clade are required to test this possibility.
Our wide search across the Chlamydomonadales led to the identification of a second clearly separate group with the Chlamydomonas type of telomeric sequence, the Chloromonadinia clade (figs. 2 and 7). Their telomeres are formed by relatively short arrays of telomeric sequences (fig. 4; supplementary fig. S2, Supplementary Material online) and contain a mixture of the Arabidopsis- and Chlamydomonas-type variants (fig. 3) in all Chloromonadinia strains investigated. Our observation is in agreement with previous findings that telomeres of unicellular organisms are usually short, for instance the approximately 300-bp long telomeres of the alga C. vulgaris (Higashiyama et al. 1995) or of the ciliate Tetrahymena (Shampay and Blackburn 1989). Telomerase maintains only the distal part of telomeres, so the genomic presence of the ancestral telomeric type could be attributed to more proximal DNA of telomeric and subtelomeric regions and also to enzymatic properties of telomerase. Telomerases in various species differ in their accuracy of synthesis. In particular, plant telomerases have been described showing a high error rate under in vitro and in vivo conditions (Fitzgerald et al. 1996; Sykorova et al. 2003b; Weiss-Schneeweiss et al. 2004). We found that the Chloromonadinia species possess telomerase that synthesizes of a large portion of variant telomeric repeats resulting from T-slippage errors (9–25%, table 2).
Complex Telomere Evolution in the Dunaliellinia and the Stephanosphaeria
Special attention was paid to telomeres of two chlamydomonadalean clades that showed unclear dot-blot and TRAP results, the Dunaliellinia and the Stephanosphaeria. We investigated the possibility that the telomeric sequence in this clade has changed to a minisatellite type derived from the ancestral TTTAGGG by a single nucleotide change, for example, to the TCTAGGG variant found in the chlorarachniophyte nucleomorph (Gilson and McFadden 1995), or minisatellite types TTCAGGG and TTTAGGC. However, none of these variants seem to form telomeres in the Dunaliellinia or Stephanosphaeria. Finally, cloning of the TRAP products suggested telomerase synthesizing the human-type telomeric repeat in four species representing a distinct subclade within the Dunaliellinia (figs. 2 and 7). The number of telomerase errors is very high with the majority of the Arabidopsis-type “errors” synthesized (table 2). Interestingly, the telomerase inaccuracy of Dunaliellinia species is similar to those of other groups with a noncanonical telomere type (see above). A Blast search of whole genome shotgun data from an ongoing Dunaliella salina genome sequencing project available in Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/) revealed the presence of short stretches of telomere-like sequences and also of long tracts consisting of the Arabidopsis-type variant or of a mixture of the human-type and the Arabidopsis-type variants (not shown). The terminal position of these sequences cannot be assessed without genome/scaffold assembly, but they leave open the possibility that the human-type variant contributes to telomeres also in other species from this clade.
The Stephanosphaeria clade displayed divergent results showing the Arabidopsis type of telomeric repeats synthesized by telomerase of C. perforata (TEL157, table 2) and an abundance of the human-type minisatellite in the genome of TEL157 and in the other two species C. sphacosum (TEL106, fig. 3) and N. gelatinosum (TEL180, fig. 6). The terminal position of the Arabidopsis- and the human-type minisatellites in chromosomes of TEL157 and TEL180 was also confirmed by BAL-31 digestion, suggesting the co-occurrence of both telomeric variants in mixed arrays (fig. 6). These genomic features are shared by all Stephanosphaeria species investigated as well as by TEL168 from the Dunaliellinia clade (see above). Due to a low telomerase activity/processivity resulting in an unclear TRAP pattern (table 2 and fig. 5E–G) and a failure in cloning of the TRAP products (except TEL157), the question about the steps in telomere evolution of the Stephanosphaeria remains open.
Nevertheless, according to the 18S rDNA phylogeny, Dunaliellinia and Stephanosphaeria, together with several other chlamydomonadalean clades, constitute a higher order clade dubbed Caudivolvoxa (Nakada et al. 2008; fig. 2). It is, therefore, possible that the telomere synthesis started to depart from the ancestral state early in the Caudivolvoxa evolution, before the split of the Dunaliellinia and Stephanosphaeria lineages. Obviously, a much broader survey of telomere sequences of Caudivolvoxa coupled with a more precise phylogenetic scheme for this group are required to shed more light on this issue.
Implications for the General Mode of Telomere Evolution
The data about telomeric types currently available have come mostly from common model organisms. A majority of eukaryotic groups have telomeres formed by minisatellite repeats synthesized by telomerase. By now, several groups with more than one minisatellite telomeric type have been described, for example, ciliates (TTGGGG in Tetrahymena and TTTTGGGG in Oxytricha), plants (TTTAGGG as the major type, TTAGGG in several families of Asparagales), or fungi (TTAGGG as a major type, different repeats in yeasts; for details, see the Telomerase database telomerase.asu.edu/sequences.html; Podlevsky et al. 2008). A specific group of organisms with two telomeric types is represented by chlorarachniophytes and cryptomonads with different telomeres of the nuclear and the nucleomorph chromosomes (Gilson and McFadden 1995; Zauner et al. 2000). In this study, we have substantially expanded the diversity of telomeric sequences known for green algae (fig. 7), showing evidence for two evolutionarily independent transitions from the ancestral TTTAGGG (Arabidopsis type) motif to the derived TTTTAGGG (Chlamydomonas type) motif in a Reinhardtinia subclade and in the Chloromonadinia clade, and for the emergence of the TTAGGG (human type) motif in Dunaliellinia and Stephanosphaeria clades. It should be mentioned that there is also an increasing number of reports about species or groups of organisms with an “unknown” telomeric type or at least with the lack of typical telomeric types. Some of these cases have later been reevaluated, for example, in insects, where the originally described multiple loss of a typical telomeric sequence (TTAGG) in some Coleoptera (Frydrychova and Marec 2002; Frydrychova et al. 2004) was explained for the Tenebrionoidea superfamily by a switch to the novel telomere type TCAGG first identified in Tribolium (Mravinac et al. 2011).
The sequence of the telomeric repeat synthesized by telomerase is determined by the sequence in the template region of its RNA subunit (TR). The simplest hypothetical event resulting in a change to a different telomeric minisatellite repeat would be a mutation in this template region. Such a hypothesis is supported by our observation of recently diverged telomere variants in green algae (TTTTAGGG, Chlamydomonas type; TTAGGG, human type), which differ very little from the ancestral telomere-type variant (TTTAGGG). This divergence likely arises from a single nucleotide change (insertion or deletion) in the TR template region. This mode of TR evolution is the most common process in the evolution of telomere motifs, as has been suggested for the evolution of land plant and insect telomerases. Another possible cause of variant repeat synthesis could be the change in telomerase template usage caused by mutation in the catalytic telomerase subunit TERT (Sykorova, Leitch, et al. 2006).
The change of the minisatellite type forming the telomeres probably influences other aspects of telomere function, for example, the DNA-binding activity of telomere-associated proteins. However, a high flexibility of plant telomeric proteins has been described in land plants, where 1) the Arabidopsis-type variant of telomeric motif was changed to the human type in several families of Asparagales and proteins binding both telomeric motifs were found (Rotkova et al. 2004, 2007); 2) the human type was lost in the genus Allium of the Alliaceae family and similar proteins binding both telomere variants were detected in vitro (Fajkus et al. 2005); 3) the typical Arabidopsis-type telomeres were lost in three plant genera of the Solanaceae family and the DNA-binding activity of Cestrum proteins to the ancestral Arabidopsis-type variant was reported (Peska et al. 2008). The telomere-binding properties may also be a subject of rapid adaptive coevolution of function as occurs with another essential structure, centromeric repeats and specific centromeric histone types (Malik and Henikoff 2001). How a mutation in a telomere sequence synthesized by telomerase becomes fixed is unknown, but we would presume that the inner parts of the telomere formed by ancestral sequences (still binding telomere-associated proteins) could form a buffer zone that may help to deal with the new situation. Also the observed high number of errors in telomere synthesis, especially in groups with “recently” changed telomere types, could participate and make the passage to new telomere type easier.
Supplementary Material
Supplementary tables S1 and S2 and figure S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Grant Agency of the Czech Republic (521/09/1912 to E.S.), by the project ‘‘CEITEC—Central European Institute of Technology’’ (CZ.1.05/1.1.00/02.0068 to J.F.) from the European Regional Development Fund, by the Research and Development for Innovations Operational Programme (EU Structural Funds, project no. CZ.1.05/2.1.00/03.0100 to M.E.), and by institutional funding (MSM0021622415 to Masaryk University, (AV0Z50040702 to the Institute of Biophysics, and AV0Z60660521 to the Institute of Soil Biology). We would like to thank Mrs Jedličková, Mrs Šipková (Institute of Biophysics), Mrs Hrčková, Mrs Bohunická, Ms Pažoutová, Ms Hesounová, and Mrs Potclanová (Institute of Soil Biology) for culture cultivation and excellent technical help.
References
- Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Biessmann H, Mason JM. Telomerase-independent mechanisms of telomere elongation. Cell Mol Life Sci. 2003;60:2325–2333. doi: 10.1007/s00018-003-3247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010;22:2943–2955. doi: 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cock JM, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- Coleman AW, Mai JC. Ribosomal DNA ITS-1 and ITS-2 sequence comparisons as a tool for predicting genetic relatedness. J Mol Evol. 1997;45:168–177. doi: 10.1007/pl00006217. [DOI] [PubMed] [Google Scholar]
- Collard BCY, Das A, Virk PS, Mackill DJ. Evaluation of “quick” and “dirty” DNA extraction methods for marker-assisted selection in rice (Oryza sativa L.) Plant Breed. 2007;126:47–50. [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Derelle E, et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci U S A. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajkus J, et al. Plant cells express telomerase activity upon transfer to callus culture, without extensively changing telomere lengths. Mol Gen Genet. 1998;260:470–474. doi: 10.1007/s004380050918. [DOI] [PubMed] [Google Scholar]
- Fajkus J, Sykorova E, Leitch AR. Telomeres in evolution and evolution of telomeres. Chromosome Res. 2005;13:469–479. doi: 10.1007/s10577-005-0997-2. [DOI] [PubMed] [Google Scholar]
- Fitzgerald MS, McKnight TD, Shippen DE. Characterization and developmental patterns of telomerase expression in plants. Proc Natl Acad Sci U S A. 1996;93:14422–14427. doi: 10.1073/pnas.93.25.14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fojtova M, Fulneckova J, Fajkus J, Kovarik A. Recovery of tobacco cells from cadmium stress is accompanied by DNA repair and increased telomerase activity. J Exp Bot. 2002;53:2151–2158. doi: 10.1093/jxb/erf080. [DOI] [PubMed] [Google Scholar]
- Fojtova M, et al. Telomere maintenance in liquid crystalline chromosomes of dinoflagellates. Chromosoma. 2010;119:485–493. doi: 10.1007/s00412-010-0272-y. [DOI] [PubMed] [Google Scholar]
- Frydrychova R, et al. Phylogenetic distribution of TTAGG telomeric repeats in insects. Genome. 2004;47:163–178. doi: 10.1139/g03-100. [DOI] [PubMed] [Google Scholar]
- Frydrychova R, Marec F. Repeated losses of TTAGG telomere repeats in evolution of beetles (Coleoptera) Genetica. 2002;115:179–187. doi: 10.1023/a:1020175912128. [DOI] [PubMed] [Google Scholar]
- Fuchs J, Brandes A, Schubert I. Telomere sequence localization and karyotype evolution in higher plants. Plant Syst Evol. 1995;196:227–241. [Google Scholar]
- Fuchs J, Schubert I. Arabidopsis-type telomere sequences on chromosome termini of Selaginella martensii Spring (Pteridophyta) Biol Zbl. 1996;115:260–265. [Google Scholar]
- Gilson P, McFadden GI. The chlorarachniophyte: a cell with two different nuclei and two different telomeres. Chromosoma. 1995;103:635–641. doi: 10.1007/BF00357690. [DOI] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas sourcebook. San Diego (CA): Academic Press; 1989. [Google Scholar]
- Higashiyama T, Maki S, Yamada T. Molecular organization of Chlorella vulgaris chromosome I: presence of telomeric repeats that are conserved in higher plants. Mol Gen Genet. 1995;246:29–36. doi: 10.1007/BF00290130. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Katana A, et al. Phylogenetic position of Koliella (Chlorophyta) as inferred from nuclear and chloroplast small subunit rDNA. J Phycol. 2001;37:443–451. [Google Scholar]
- Leliaert F, et al. Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci. 2012;31(1):1–46. [Google Scholar]
- Malik HS, Henikoff S. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- McClintock B. The fusion of broken chromosome ends of sister half-chromatids following chromatid breakage at meiotic anaphases. Mo Agric Exp Stn Res Bull. 1938;290:1–48. [Google Scholar]
- Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyne J, Ratliff RL, Moyzis RK. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci U S A. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Proceedings of the Gateway Computing Environments Workshop (GCE) 2010 Nov 14; New Orleans (LA); New Orleans, LA: Institute of Electrical and Electronics Engineers (IEEE): 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; pp. 1–8. [Google Scholar]
- Mravinac B, Mestrovic N, Cavrak VV, Plohl M. TCAGG, an alternative telomeric sequence in insects. Chromosoma. 2011;120:367–376. doi: 10.1007/s00412-011-0317-x. [DOI] [PubMed] [Google Scholar]
- Nakada T, Misawa K, Nozaki H. Molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18S rRNA phylogenetic analyses. Mol Phylogenet Evol. 2008;48:281–291. doi: 10.1016/j.ympev.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Nemcova Y, et al. Jenufa gen. nov.: a new genus of coccoid green algae (Chlorophyceae, Incertae Sedis) previously recorded by environmental sequencing. J Phycol. 2011;47:928–938. doi: 10.1111/j.1529-8817.2011.01009.x. [DOI] [PubMed] [Google Scholar]
- Neplechova K, Sykorova E, Fajkus J. Comparison of different kinds of probes used for analysis of variant telomeric sequences. Biophys Chem. 2005;117:225–231. doi: 10.1016/j.bpc.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Neustupa J, et al. Xylochloris irregularis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta), a novel subaerial coccoid green alga. Phycologia. 2011;50:57–66. [Google Scholar]
- Okazaki S, et al. Identification of a pentanucleotide telomeric sequence, (TTAGG)n, in the silkworm Bombyx mori and in other insects. Mol Cell Biol. 1993;13:1424–1432. doi: 10.1128/mcb.13.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peska V, Sykorova E, Fajkus J. Two faces of Solanaceae telomeres: a comparison between Nicotiana and Cestrum telomeres and telomere-binding proteins. Cytogenet Genome Res. 2008;122:380–387. doi: 10.1159/000167826. [DOI] [PubMed] [Google Scholar]
- Petracek ME, Lefebvre PA, Silflow CD, Berman J. Chlamydomonas telomere sequences are A+T-rich but contain three consecutive G-C base pairs. Proc Natl Acad Sci U S A. 1990;87:8222–8226. doi: 10.1073/pnas.87.21.8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlevsky JD, et al. The telomerase database. Nucleic Acids Res. 2008;36:D339–D343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik SE, et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329:223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proschold T, Harris EH, Coleman AW. Portrait of a species: Chlamydomonas reinhardtii. Genetics. 2005;170:1601–1610. doi: 10.1534/genetics.105.044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proschold T, Marin B, Schlosser UG, Melkonian M. Molecular phylogeny and taxonomic revision of Chlamydomonas (Chlorophyta). I. Emendation of Chlamydomonas Ehrenberg and Chloromonas Gobi, and description of Oogamochlamys gen. nov. and Lobochlamys gen. nov. Protist. 2001;152:265–300. doi: 10.1078/1434-4610-00068. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Ausubel FM. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- Rotkova G, et al. An evolutionary change in telomere sequence motif within the plant section Asparagales had significance for telomere nucleoprotein complexes. Cytogenet Genome Res. 2004;107:132–138. doi: 10.1159/000079584. [DOI] [PubMed] [Google Scholar]
- Rotkova G, Sykorova E, Fajkus J. Characterization of nucleoprotein complexes in plants with human-type telomere motifs. Plant Physiol Biochem. 2007;45:716–721. doi: 10.1016/j.plaphy.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning—a laboratory manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shampay J, Blackburn EH. Tetrahymena micronuclear sequences that function as telomeres in yeast. Nucleic Acids Res. 1989;17:3247–3260. doi: 10.1093/nar/17.8.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Suzuki K. Characterization of telomere DNA among five species of pteridophytes and bryophytes. J Bryol. 2004;26:175–180. [Google Scholar]
- Sykorova E, et al. Minisatellite telomeres occur in the family Alliaceae but are lost in Allium. Am J Bot. 2006;93:814–823. doi: 10.3732/ajb.93.6.814. [DOI] [PubMed] [Google Scholar]
- Sykorova E, Leitch AR, Fajkus J. Asparagales telomerases which synthesize the human type of telomeres. Plant Mol Biol. 2006;60:633–646. doi: 10.1007/s11103-005-5091-9. [DOI] [PubMed] [Google Scholar]
- Sykorova E, et al. The absence of Arabidopsis-type telomeres in Cestrum and closely related genera Vestia and Sessea (Solanaceae): first evidence from eudicots. Plant J. 2003a;34:283–291. doi: 10.1046/j.1365-313x.2003.01731.x. [DOI] [PubMed] [Google Scholar]
- Sykorova E, et al. Telomere variability in the monocotyledonous plant order Asparagales. Proc R Soc Lond B Biol Sci. 2003b;270:1893–1904. doi: 10.1098/rspb.2003.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, et al. Deep division in the Chlorophyceae (Chlorophyta) revealed by chloroplast phylogenomic analyses. J Phycol. 2008;44:739–750. doi: 10.1111/j.1529-8817.2008.00510.x. [DOI] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, et al. Chromosome termini of the monocot plant Othocallis siberica are maintained by telomerase, which specifically synthesises vertebrate-type telomere sequences. Plant J. 2004;37:484–493. doi: 10.1046/j.1365-313x.2003.01974.x. [DOI] [PubMed] [Google Scholar]
- Zauner S, et al. Chloroplast protein and centrosomal genes, a tRNA intron, and odd telomeres in an unusually compact eukaryotic genome, the cryptomonad nucleomorph. Proc Natl Acad Sci U S A. 2000;97:200–205. doi: 10.1073/pnas.97.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]