Abstract
The ability of Streptococcus mutans to produce and tolerate organic acids from carbohydrate metabolism represents a major virulence factor responsible for the formation of carious lesions. Pyruvate is a key metabolic intermediate that, when rerouted to other metabolic pathways such as amino acid biosynthesis, results in the alleviation of acid stress by reducing acid end products and aiding in maintenance of intracellular pH. Amino acid biosynthetic genes such as ilvC and ilvE were identified as being upregulated in a proteome analysis of Streptococcus mutans under acid stress conditions (A. C. Len, D. W. Harty, and N. A. Jacques, Microbiology 150:1353–1366, 2004). In Lactococcus lactis and Staphylococcus carnosus, the ilvE gene product is involved with biosynthesis and degradation of branched-chain amino acids, as well as in the production of branched-chain fatty acids (B. Ganesan and B. C. Weimer, Appl. Environ. Microbiol. 70:638–641, 2004; S. M. Madsen et al., Appl. Environ. Microbiol. 68:4007–4014, 2002; and M. Yvon, S. Thirouin, L. Rijnen, D. Fromentier, and J. C. Gripon, Appl. Environ. Microbiol. 63:414–419, 1997). Here we constructed and characterized an ilvE deletion mutant of S. mutans UA159. Growth experiments revealed that the ilvE mutant strain has a lag in growth when nutritionally limited for branched-chain amino acids. We further demonstrated that the loss of ilvE causes a decrease in acid tolerance. The ilvE strain exhibits a defect in F1-Fo ATPase activity and has reduced catabolic activity for isoleucine and valine. Results from transcriptional studies showed that the ilvE promoter is upregulated during growth at low pH. Collectively, the results of this investigation show that amino acid metabolism is a component of the acid-adaptive repertoire of S. mutans.
INTRODUCTION
The human oral cavity contains a large and complex bacterial community, with estimates of approximately 200 species in individuals and perhaps as many as 700 species distributed across the human population (1). Among oral bacteria, Streptococcus mutans is well established as a causative agent of human dental caries (44). Production of organic acids as a consequence of sugar fermentation by S. mutans results in a dramatic decrease in overall pH within dental plaque (47), which, upon long-term exposure, leads to damaged tooth surfaces and development of caries. Such an environment is not only detrimental to the host but also represents a challenge for microbial survival. Unlike other members of the oral microflora, which are generally sensitive to acidic pHs, Streptococcus mutans is both acidogenic and aciduric. As a result, the organism has evolved a variety of adaptive mechanisms that contribute to its ability to tolerate and survive in low-pH conditions (29, 37, 44, 47). The acid tolerance response includes at least the following elements: upregulation of the membrane-bound, proton-pumping F1-Fo ATPase (9, 33), production of alkali via the agmatine deiminase system (26), alterations in metabolic pathways (37), and low-pH-dependent changes in membrane fatty acid composition (20).
Studies with lactic acid bacteria, including S. mutans, have indicated that when representative organisms encounter stress as a consequence of their own acid secretion, metabolic products that would be used to produce organic acids can be rerouted or modulated, with the effect of alleviating the acid burden (29). For example, Lactobacillus bulgaricus generates lactic acid from pyruvate, and as part of its acid tolerance response, it alleviates acidic conditions by rerouting pyruvate to fatty acid biosynthesis through the induction of biosynthetic genes involved in lipid metabolism (17). Furthermore, proteomic analysis of the acid tolerance response in S. mutans showed that expression of several metabolic proteins involved in central carbon metabolism was altered under acidic growth conditions (40). Among these were elevated levels of proteins involved in branched-chain amino acid (bcAA) biosynthesis. Jacques et al. proposed that S. mutans could survive under low-pH conditions by adjusting end products of carbon utilization, including the rerouting of carbon rather than pyruvate into amino acid biosynthesis to maintain intracellular pH (39). Alteration of carbon flux from acid production to branched-chain amino acid synthesis may be an important physiological alternative that contributes to acid tolerance and the ability of S. mutans to outcompete other oral species (43).
The initial reaction in the synthesis of the branched-chain amino acids leucine and valine is the condensation of two molecules of pyruvate to acetolactate via acetolactate synthase (16). In the process, pyruvate is converted to α-keto acid intermediates and, by a series of sequential reactions, finally converted by aminotransferases (ATases) into branched-chain amino acids. During branched-chain amino acid synthesis, both pyruvate and NADPH are consumed, thereby reducing the amount of carbon flowing into lactic acid. Branched-chain amino acid biosynthesis of isoleucine, on the other hand, also produces NH3+, which could ameliorate acidification of the cytoplasm by reacting with protons to form ammonia (40). In Streptococcus thermophilus, ilvC, a branched-chain amino acid biosynthetic gene, is thought to play a role in intracellular pH homeostasis (24).
In studies involving S. mutans, differential display reverse transcription-PCR was used to identify genes involved in the acid stress response of Streptococcus mutans GS-5 (13). The results indicated that a gene identified as AP-185 was induced upon acid treatment. A mutant strain defective in AP-185 exhibited decreased growth, with susceptibility to acid killing at low pH. The gene was later shown to share homology with branched-chain amino acid aminotransferases of Bacillus subtilis (35).
Proteome analysis of Streptococcus mutans LT11 grown in continuous culture identified a branched-chain amino acid aminotransferase homologous to Streptococcus pneumoniae Sp086, and also to ilvC, in both the cellular and extracellular milieu (39). In that proteome analysis, performed by Len et al. (39) and using conditions of acid stress, several genes involved in branched-chain amino acid biosynthesis were upregulated at pH 5.0, including ilvC, encoding a keto acid reductoisomerase, and ilvE, encoding a branched-chain amino acid aminotransferase. Two-dimensional fluorescence gel analysis of S. mutans biofilms also identified ilvE as being expressed differentially (48). Taken together, these findings suggest a role for amino acid biosynthetic genes in the acid stress response and could represent a pathway for the modulation of acid end products of fermentation. IlvC also links amino acid biosynthesis to acetolactate-acetoin metabolism, where pyruvate can be transformed into acetoin, thus allowing for the production of more neutral compounds with a higher pKa than that of lactic acid. Branched-chain amino acid ATases catalyze the final step of amino acid biosynthesis; however, due to their dual function, they can play a role in both the biosynthesis and catabolism of branched-chain amino acids (6, 23). In their biosynthetic capacity, ATases catalyze the final step in the generation of bcAAs. In their catabolic capacity, they initiate degradation of bcAAs for the production of α-keto acids, which are intermediates in branched-chain fatty acid (bcFA) production (31, 32). The ilvE gene, encoding a branched-chain amino acid aminotransferase, has been shown to play an important role in bcFA synthesis by providing precursors for the fatty acid biosynthetic machinery. It has also been shown that a mutation in ilvE leads to a nutritional requirement for amino acids and a change in the catabolites produced from amino acid degradation (6). For Staphylococcus carnosus, an ilvE mutant was shown to lose ATase activity for branched-chain amino acids (42), demonstrating its role in amino acid catabolism.
In the present study, we focused on the role of a putative ilvE ortholog, SMU.1203c, annotated as a branched-chain amino acid aminotransferase gene in the UA159 genome (2). The study was based on the hypothesis that branched-chain amino acids, as precursors of branched-chain fatty acids, could be metabolized by IlvE, which could potentially contribute to acid resistance in S. mutans. Results from physiological studies revealed that the loss of ilvE led to a nutritional requirement for branched-chain amino acids, supporting the role of the gene in amino acid biosynthesis. Deletion of ilvE also led to an acid-sensitive phenotype, as demonstrated by a decrease in survival upon acid challenge. Promoter fusion experiments showed that the ilvE promoter is responsive to acidic external pH values. Taken together, the results presented in this report establish that IlvE is a significant contributor to the acid-adaptive mechanism of S. mutans.
MATERIALS AND METHODS
Bacterial strains.
Strains in this study are detailed in Table 1. Streptococcus mutans UA159, the genomic type strain, was the parent strain used in this study (2, 45). Mutant strains of S. mutans UA159 were deleted for the entire coding regions of ilvE (SMU.1203c) and aspC (SMU.24), using a PCR-based, ligation-independent cloning method (LIC mutagenesis) as described previously (3, 36, 51). Flanking regions of approximately 400 bp upstream and downstream were used to mediate replacement of genomic copies of genes, resulting in S. mutans ΔilvE (SMU.1203c) and ΔaspC (SMU.24) strains. Appropriate constructs were verified using specific primers for each strain. The ΔilvE strain was verified using primers MU1099 P7UPF (5′-TGAGCACCTCAGCCAAAACATTAC-3′) and MU1099 P8DNR (5′-ACAGAAAGTCCACCCTTAACAGCA-3′). The ΔaspC strain was verified by using primers MU0020 P7UPF (5′-GCAGAGAAAGTGGCTAAGACTATTGG-3′) and MU0020 P8DNR (5′-TTGTCTCCACCCATAGCATCAACT-3′).
Table 1.
Strains and plasmids used for this study
| Strain or plasmid | Relevant feature(s) | Source or reference |
|---|---|---|
| S. mutans strains | ||
| UA159 | Parent strain | 2 |
| ΔilvE strain | SMU.1203c (ΔilvE) mutant strain of UA159; Ermr | This study |
| ΔaspC strain | SMU.24 (ΔaspC) mutant strain of UA159; Ermr | This study |
| UR287 | ilvE complement strain (ilvEc-); Ermr Kanr | This study |
| UR212 | UA159 transformed with pJL84, carrying a promoterless CAT fusion; Kanr | This study |
| UR213 | UA159 transformed with pJL1099, carrying an ilvE-CAT fusion; Kanr | This study |
| Plasmids | ||
| pCRBlunt | Cloning vector | Invitrogen |
| pCRilvEc | ilvE coding region and cognate promoter in pCRBlunt; Kanr | This study |
| pSUGKBgl | Integration vector for complementation into gtfA locus; Kanr | 15 |
| pSUGBilvE | Complementation vector used to create UR287 | This study |
| pJL84 (VC) | Promoterless CAT reporter plasmid; Kanr | Jose Lemos |
| pJL1099 | ilvE promoter-CAT reporter fusion; Kanr | This study |
Generation of ilvE complement strain.
A genetic complement strain of S. mutans was created using primers to amplify the ilvE coding and intergenic regions from UA159 chromosomal DNA. Primer 1099BglIIUp (5′-CATAGATCTGATAACATTTCTTACAAAAAG-3′) binds upstream of the ilvE promoter region and contains a BglII restriction site (underlined). Primer 1099BglIIDown (5′-CAGTAGATCTGCTTGCAAGAGCAAGCTCGC-3′) binds downstream of the ilvE coding region and contains a BglII restriction site (underlined). The resulting PCR product was cloned into pCRBlunt (Invitrogen, Carlsbad, CA), and clones were verified by nucleotide sequencing. A positive clone, pCRilvEc, containing the ilvE coding region and cognate promoter, was digested with BglII, and the insert was gel purified using a QIAquick gel extraction kit (Qiagen, Valencia, CA). Purified fragments were cloned into the integration vector pSUGKBgl (15), and presumptive clones were screened by colony PCR. The ΔilvE mutant strain was transformed with pSUGBilvE, and colonies were selected on brain heart infusion (BHI) agar medium containing kanamycin. Positive transformants were identified by colony PCR. One strain, containing the ilvE coding region and promoter in the gtfA locus (SMU.0881), was named UR287.
Bacterial growth conditions.
S. mutans batch cultures were grown in BHI broth (Becton Dickinson/Difco, Franklin Lakes, NJ), tryptone-yeast extract (TY) medium plus 1% (wt/vol) glucose, or FMC minimal medium (56) plus 1% (wt/vol) glucose. Nutritional supplements were exogenously supplied amino acids or fatty acids. Final concentrations of additives were as follows: 1 mM leucine (L), 1 mM isoleucine (I), and 1 mM valine (V), individually or mixed (ILV), as well as 1 mM isovaleric acid (IA). For ATase assays, cultures of desired strains were grown overnight in BHI broth (Becton Dickinson/Difco). Overnight cultures were diluted 1:20 (in triplicate) in fresh BHI and grown to an optical density at 600 nm (OD600) of 0.4 to 0.6. Cells were harvested and pellets stored frozen until assayed for enzymatic activity.
Bioscreen growth curves.
Comparative growth data were obtained using a Bioscreen C plate (Growth Curves USA, Haverhill, MA) as previously described (30). Briefly, overnight cultures were diluted 1:20 in fresh BHI medium or FMC minimal medium and allowed to grow to an OD600 of approximately 0.3. Media were prepared for each specific growth condition tested, and 300 μl was loaded into the wells of a Bioscreen plate (Growth Curves USA, Haverhill, MA). For FMC minimal medium (55, 56), the final concentration of additives was 1 mM isoleucine, 1 mM isovaleric acid, or a 1 mM concentration of each amino acid (ILV), combined. The plates were inoculated with 10 μl of cell suspension per well, and growth at 37°C was monitored for 24 h, with 15 s of shaking before each read.
Gas chromatography-fatty acid methyl ester (GC-FAME) analysis of fatty acid composition.
Cells were harvested from steady-state chemostat cultures of the ilvE mutant strain grown in TY medium plus 1% (wt/vol) glucose at a pH of 5 or 7. Two independent cultures were sent for analysis. Membrane fatty acid analysis was performed by Microbial ID (Newark, DE) per the method of Bligh and Dyer (10).
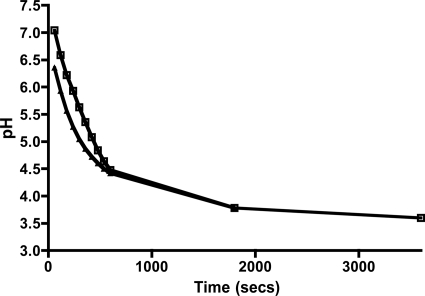
Minimum glycolytic pH determination.
Minimum glycolytic pH values were determined following an established protocol (8). Briefly, the ilvE mutant strain was grown overnight in BHI medium supplemented with 1% (wt/vol) glucose. Cells were harvested, washed, and resuspended in 50 mM KCl, 1 mM MgCl2. A 5-ml aliquot of cell suspension was stirred at room temperature, and the pH was adjusted to 7.2 using KOH. Glucose was added to the cell suspension at 55.5 mM (final concentration), and the pH value was measured every 15 s for 30 min. The experiment was performed in duplicate for two independent cultures.
Acid sensitivity determination.
Acid challenges were performed as described previously (20). Briefly, 10 ml of cells was harvested, in duplicate, from steady-state cultures grown at pH 5 or 7 and resuspended in 0.1 M glycine, pH 2.5. Cells were removed at the time points indicated, serially diluted, and plated on BHI agar medium. Results are presented as log(N/N0) values. Assays were performed on two independent cultures, plated in duplicate.
Aminotransferase assays.
Cell lysates were prepared and ATase assays performed as previously described (58). Briefly, cells grown to late logarithmic phase were collected by centrifugation and washed with 50 mM β-glycerophosphate buffer, pH 7.0. Cell pellets were resuspended in 50 mM KPO4, pH 7.5, 2 mM 2-mercaptoethanol, 1 mM EDTA, and 0.1 mM pyridoxal-5′-phosphate (PLP; Sigma-Aldrich, St. Louis, MO) to a final volume of 2 ml and lysed using a mini-bead beater (Biospec Products, Bartlesville, OK). After centrifugation (14,000 × g for 20 min at 4°C), cell lysates were filtered through a 0.45-μm filter (Millipore, Billerica, MA) and used for enzymatic assays. Total protein concentrations were estimated by the method of Bradford (11). Aminotransferase activity was determined using an l-glutamic acid detection kit (R Biopharm Inc., Marshall, MI). The individual amino acid donors, isoleucine, leucine, and valine, were tested in each cell extract at a final concentration of 3 mM, while the acceptor, α-ketoglutarate (Sigma-Aldrich, St. Louis, MO), was used at a 10 mM final concentration. ATase activity is presented as nmol of glutamate detected per mg of protein. Each determination was performed in triplicate for each condition and strain. A reaction mixture without cell lysate was used as a background control.
ATPase assays.
Permeabilization of cells and ATPase assays were performed as previously described (7, 46). Permeabilized cells were incubated and aliquots removed at timed intervals. Inorganic phosphate released from ATP in permeabilized cells was measured using Fiske and Subbarow reagent (Sigma-Aldrich) (18). Assays were performed on cells harvested from three independent steady-state cultures per condition, performed in duplicate. Data are expressed as millimoles PO4 released per μg protein.
Proton permeability assays.
Proton permeability assays were performed as described previously (9, 21, 41). Briefly, 200-ml cultures of the ilvE mutant strain were grown in TY plus 1% (wt/vol) glucose at 37°C in 5% (vol/vol) CO2–95% (vol/vol) air overnight. Cells were harvested by centrifugation, washed in 5 mM MgCl2, resuspended in 20 mM KPO4 buffer, pH 7.2, 50 mM KCl, 1 mM MgCl2, and incubated for 2 h. After incubation, cells were harvested and resuspended to 20 mg/ml. A 1-ml aliquot was titrated to pH 4.7 by the addition of 10 mM HCl–50 mM KCl, and the pH value was recorded. At the 50-min time point, 10% (vol/vol) butanol was added to disrupt the cell membrane and allow the cytoplasmic and external pHs to equilibrate. At 80 min, final pH values were recorded. These determinations were performed in triplicate using three independent cultures.
ilvE promoter-CAT reporter fusion construction.
For chloramphenicol acetyltransferase (CAT) transcriptional fusions, the ilvE intergenic region was amplified via PCR using the primers 1099FWD (5′-CATGAGCTCGATAACATTTCTTACAAAAAGACAGG-3′) (SacI site is underlined) and 1099REV (5′-CATGGATCCTTTAATACCTCTTTCCTCG-3′) (BamHI site is underlined). The ilvE intergenic region PCR product was cloned directly into the Zero Blunt vector (Invitrogen). The ilvE promoter Zero Blunt clone was digested with the BamHI and SacI restriction enzymes. Digested fragments were gel purified and cloned into the promoterless chloramphenicol acetyltransferase reporter plasmid pJL84 digested with BamHI and SacI. Transformants were selected on LB-kanamycin (LB-Kan) (50 μg/ml) plates. Clones were screened by colony PCR, and a correct construct was named pJL1099. The S. mutans parent strain UA159 was transformed with promoterless pJL84 (vector control [VC]) and pJL1099 to generate a single-copy integration reporter fusion. Clones were screened for proper integration into the chromosome by PCR. Transformants for each construct were named as described in Table 1.
CAT assay.
CAT assays were performed following a previously described protocol (34, 50). Briefly, cell pellets were resuspended in 1 ml 10 mM Tris-Cl, pH 7.8. Resuspended cells were lysed using a mini-bead beater and 0.1-mm glass beads (Biospec Products, Bartlesville, OK). Cell debris was removed by centrifugation at 10,000 rpm for 10 min. Lysates were removed and used for total protein quantification (11) and CAT assays. CAT assay reaction mixtures consisted of 50 μl of whole-cell lysate, 100 mM Tris-Cl, pH 7.8, 0.1 mM acetyl-coenzyme A (acetyl-CoA), and 0.4 mg/ml 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB) in a total volume of 1 ml. Reactions were initiated by addition of 0.1 mM chloramphenicol. Optical density measurements at 412 nm were monitored over 3 min. Reaction rate and total protein quantifications were used to determine CAT activity, represented as μmol chloramphenicol acetylated/min/mg of total protein.
RESULTS
Identification of ilvE, a branched-chain amino acid aminotransferase in S. mutans.
Homology searches using identified branched-chain aminotransferase genes from B. subtilis and Lactococcus lactis (4) showed that SMU.1203c (2) (or bcaT/ilvE) in the Streptococcus mutans genome shared a high level of homology at the amino acid level with genes from these organisms. The presumptive IlvE reading frame is annotated as a branched-chain amino acid aminotransferase (ATase), and through SIM sequence alignment (http://ca.expasy.org/tools), it was shown to share 74.1% identity with the L. lactis branched-chain aminotransferase BcaT (Fig. 1) and homology, to a lesser extent, with the B. subtilis YbgE and Salmonella enterica serovar Typhimurium CT18 IlvE proteins (data not shown). The S. mutans IlvE protein sequence contains the distinctive class IV signature pattern consistent with pyridoxal-5′-phosphate-dependent enzymes such as aminotransferases (5). The SMU.1203c gene described in this study is referred to as ilvE from this point forward.
Fig 1.
Alignment of aminotransferase orthologs. Alignment of the SMU.1203c (ilvE) branched-chain amino acid aminotransferase and the L. lactis BcaT 0086 branched-chain aminotransferase. Alignment was done using the Expasy (http://ca.expasy.org/tools/) SIM protein alignment tool. Alignment was generated after querying the S. mutans IlvE protein sequence (http://www.oralgen.lanl.gov) with the L. lactis BcaT protein sequence. The boxed region indicates the canonical class IV signature pattern consistent with pyridoxal-5′-phosphate-dependent enzymes.
Growth of the ilvE mutant strain is impaired in minimal medium.
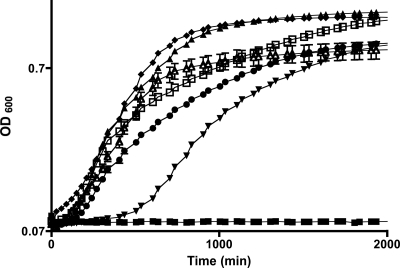
To examine the role of IlvE in the physiology of S. mutans, a deletion mutant strain was created as described in Materials and Methods. As an aminotransferase gene, ilvE is part of the branched-chain amino acid biosynthetic pathway, and loss of the gene could confer a nutritional requirement. Therefore, the mutant strain was examined for growth defects by monitoring growth in rich and minimal media. The ilvE mutant strain was able to grow in rich medium, exhibiting an extended lag phase and a reduced rate of exponential-phase growth; however, the final growth yield was similar to that of the parent strain (data not shown). The ilvE mutant strain was then assessed for growth under nutrient-limiting conditions. Inoculation of FMC medium with the ilvE mutant strain from a plate colony generally resulted in no growth (data not shown). The ilvE mutant strain was able to grow in minimal medium lacking the branched-chain amino acids isoleucine, leucine, and valine, possibly owing to carryover of nutrients from media in seed cultures, although there was a pronounced delay in lag phase and a decreased exponential-phase growth rate compared to those of the parent strain (Fig. 2). Addition of bcAAs to minimal medium restored growth of the ilvE mutant strain to levels similar to those of the parent strain. An exogenous supply of isovaleric acid (IA), an end product of leucine degradation, also aided growth of the mutant strain. To confirm that the growth defect was caused by the loss of ilvE, a genetic complement strain was created (UR287). The ilvE complement strain grew without the addition of bcAAs, confirming that the ilvE defect had been restored in the complement strain (Fig. 2). The complement strain was restored to a lag phase similar to that of the parent strain, though the final growth yield was somewhat lower than that of the parent. The source of the differences between the parent and complement strains is not clear, though it may be related to the ectopic location of the ilvE gene in the gtfA locus. These results indicate that the growth defect of the ilvE mutant strain can be overcome in part by exogenous supply of either bcAAs or by-products derived from bcAAs.
Fig 2.
Growth of S. mutans ilvE mutant in FMC minimal medium. The parent strain, UA159 (triangles), the ilvE mutant strain (inverted triangles), and the ilvE complement strain (open triangles) were grown in FMC minimal medium. Growth medium for the ilvE mutant was supplemented with branched-chain amino acids, using a final concentration of 1 mM isoleucine (circles) or a mixture of 1 mM (each) isoleucine, leucine, and valine (ILV) (diamonds). Cultures of the ilvE mutant strain were also supplemented with isovaleric acid (IA) (open squares), an end product of leucine degradation. Filled squares, uninoculated growth medium. Error bars represent standard deviations.
Loss of ilvE results in a lag in growth at low pH.
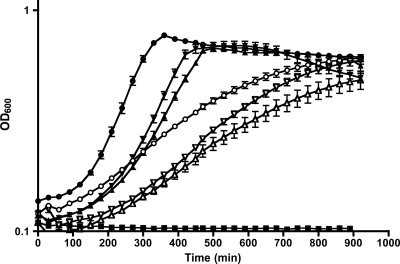
In order to determine whether the absence of a biosynthetic gene such as ilvE would have an effect on growth at low pH, the growth kinetics of the ilvE mutant at pH 5.4 were determined. Results from growth experiments with the ilvE mutant and parent strains in BHI medium buffered to pH 7 and BHI medium titrated to pH 5.4 showed that the ilvE mutant strain exhibited diminished growth kinetics and reduced final growth yields compared to those of the parent strain at pH 5.4 (Fig. 3). The results support the concept that bcAA metabolism is part of acid resistance in S. mutans.
Fig 3.
Growth of S. mutans ilvE mutant at low pH. The parent strain, UA159 (circles), the ilvE mutant strain (triangles), and the ilvE complement strain (inverted triangles) were grown in BHI medium titrated to pH 5.4 with HCl (open symbols) or buffered to pH 7 with KPO4 (closed symbols) (n = 10 for each strain). Error bars represent standard deviations.
The ilvE mutant strain is acid sensitive.
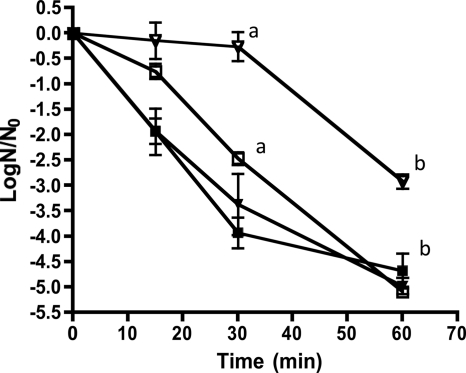
The lag in growth of the ilvE deletion strain at low pH suggested an inability to adapt to acid stress and to survive under acidic conditions. Typically, S. mutans grown at low pH exhibits a robust adaptive response resulting in a substantial ability to survive subsequent acidic insult. We performed an acid challenge experiment to determine whether the ilvE mutant strain was defective in its acid-adaptive response. Steady-state cultures of the ilvE mutant strain grown to pH 5 or 7 were exposed to an acid stress with 0.1 M glycine, pH 2.5. Compared to the parent strain grown under the same conditions and exposed to acid stress, the ilvE mutant strain was less able to survive the challenge, as evidenced by a 2-log difference in the number of viable organisms (Fig. 4). Thus, deletion of ilvE presented a significant defect in the ability of the organism to adapt to acidification.
Fig 4.
The ilvE mutant is acid sensitive. S. mutans UA159 (inverted triangles) and the ilvE mutant (squares) were grown to steady state in chemostat cultures at a pH of 5 (open symbols) or 7 (closed symbols) and then exposed to acid challenge (pH 2.5). Data are from two independent cultures performed in duplicate and are represented as log(N/N0) values. Error bars represent standard deviations. Statistically significant pairwise comparisons are indicated by shared letters (a and b) for open symbols. Statistical significance was determined by Student's t test. a, P ≤ 0.001; b, P ≤ 0.001.
Membrane fatty acid composition of the ilvE mutant strain is responsive to acid stress.
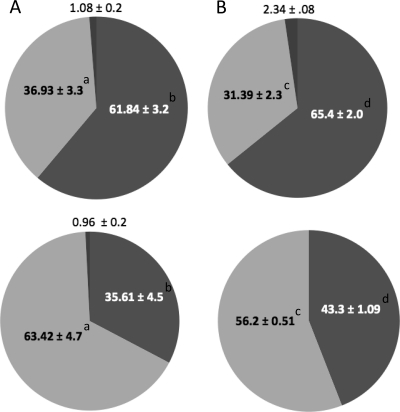
Previously, we reported that during growth under acidic conditions, S. mutans alters its membrane fatty acid composition by increasing the proportion of unsaturated fatty acids (19, 20). Changes in the proportion and type of membrane fatty acid are a common bacterial response to environmental stress (25, 27, 49). The changes in membrane composition in S. mutans are essential for survival of acid stress, since the inability to incorporate unsaturated fatty acids leads to extreme acid sensitivity and decreased virulence in the rat model of caries (22). In order to rule out a defect in adaptation of the membrane fatty acid content as the cause of the sensitivity of the ilvE mutant, GC-FAME analysis was performed using cells from cultures grown to steady state. Compared to the parent strain, the ilvE mutant showed no difference in membrane fatty acid composition when it was grown at low pH (Fig. 5). The ilvE mutant was able to shift its fatty acid composition and increase its overall proportion of unsaturated fatty acids to approximately 55% when it was grown at pH 5. The data demonstrate that changes in membrane composition occurred in the ilvE mutant strain; however, they were not sufficient for its acid survival.
Fig 5.
Membrane fatty acid composition of the ilvE mutant strain. GC-FAME analysis was used to determine membrane fatty acid content for cultures of the parental strain, UA159 (A), or the ilvE mutant (B) grown to a steady-state pH of 7 (top graphs) or 5 (bottom graphs). Samples for GC-FAME analyses were prepared as described in Materials and Methods. Percentages are shown for unsaturated fatty acids (light gray), saturated fatty acids (medium gray), and branched-chain/“other” fatty acids (dark gray). Values represent percentages of fatty acids recovered from the total fatty acid content (means ± standard deviations [SD]). Statistically significant pairwise comparisons (P ≤ 0.05) are indicated by shared letters (a, b, c, and d).
The ilvE mutant strain does not have a defect in glycolytic output.
Previously, we had shown that mutation of the fabM gene in S. mutans leads to a reduction in the ability of the organism to lower pH values via production of organic acids. Here we examined the ability of the ilvE mutant strain to metabolize glucose. Our interest was to determine whether the loss of IlvE, an amino acid biosynthetic enzyme, could lead to a loss of the ability to direct pyruvate into amino acid biosynthesis and thus result in excessive acid production and increased acid stress. However, the ilvE mutant strain did not show a detectable defect in the glycolytic machinery, as its minimum glycolytic pH did not differ from that of the parent strain, UA159 (Fig. 6). The observation indicated that the inability of the ilvE mutant strain to survive acid challenge was not due to a defect in its glycolytic machinery.
Fig 6.
The ilvE mutant is able to perform glycolysis at low pH. The minimum glycolytic pH under conditions of excess glucose was determined as outlined in Materials and Methods for the parent strain, UA159 (open squares), and the ilvE mutant strain (triangles). The experiment was performed in duplicate for two independent cultures of each strain.
The ilvE mutant strain exhibits impaired F1-Fo ATPase activity.
An important contributor to the known acid response mechanism of S. mutans is the membrane-bound F1-Fo ATPase (34). Extensive work on the S. mutans F1-Fo ATPase has demonstrated its importance in maintenance of proper intracellular pH conditions by means of its proton-pumping activity, which increases under conditions of acid stress (9, 33, 47). Moreover, we have shown that rates of transcription for the F-ATPase operon are directly related to external pH (34). The ilvE mutant strain was able to alter membrane fatty acids in response to acid stress conditions, indicating that acid adaptation occurs to some extent in the deletion strain. In order to determine the mechanism of the acid-sensitive phenotype of the ilvE mutant strain, F1-Fo ATPase assays were performed using cells grown under steady-state conditions at pHs 7 and 5. The results indicated that the ilvE mutant strain had deficiencies in ATPase activity compared to the parent strain. The ilvE mutant strain exhibited a lower ATPase activity than the parent strain at both culture pHs (7 and 5) (Fig. 7). Furthermore, no significant increase in ATPase level was observed in cells grown at low pH, suggesting a defect in one of the major acid-adaptive mechanisms for the organism. The reduced ability to effectively pump protons out of the cytoplasm could result in acid sensitivity of the mutant strain.
Fig 7.
F1-Fo ATPase activity in the ilvE mutant strain is defective. Cultures of S. mutans UA159 (inverted triangles) and the ilvE mutant (squares) were grown to a steady-state pH of 5 (open symbols) or 7 (closed symbols). ATPase activity is expressed as a measure of the inorganic phosphate released from ATP per μg of total protein. Values represent average ATPase activities for three independent cultures assayed twice. Statistically significant pairwise comparisons are indicated as follows: *, P ≤ 0.05; #, P ≤ 0.05.
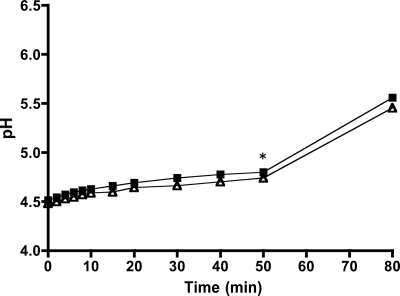
Proton permeability is unaffected in the ilvE mutant strain.
The reduction in F1-Fo ATPase activity led us to examine the ability of the ilvE mutant strain to maintain the ΔpH across the membrane. Intracellular pH in S. mutans is normally basic with respect to external conditions, and a ΔpH of approximately 1.4 has been measured for an external pH of 5.0 for this organism (14). Several glycolytic enzymes are sensitive to pH and therefore could be inhibited under acid stress (9). If the ilvE mutant was unable to maintain the proper ΔpH, this could contribute to its acid-sensitive phenotype. The proton permeability of the membrane of the ΔilvE strain was determined; however, the data suggest that proton diffusion across the membrane of the mutant strain did not differ from that of the parental strain (Fig. 8). We had previously shown that changes in membrane fatty acid composition caused by the loss of unsaturated fatty acids could alter the permeability of the Streptococcus mutans membrane (21). As the GC-FAME results demonstrated, the ilvE mutant strain exhibited a membrane fatty acid composition similar to that of the parent strain, consistent with the observation that membrane permeability was uncompromised in the mutant strain.
Fig 8.
Proton permeability of the ilvE mutant strain. Proton permeability was measured as described in Materials and Methods. At 50 min, butanol was added (*). The average values shown are for three independent cultures of the parent strain, UA159 (squares), and the ilvE mutant strain (triangles).
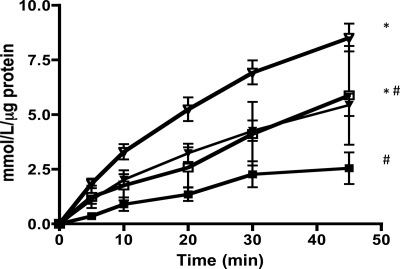
The ilvE mutant strain has altered aminotransferase activity for branched-chain amino acids.
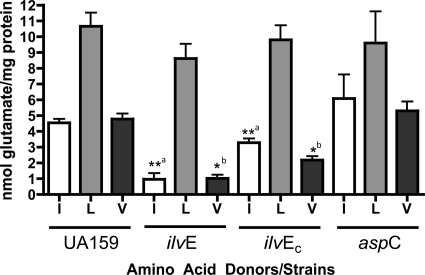
The requirement for branched-chain amino acids under nutrient-limiting conditions suggested a role for IlvE in biosynthesis of branched-chain amino acids. Because ilvE is annotated as an aminotransferase gene, it was essential to verify its biochemical activity. Like other ATases, IlvE in S. mutans could have dual functions, thereby also playing a role in amino acid catabolism. An aminotransferase activity assay was performed for three bcAAs for each strain examined to measure l-glutamic acid released from cell extracts (58, 59).
We predicted that the aminotransferase activity would be altered in the ilvE mutant strain compared to the parent strain. The ATase activity in extracts of the parent strain, UA159, was highest for leucine, followed by isoleucine and valine. Deletion of ilvE resulted in a decrease in ATase activity for isoleucine and valine, but not leucine, compared to that for the parent strain, suggesting that IlvE in S. mutans has ATase activity for isoleucine and valine (Fig. 9). Leucine is derived from a biosynthetic pathway separate from that for isoleucine and valine, and IlvE may be more specific for other branched-chain amino acids. Activity for leucine in extracts of the ilvE mutant and parent strains suggests that S. mutans contains another, as yet uncharacterized ATase, specific for leucine, whose function is independent of IlvE.
Fig 9.
Aminotransferase activities of parent and ilvE mutant strains. ATase activities for branched-chain amino acids are shown for cultures of the parent strain (UA159), the ilvE mutant strain, the ilvE complement strain (ilvEc), and the aspC mutant strain. α-Ketoglutarate (10 mM) was used as an amino acid acceptor. The concentration of the amino acid donors isoleucine (I), leucine (L), and valine (V) was 3 mM. All enzymatic determinations were performed in triplicate for three independent cultures. Statistically significant pairwise comparisons are indicated by shared letters (a and b). Statistical significance was determined by Student's t test (n = 3). *, P ≤ 0.05; **, P ≤ 0.001.
AspC, an aromatic amino acid aminotransferase, does not show catabolic activity for branched-chain amino acids.
In many cases, ATase genes are redundant within the genome of an organism, and as many as 11 different genes encoding ATases can be found in a single organism (23). In L. lactis, two ATases were identified, and only in a double mutant strain where both ATases were absent was the activity completely abolished (58). It has been demonstrated for other organisms that deletion of one ATase does not completely eliminate detectable ATase activity for its cognate amino acid, because other enzymes can perform the same reactions due to redundancy within the genome (12). Using homology searches, we identified an AraT (42) homolog in S. mutans, namely, aspC (SMU.24), which shares approximately 80% homology with lactococcal AraT and is annotated as an aromatic amino acid aminotransferase gene (aspC) (data not shown). A deletion mutant for aspC was created and examined for ATase activity in the presence of bcAA substrates. AspC should not have ATase activity for bcAAs; however, loss of this gene (such as in the case of the lactococcal deletion strain) could have an effect on other ATases and their activities. In S. mutans UA159, loss of aspC did not cause a significant change in ATase activity for the branched-chain amino acids tested (Fig. 9). It appears that AspC has specific activity for aromatic amino acids and does not catabolize branched-chain amino acids under the conditions tested.
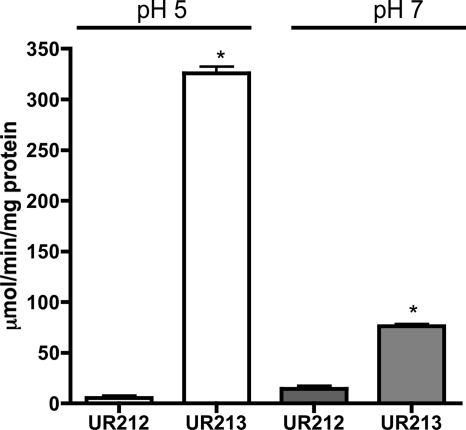
The ilvE promoter is upregulated at acidic pHs.
Proteomic studies identified IlvE as one of the proteins with elevated expression following growth at low pH (39, 40). Our physiological characterization of the ilvE mutant strain demonstrated that ilvE was important for S. mutans acid tolerance and therefore would most likely be induced upon growth at low pH. An ilvE promoter-CAT reporter fusion was constructed to study the effects of low pH on ilvE transcription. When the S. mutans parent strain (UA159) carrying the ilvE-CAT reporter (UR213) and a vector control strain (UR212) were grown at pH 5.0, the ilvE transcript was elevated approximately 7-fold compared to that for growth at pH 7.0 (Fig. 10). The elevated transcription of the ilvE promoter during growth at low pH correlated well with the physiological data showing acid sensitivity of the ΔilvE strain.
Fig 10.
ilvE promoter activity is increased during growth at low pH. Chloramphenicol acetyltransferase activities are shown for S. mutans UR212 (VC) and UR213 (ilvE promoter-CAT reporter fusion). Transcriptional activity was determined for three independent cultures in duplicate for each growth condition as described in Materials and Methods. Statistical significance was determined by Student's t test (n = 3). *, P ≤ 0.00001.
DISCUSSION
Acid adaptation is a major factor in the ability of S. mutans to thrive in dental plaque and to outcompete commensal organisms found within the oral cavity. Many of the adaptive mechanisms that allow the organism to survive in dental plaque have been studied in detail (9, 21, 37, 39). Proteomic studies of S. mutans grown under acid stress conditions suggested that the bacterium may use biosynthesis of amino acids as a way to reroute pyruvate away from lactic acid and also as a way to maintain proper intracellular pH conditions (40). Modulation of the intermediates of glycolysis, as well as removal of protons from the cytoplasm through degradation/biosynthesis of amino acids, can be important for virulence. IlvE was among the proteins identified by Len et al. as being upregulated following growth at low pH (39). Protein sequence homology of IlvE with other known ATases supported its role as a branched-chain amino acid aminotransferase.
Initial characterization of the ilvE mutant strain created in this study revealed a requirement for branched-chain amino acids, which could be restored by supplementing the medium with amino acids or amino acid-derived products such as isovaleric acid. Due to the catabolic and biosynthetic enzymatic activities inherent to ATases, it is possible that the loss of IlvE also had an effect on the end products of amino acid degradation and, for this reason, an exogenous supply of isovaleric acid also aided its growth.
The role of ilvE during growth at low pH was also explored. Growth of the ilvE mutant strain at pH 5.4 showed a lag during exponential phase, with a lower final yield than that of UA159. The defect was specific for acidic growth conditions, since growth at neutral pH was unaffected unless nutritional limitation was encountered.
Since acid adaptation is a key virulence component of S. mutans, the loss of ilvE could alter the ability of the organism to form reducing equivalents from ammonia or to reroute end products such as pyruvate for amino acid biosynthesis. Hence, the acid tolerance of the mutant strain was of interest. We found that the ilvE mutant strain was defective in its acid tolerance compared to the parental strain, in stark contrast to our previous reports concerning acid-adaptive attributes of the parental strain. Our data demonstrate that the loss of ilvE is sufficient to cause a change in acid adaptation and support the contribution of IlvE to acid tolerance.
To clarify the mechanism(s) underlying the acid-sensitive phenotype of the ilvE mutant strain, we first analyzed the membrane fatty acid content by GC-FAME analysis to determine if changes in composition could account for the acid sensitivity. Previous studies have shown that the inability to elevate unsaturated fatty acid content in response to external acidification leads to acid sensitivity (21, 22). GC-FAME analysis of the ilvE mutant strain grown at steady-state culture pHs of 5 and 7 revealed a fatty acid composition similar to that of the parent strain. In agreement with our previous reports (19, 20), the ilvE mutant produced elevated levels of unsaturated membrane fatty acids when it was grown at pH 5. Thus, the mutant exhibited membrane composition changes in response to acid exposure. In unison with the membrane composition data, proton permeability was also relatively unaffected in the mutant. Even though these characteristics are not interchangeable, membrane composition can contribute to and partly explain the permeability characteristics of the mutant. The acid sensitivity displayed by the ilvE mutant strain was not due to an inability to modulate its membrane fatty acid composition, suggesting that the loss of ilvE causes a defect in acid adaptation that is independent of membrane composition. The data also show that the acid-adaptive changes occurring in the membrane are insufficient to compensate for the loss of ilvE.
Results from minimum glycolytic pH assays demonstrated that even though ilvE was absent, the amount of acid produced by the mutant was comparable to that of the parent strain. This was unexpected, because we had hypothesized that the loss of ilvE could potentially have an effect on acid production. Redundancy within the genome could account for the glycolytic profile of the mutant if other ATases are induced in the absence of IlvE.
In experiments with the ilvE mutant and parent strains, we established that IlvE exhibits enzymatic activity on isoleucine and valine. Furthermore, AspC, an aromatic amino acid aminotransferase, does not participate in branched-chain amino acid catabolism, since deletion of the gene had no significant effect on ATase activity compared to that of the parent strain. The data also support an argument that there is another, as yet undefined ATase with activity for leucine.
The biosynthetic role of branched-chain amino acid aminotransferases is vital for the synthesis of important signaling molecules, i.e., the branched-chain amino acids. Under conditions of acid stress, S. mutans requires bcAAs to signal known acid stress responses reliant upon branched-chain amino acids as sensors of overall fitness (38). CodY, a global regulator of S. mutans, uses intracellular branched-chain amino acid pools as coeffectors for regulation of its target genes (53, 54). Loss of ilvE as a branched-chain amino acid aminotransferase could have global effects on the regulatory network of S. mutans. A recent study has shown that CodY activity is affected by ATases that modulate the intracellular pools of amino acids (12). The loss of ilvE could have a much larger effect on the regulatory capabilities of CodY and its targets, which could partly explain the acid sensitivity displayed by the ilvE mutant, particularly if important acid-adaptive processes are disregulated due to inadequate CodY function in the absence of branched-chain amino acids.
The proton-pumping ATPase is essential for maintenance of intracellular pH conditions that are favorable for S. mutans. Levels of ATPase activity were diminished in the ilvE mutant compared to those in the parent strain, reflecting the impact of the mutation on another key adaptive mechanism that was altered in the absence of ilvE.
A mechanism to explain the downregulation of F-ATPase activity in the ilvE mutant strain is not yet clear at this point. We have shown previously that F-ATPase operon transcription is upregulated during growth at low pH (34) and that if the ΔpH is disrupted via the fabM mutation, the cytoplasm becomes acidified and ATPase production is elevated in response (21). Because of the presence of a cre site for potential CcpA binding at position −160 in the F-ATPase operon, it is appealing to interpret reduced F-ATPase activity in the ilvE mutant strain in light of potential CodY and CcpA interactions. Previous work with B. subtilis has shown that the global regulators of transcription CcpA, CodY, and TnrA can participate in regulating branched-chain amino acid biosynthesis (57). Studies with other bacterial model systems, including S. mutans (38) and L. lactis (28), have shown that CodY regulates ilvE in addition to many other genes and that, for S. mutans, a codY strain is acid sensitive (38). Moreover, CodY and CcpA can act as negative and positive regulators of the same gene(s), or cooperatively as positive regulators, in controlling carbon flow through acetate kinase in B. subtilis (52). Experiments are under way to determine potential relationships between CcpA, CodY, and isoleucine and their effects on the expression of the F-ATPase.
The acid-sensitive phenotype of the ilvE mutant strain suggested that the ilvE gene could be under external pH regulation. ilvE promoter fusions showed that transcription of the gene is increased under low-pH conditions. Currently, we are studying the transcriptional regulation of ilvE and its interaction with CodY with respect to its effects on membrane composition and acid tolerance in S. mutans. Transcriptional as well as physiological data indicate that ilvE is part of the acid tolerance response of S. mutans.
ACKNOWLEDGMENTS
This study was supported by NIH/NIDCR grants DE-17157 (R.G.Q.) and DE-17425 (R.G.Q.) and by the Training Program in Oral Sciences at the University of Rochester School of Medicine and Dentistry (T32 DE-07165) (B.S.).
Footnotes
Published ahead of print 10 February 2012
REFERENCES
- 1. Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43: 5721– 5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ajdic D, et al. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99: 14434– 14439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aslanidis C, de Jong PJ. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18: 6069– 6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atiles MW, Dudley EG, Steele JL. 2000. Gene cloning, sequencing, and inactivation of the branched-chain aminotransferase of Lactococcus lactis LM0230. Appl. Environ. Microbiol. 66: 2325– 2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bairoch A. 1993. The PROSITE dictionary of sites and patterns in proteins, its current status. Nucleic Acids Res. 21: 3097– 3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beck HC. 2005. Branched-chain fatty acid biosynthesis in a branched-chain amino acid aminotransferase mutant of Staphylococcus carnosus. FEMS Microbiol. Lett. 243: 37– 44 [DOI] [PubMed] [Google Scholar]
- 7. Belli WA, Marquis RE. 1991. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl. Environ. Microbiol. 57: 1134– 1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Belli WA, Marquis RE. 1994. Catabolite modification of acid tolerance of Streptococcus mutans GS-5. Oral Microbiol. Immunol. 9: 29– 34 [DOI] [PubMed] [Google Scholar]
- 9. Bender GR, Sutton SV, Marquis RE. 1986. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infect. Immun. 53: 331– 338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911– 917 [DOI] [PubMed] [Google Scholar]
- 11. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248– 254 [DOI] [PubMed] [Google Scholar]
- 12. Chambellon E, Yvon M. 2003. CodY-regulated aminotransferases AraT and BcaT play a major role in the growth of Lactococcus lactis in milk by regulating the intracellular pool of amino acids. Appl. Environ. Microbiol. 69: 3061– 3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chia JS, Lee YY, Huang PT, Chen JY. 2001. Identification of stress-responsive genes in Streptococcus mutans by differential display reverse transcription-PCR. Infect. Immun. 69: 2493– 2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dashper SG, Reynolds EC. 1992. pH regulation by Streptococcus mutans. J. Dent. Res. 71: 1159– 1165 [DOI] [PubMed] [Google Scholar]
- 15. Derr AD, et al. 2012. Mutation of NADH oxidase (nox) reveals an overlap of the oxygen- and acid-mediated stress responses in Streptococcus mutans. Appl. Environ. Microbiol. 78: 1215– 1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Epelbaum S, et al. 1998. Branched-chain amino acid biosynthesis in Salmonella typhimurium: a quantitative analysis. J. Bacteriol. 180: 4056– 4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fernandez A, et al. 2008. Rerouting of pyruvate metabolism during acid adaptation in Lactobacillus bulgaricus. Proteomics 8: 3154– 3163 [DOI] [PubMed] [Google Scholar]
- 18. Fiske CH, Subbarow Y. 1925. The colorimetric determination of phosphorus. J. Biol. Chem. 66: 375– 400 [Google Scholar]
- 19. Fozo EM, Kajfasz JK, Quivey RG., Jr 2004. Low pH-induced membrane fatty acid alterations in oral bacteria. FEMS Microbiol. Lett. 238: 291– 295 [DOI] [PubMed] [Google Scholar]
- 20. Fozo EM, Quivey RG., Jr 2004. Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl. Environ. Microbiol. 70: 929– 936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fozo EM, Quivey RG., Jr 2004. The fabM gene product of Streptococcus mutans is responsible for the synthesis of monounsaturated fatty acids and is necessary for survival at low pH. J. Bacteriol. 186: 4152– 4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fozo EM, Scott-Anne K, Koo H, Quivey RG., Jr 2007. Role of unsaturated fatty acid biosynthesis in virulence of Streptococcus mutans. Infect. Immun. 75: 1537– 1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ganesan B, Weimer BC. 2004. Role of aminotransferase IlvE in production of branched-chain fatty acids by Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 70: 638– 641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garault P, Letort C, Juillard V, Monnet V. 2000. Branched-chain amino acid biosynthesis is essential for optimal growth of Streptococcus thermophilus in milk. Appl. Environ. Microbiol. 66: 5128– 5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Giotis ES, McDowell DA, Blair IS, Wilkinson BJ. 2007. Role of branched-chain fatty acids in pH stress tolerance in Listeria monocytogenes. Appl. Environ. Microbiol. 73: 997– 1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Griswold AR, Jameson-Lee M, Burne RA. 2006. Regulation and physiologic significance of the agmatine deiminase system of Streptococcus mutans UA159. J. Bacteriol. 188: 834– 841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guckert JB, Hood MA, White DC. 1986. Phospholipid ester-linked fatty acid profile changes during nutrient deprivation of Vibrio cholerae: increases in the trans/cis ratio and proportions of cyclopropyl fatty acids. Appl. Environ. Microbiol. 52: 794– 801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guedon E, Serror P, Ehrlich SD, Renault P, Delorme C. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 40: 1227– 1239 [DOI] [PubMed] [Google Scholar]
- 29. Hamilton IR, Buckley ND. 1991. Adaptation by Streptococcus mutans to acid tolerance. Oral Microbiol. Immunol. 6: 65– 71 [DOI] [PubMed] [Google Scholar]
- 30. Kajfasz JK, et al. 2010. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J. Bacteriol. 192: 2546– 2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaneda T. 1977. Fatty acids of the genus Bacillus: an example of branched-chain preference. Bacteriol. Rev. 41: 391– 418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaneda T. 1963. Valine as a source of the branched short chain precursor in the biosynthesis of iso-C14, iso-C15, iso-C16 and iso-C17 fatty acids by Bacillus subtilis. Biochem. Biophys. Res. Commun. 10: 283– 287 [DOI] [PubMed] [Google Scholar]
- 33. Kuhnert WL, Quivey RG., Jr 2003. Genetic and biochemical characterization of the F-ATPase operon from Streptococcus sanguis 10904. J. Bacteriol. 185: 1525– 1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuhnert WL, Zheng G, Faustoferri RC, Quivey RG., Jr 2004. The F-ATPase operon promoter of Streptococcus mutans is transcriptionally regulated in response to external pH. J. Bacteriol. 186: 8524– 8528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kunst F, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390: 249– 256 [DOI] [PubMed] [Google Scholar]
- 36. Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49: 193– 205 [DOI] [PubMed] [Google Scholar]
- 37. Lemos JA, Burne RA. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154: 3247– 3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lemos JA, Nascimento MM, Lin VK, Abranches J, Burne RA. 2008. Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J. Bacteriol. 190: 5291– 5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Len AC, Cordwell SJ, Harty DW, Jacques NA. 2003. Cellular and extracellular proteome analysis of Streptococcus mutans grown in a chemostat. Proteomics 3: 627– 646 [DOI] [PubMed] [Google Scholar]
- 40. Len AC, Harty DW, Jacques NA. 2004. Proteome analysis of Streptococcus mutans metabolic phenotype during acid tolerance. Microbiology 150: 1353– 1366 [DOI] [PubMed] [Google Scholar]
- 41. Ma Y, Marquis RE. 1997. Thermophysiology of Streptococcus mutans and related lactic-acid bacteria. Antonie Van Leeuwenhoek 72: 91– 100 [DOI] [PubMed] [Google Scholar]
- 42. Madsen SM, et al. 2002. Cloning and inactivation of a branched-chain-amino-acid aminotransferase gene from Staphylococcus carnosus and characterization of the enzyme. Appl. Environ. Microbiol. 68: 4007– 4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Matsui R, Cvitkovitch D. Acid tolerance mechanisms utilized by Streptococcus mutans. Future Microbiol. 5: 403– 417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mitchell TJ. 2003. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat. Rev. Microbiol. 1: 219– 230 [DOI] [PubMed] [Google Scholar]
- 45. Murchison HH, Barrett JF, Cardineau GA, Curtiss R., III 1986. Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect. Immun. 54: 273– 282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nascimento MM, Lemos JA, Abranches J, Goncalves RB, Burne RA. 2004. Adaptive acid tolerance response of Streptococcus sobrinus. J. Bacteriol. 186: 6383– 6390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Quivey RG, Jr, Kuhnert WL, Hahn K. 2000. Adaptation of oral streptococci to low pH. Adv. Microb. Physiol. 42: 239– 274 [DOI] [PubMed] [Google Scholar]
- 48. Rathsam C. 2005. Two-dimensional fluorescence difference gel electrophoretic analysis of Streptococcus mutans biofilms. J. Proteome Res. 4: 2161– 2173 [DOI] [PubMed] [Google Scholar]
- 49. Shabala L, Ross T. 2008. Cyclopropane fatty acids improve Escherichia coli survival in acidified minimal media by reducing membrane permeability to H+ and enhanced ability to extrude H+. Res. Microbiol. 159: 458– 461 [DOI] [PubMed] [Google Scholar]
- 50. Shaw WV, Hopwood DA. 1976. Chloramphenicol acetylation in Streptomyces. J. Gen. Microbiol. 94: 159– 166 [DOI] [PubMed] [Google Scholar]
- 51. Sheng J, Baldeck JD, Nguyen PT, Quivey RG, Jr, Marquis RE. 2010. Alkali production associated with malolactic fermentation by oral streptococci and protection against acid, oxidative, or starvation damage. Can. J. Microbiol. 56: 539– 547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shivers RP, Dineen SS, Sonenshein AL. 2006. Positive regulation of Bacillus subtilis ackAby CodY and CcpA: establishing a potential hierarchy in carbon flow. Mol. Microbiol. 62: 811– 822 [DOI] [PubMed] [Google Scholar]
- 53. Shivers RP, Sonenshein AL. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53: 599– 611 [DOI] [PubMed] [Google Scholar]
- 54. Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8: 203– 207 [DOI] [PubMed] [Google Scholar]
- 55. Terleckyj B, Shockman GD. 1975. Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect. Immun. 11: 656– 664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Terleckyj B, Willett NP, Shockman GD. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect. Immun. 11: 649– 655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tojo S, et al. 2005. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol. Microbiol. 56: 1560– 1573 [DOI] [PubMed] [Google Scholar]
- 58. Yvon M, Chambellon E, Bolotin A, Roudot-Algaron F. 2000. Characterization and role of the branched-chain aminotransferase (BcaT) isolated from Lactococcus lactis subsp. cremoris NCDO 763. Appl. Environ. Microbiol. 66: 571– 577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yvon M, Thirouin S, Rijnen L, Fromentier D, Gripon JC. 1997. An aminotransferase from Lactococcus lactis initiates conversion of amino acids to cheese flavor compounds. Appl. Environ. Microbiol. 63: 414– 419 [DOI] [PMC free article] [PubMed] [Google Scholar]