Abstract
The molecular alarmone (p)ppGpp functions as a global regulator of gene expression in bacteria. In Streptococcus mutans, (p)ppGpp synthesis is catalyzed by three gene products: RelA, RelP, and RelQ. RelA is responsible for (p)ppGpp production during a stringent response, and RelP is the primary source of (p)ppGpp during exponential growth, but the role of RelQ has not been thoroughly investigated. In this study, we analyzed the four-gene relQ operon to establish how these gene products may affect homeostasis and stress tolerance in the dental caries pathogen S. mutans. Northern blotting and reverse transcriptase PCR demonstrated that relQ is cotranscribed with the downstream genes ppnK (NAD kinase), rluE (pseudouridine synthase), and pta (phosphotransacetylase). In addition, a promoter located within the rluE gene was shown to drive transcription of pta. Inactivation of relQ, ppnK, or rluE did not significantly affect growth of or stress tolerance by S. mutans, whereas strains lacking pta were more sensitive to acid and oxidative stresses. Interestingly, introduction of an rluE deletion into the pta mutant reversed the deleterious effects of the pta mutation on growth and stress tolerance. Accumulation of (p)ppGpp was also decreased in a pta mutant strain, whereas inactivation of relQ caused enhanced (p)ppGpp synthesis in exponential-phase cells. The results reveal an important role for the relQ operon in the expression of traits that are essential for persistence and pathogenesis by S. mutans and provide evidence for a molecular connection of acetate and (p)ppGpp metabolism with tolerance of acid and oxidative stresses.
INTRODUCTION
Streptococcus mutans is a Gram-positive, facultatively anaerobic bacterium found almost exclusively in the oral cavity that is the primary causative agent of dental caries in humans. The pathogenic potential of S. mutans is associated with its ability to form biofilms on teeth, to produce organic acids via the fermentation of carbohydrates, and to tolerate a variety of environmental stresses. In addition, S. mutans adapts well to the fluctuating environmental conditions in the human mouth associated with diurnal rhythms and intermittent feeding by the host. Notably, those environmental factors that vary widely in the oral cavity and have the most profound impact on oral biofilm ecology—pH, nutrient availability and source, and degree of anaerobiosis—strongly affect the expression of virulence attributes by S. mutans (6, 9, 28).
Microbes colonizing the teeth are exposed to a spectrum of oxygen levels and redox potentials (33), and a variety of reactive oxygen species (ROS), including superoxide ions and hydrogen peroxide, are generated by host defenses and by bacterial metabolism of oxygen. It is becoming well recognized that exposure to oxygen and reactive oxygen species is a major challenge for S. mutans and that redox represents an important ecological determinant balancing the growth of health-associated commensals against that of particular pathogens (3, 23, 24, 33). The primary defenses utilized by S. mutans against oxygen are NADH oxidases that reduce environmental oxygen levels by metabolizing it to yield H2O or H2O2 (18, 19).
A significant change in the flow of carbohydrate through metabolic pathways in response to oxygen availability also occurs in S. mutans (23). In an anaerobic, carbohydrate-rich environment, the major product of fermentation by S. mutans is lactic acid produced via an NAD-dependent lactate dehydrogenase (LDH) that is activated by fructose-1,6-bisphosphate (8, 26). Under carbohydrate-limiting conditions, fermentation shifts from lactate to the production of formate and acetate, mediated principally through a pyruvate formate lyase (PFL) enzyme (50). The PFL enzyme is responsible for the conversion of pyruvate and conenzyme A (CoA) into formate and acetyl-CoA, but the PFL enzyme of S. mutans is inactivated under aerobic conditions (50). When S. mutans is growing in the presence of oxygen, the pyruvate dehydrogenase (PDH) pathway is activated, facilitating the conversion of pyruvate to acetyl-CoA (10). Expression profiling has shown that the genes for the PDH complex and for the partial tricarboxylic acid (TCA) cycle of S. mutans are upregulated during aerobic growth, which would allow the cells to regenerate sufficient quantities of NADH to power the NADH oxidase enzymes (2, 3, 5). In addition to fueling the TCA cycle and certain anabolic processes, the production of acetyl-CoA by PFL or PDH is advantageous from a bioenergetic perspective. Specifically, acetyl-CoA can be acted on by a phosphotransacetylase (Pta) enzyme to yield acetyl phosphate, which can be utilized by acetate kinase (Ack) for ATP production. By using the Pta/Ack pathway, the organisms gain an additional ATP compared to shunting pyruvate through Ldh. Acetyl phosphate also plays critical roles in gene regulation and signaling in bacteria (31, 35), so the levels of this high energy compound need to be tightly controlled.
One bacterial strategy for adaptation to environmental changes is the stringent response, which is mediated by accumulation of (p)ppGpp GDP- and GTP-derived molecular alarmones (31, 36). Accumulation of (p)ppGpp occurs when aminoacyl tRNA pools fail to keep up with the demands of protein synthesis (20), signaling nutritional stress that leads to adjustments in gene expression and the inhibition of stable rRNA and tRNA synthesis (43). Without a proper stringent response, increases in translation errors occur and energy sources required for bacterial survival can become depleted. In Escherichia coli, (p)ppGpp metabolism is catalyzed by RelA and SpoT (21, 43); RelA has synthetase activity only, but SpoT has weak synthetase activity and efficient (p)ppGpp hydrolase activity (20, 37).
Previously, it was considered that Gram-positive bacteria possessed a single Rel enzyme that had both synthetase and hydrolase activity (20, 37); the enzyme was generally referred to as a RelA/SpoT hydrolase (RSH) enzyme. The RelA protein of S. mutans is of the RSH type and contains hydrolase, synthase, and allosteric regulation domains (27, 30). However, members of a class of small (p)ppGpp synthetases (RelP and RelQ) were first discovered in S. mutans, and subsequently in a variety of other organisms, that are able to produce (p)ppGpp but lack the domains for hydrolase activity and allosteric regulation (30). In S. mutans, relP is cotranscribed as part of the relPRS operon, which encodes RelP and a two-component signal transduction system (TCS) (30). RelP is the major source of (p)ppGpp in S. mutans under nonstressed conditions in exponentially growing cells (30). Inactivation of relP or relRS results in a lack of accumulation of (p)ppGpp in exponential phase and a corresponding increase in growth rate and yield in the presence of agents that induce oxidative stress (30, 46). Thus, it has been suggested that RelP protects the organisms from ROS-induced damage by moderating the rate of growth in the presence of oxidative stressors. Our laboratory has also shown that S. mutans coordinates (p)ppGpp accumulation with oxidative stress tolerance and genetic competence via RelPRS and the RcrR transcriptional regulator and the RcrPQ ABC transporters, which are encoded in an operon just upstream of relPRS (46).
In contrast to RelA and, to a lesser extent, RelP, little is known about the role of the small synthetase RelQ in S. mutans or its potential homologues in other organisms, although an association between an apparent RelQ homologue and antibiotic tolerance in Enterococcus faecalis has been previously described (1). The contribution of RelQ to (p)ppGpp production in S. mutans was minimal under normal growth conditions, and RelQ did not affect alarmone accumulation in response to mupirocin or serine hydroxymate (1, 30, 38, 39). Still, the four-gene relQ operon is highly conserved in a small group of commensal and pathogenic streptococci and is part of the core genome of S. mutans (unpublished data) and may have been conserved as a result of selective pressures arising from niche adaptations. Here, we characterize the contributions of the gene products of the relQ operon to growth, homeostasis, and stress tolerance in S. mutans and disclose their influence on phenotypes that directly impact the virulence of this human pathogen.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids used in this study are listed in Table 1. S. mutans UA159 and its derivatives were maintained in brain heart infusion (BHI) medium (Difco Laboratories, Detroit, MI) supplemented with spectinomycin (Sp; 1 mg ml−1), erythromycin (Erm; 10 μg ml−1), and/or kanamycin (Km; 1 mg ml−1) (Sigma-Aldrich, St. Louis, MO), when necessary. For growth measurements, overnight cultures grown in BHI medium were diluted 1:50 and grown to mid-exponential phase (optical density at 600 nm [OD600], ∼0.5) at 37°C in a 5% CO2 atmosphere. These mid-exponential-phase cultures were then diluted 1:100 into fresh medium. The optical density of cells growing at 37°C was measured at 600 nm (OD600) every 30 min using a Bioscreen C Lab system (Helsinki, Finland). For anaerobic conditions, sterile mineral oil was overlaid on cultures. When desired, the growth medium was supplemented with 25 mM paraquat–0.003% H2O2–HCl to lower the pH of the medium to 5.5 or with 50 mM sodium acetate (Sigma-Aldrich, St. Louis, MO).
Table 1.
Bacterial strains and plasmids used in this study
| Streptococcus mutans strain or plasmid | Genotype or relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| UA159 | Wild type | |
| ΔrelQ strain | ΔrelQ::NPKmr | Laboratory stock |
| ΔppnK strain | ΔppnK::NPKmr | This study |
| ΔrluE strain | ΔrluE::NPKmr | This study |
| ΔrluEΩ strain | ΔrluE::ΩKmr | This study |
| Δpta strain | Δpta::NPKmr | This study |
| ΔptaE strain | Δpta::NPErmr | This study |
| ΔrluE ΔptaE strain | ΔrluE Δpta::NPKmr NPErmr | This study |
| WT::Ppta-cat | cat gene fusion to putative pta promoter, Spr Kmr | This study |
| KB10 | WT carrying pDL278 | This study |
| KB11 | Δpta::NPKmr carrying pDL278 | This study |
| KB12 | Δpta::NPKmr carrying pBK101 | This study |
| Plasmids | ||
| pJL105 | CAT fusion integration vector based on pJL84,a Spr Kmr | Laboratory stock |
| pDL278 | E. coli-Streptococcus shuttle vector, Spr | 4, 28 |
| pKB101 | pta coding region plus promoter region cloned into pDL278 | This study |
The integration vector pJL84 is described elsewhere (53).
Construction of deletion mutants and reporter gene fusion strains.
A variety of deletion mutants in S. mutans were created as previously described (2, 48). Briefly, fragments flanking the gene of interest were amplified with gene-specific primers carrying a BamHI or SacI recognition site. The PCR products were digested with either BamHI or SacI (New England BioLabs) and ligated with T4 DNA ligase (Invitrogen) to a nonpolar kanamycin resistance (NPKm) (25)- or erythromycin resistance (NPErm) (57)-encoding gene that had been digested with the same restriction enzymes. The resulting ligation mixtures were used to transform competent S. mutans to replace the target genes. Transformants were isolated on BHI agar plates supplemented with either kanamycin or erythromycin. Mutant construction was confirmed through PCR and DNA sequencing, including confirming that the sequences flanking the gene of interest were intact. Oligonucleotides used for mutant construction in this study are listed in Table 2. For reporter gene fusions, a staphylococcal chloramphenicol acetyltransferase (cat) gene fusion strain was created as described by Zeng et al. (57). Briefly, a 291-bp fragment directly upstream of the pta translational start site was amplified using the primers listed in Table 2 and then cloned into the pJL105 integration vector (57), which carries the cat gene lacking a promoter and ribosomal binding site (RBS). The resulting construct was transformed into a wild-type (WT) strain and the promoter-cat fusion established in a single copy in the chromosome by double-crossover homologous recombination, with the mtlA-phnA genes serving as the integration site.
Table 2.
Oligonucleotides used in this study
| Name | Sequence | Description |
|---|---|---|
| ppnK-A | GCTTGCTTTGCTCGCAATAA | ppnK deletion |
| ppnK-BamHI-B | TTATCGGTAGGATCCATCTGTGTCA | ppnK deletion |
| ppnK-BamHI-C | CTTTTATCGGGATCCTCGATTCAT | ppnK deletion |
| ppnK-D | CATACCCCATTTCTCCTCCA | ppnK deletion |
| rluE-A | GCAGATTCGCGATGACATTA | rluE deletion |
| rluE-BamHI-B | TTTTACTTTGGATCCAGCAATGAAT | rluE deletion |
| rluE-BamHI-C | GTTCCTTGAGATCCACCTTGATAG | rluE deletion |
| rluE-D | TGGTCCGATAGCATCAAACA | rluE deletion |
| pta-A | GACGAAGAAGCGCTTGAAAC | pta deletion |
| pta-SacI-B | TTTTCTCTCGAGCTCCCAAATAAAG | pta deletion |
| pta-SacI-C | GCGCAAACCGAGCTCAATACTAAAT | pta deletion |
| pta-D | CAAACTCTTCGCAAGCATCA | pta deletion |
| 3′-BglII-pta_CAT | GAATACCCATAGATCTATACCCCTA | PCR for pta promoter |
| 5′-SstI-pta_CAT | TCTGGTAAAGAGCTCCATACAAGTT | PCR for pta promoter |
| pta-SphI-FP | TCATATTGGGCATGCCCTTTTAGG | Complementation |
| pta-EcoRI-RP | AAAGCAATGGAATTCTCCAAAGAG | Complementation |
| RT-pta-FP | GGCAGTGCAAAGGCTTCTCAAGTT | Real-time RT-PCR, Northern blot analysis |
| RT-pta-RP | ATTGGCTTGTCCTGCTACGTCACT | Real-time RT-PCR |
| B-pta-RP | AGCTGTTGTCGGTACAAAGGCAGCAT | Northern blot analysis |
| 16S rRNA-S | CACACCGCCCGTCACACC | Real-time RT-PCR, Northern blot analysis |
| 16S rRNA-AS | CAGCCGCACCTTCCGATACG | Real-time RT-PCR, Northern blot analysis |
Assay for promoter activity.
Chloramphenicol acetyltransferase (CAT) activity was quantified and normalized as previously described (53). Strains carrying a Ppta-cat fusion were grown in BHI broth to mid-exponential phase at 37°C in a 5% CO2 aerobic atmosphere. Cells were harvested, washed with sodium phosphate buffer (pH 7.0), and resuspended in 10 mM TE buffer (10 mM Tris, 1 mM EDTA, pH 7.5). Proteins were extracted by mechanical disruption in a Bead Beater-16 cell disrupter (Biospec Products, Inc., Bartlesville, OK) with glass beads (0.1 mm) on ice for 30 s twice, with a 2-min interval after the first 30 s. Following centrifugation at 4,000 × g for 10 min, the supernates were used to measure CAT activity. Protein concentrations were determined using the bicinchoninic acid assay (BCA) (Thermo Scientific). One unit of CAT activity was defined as the amount of enzyme needed to acetylate 1 nmol of chloramphenicol min−1 (mg protein)−1.
RNA purification and Northern blot analysis.
Cells were grown to mid-exponential phase in BHI medium at 37°C with 5% CO2, stabilized with bacterial RNAprotect reagent (Qiagen), and resuspended in 10 mM TE buffer (10 mM Tris, 1 mM EDTA, pH 7.5). Cells were then subjected to mechanical disruption in a Bead Beater-16 cell disrupter (Biospec Products, Inc., Bartlesville, OK), total RNA was isolated using an RNeasy Minikit (Qiagen), and the concentrations of RNA samples were determined spectrophotometrically. Analysis of relQ and pta mRNA by Northern blotting was performed using a NorthernMax kit (Ambion) according to the supplier's protocol. Briefly, 10 μg of total RNA per sample was loaded on a 1% agarose–formaldehyde denaturing gel in 1× MOPS (morpholinepropanesulfonic acid) buffer (Ambion). The separated RNAs were transferred onto a BrightStar-Plus membrane (Ambion), UV cross-linked to the membrane, and hybridized with DNA probes specific for relQ or pta coding sequences at 42°C overnight in ULTRAhyb buffer (Ambion). The hybridized membrane was washed in low- and high-stringency wash solutions for 5 min and 15 min, respectively, and hybridization signals were detected using a BrightStar BioDetect kit (Ambion). The 16S rRNA in each sample was used for normalization. The pta-specific probe (408 nucleotides) was generated using PCR amplification with pta probe-FP and B (biotinylated)-pta-RP primers (see Table 2).
Real-time qRT-PCR.
To measure the level of mRNA of specific genes, real-time quantitative reverse transcriptase PCR (qRT-PCR) was performed as described elsewhere (4). cDNA was synthesized using 1 μg of total RNA and a SuperScript III First-Strand Synthesis system (Invitrogen). Target-specific primers used for real-time RT-PCRs were designed with Beacon Designer 4.0 software (Premier Biosoft International, Palo Alto, CA) (see Table 2). The copy numbers of the target genes were determined using iQ SYBR green Supermix (Bio-Rad) in an iCycler iQ real-time PCR detection system (Bio-Rad) as follows: one cycle of 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All expression data were normalized to the copy number of 16S rRNA in each sample.
Quantification of biofilm formation.
Biofilm assays were performed as previously described (2). Briefly, overnight cultures grown in BHI medium were diluted 1:50 in fresh BHI medium and grown to an OD600 of 0.5 at 37°C in a 5% CO2 aerobic atmosphere. The cultures were then diluted 1:100 in semidefined biofilm medium (BM) supplemented with either 20 mM glucose or 10 mM sucrose as the carbohydrate source. Cell cultures were grown in a 96-well polystyrene plate (Costar 3595; Corning Inc., Corning, NY) at 37°C in a 5% CO2 aerobic atmosphere for 48 h. Cells were then washed and stained with 60 μl of 0.1% crystal violet for 15 min. The stained bacteria were washed twice, the retained dye was eluted from cells using ethanol/acetone (8:2 [vol/vol]), and biofilm levels was quantified by measuring absorbance at a 575-nm wavelength.
Complementation analysis.
A 1,356-bp fragment that included the pta gene and promoter region was amplified by PCR using pta-SphI-FP (TCATATTGGGCATGCCCTTTTAGG) and pta-EcoRI-RP (AAAGCAATGGAATTCTCCAAAGAG) primers. Following gel purification of the PCR products, the DNA fragments were digested with SphI and EcoRI and cloned into pDL278 shuttle vector (4, 28) that had been digested with the same restriction enzymes. The recombinant plasmid carrying the pta gene was introduced into the Δpta strain by competent transformation.
Measurement of (p)ppGpp accumulation.
Measurements of (p)ppGpp were done as previously described (30). Overnight cultures were inoculated 1:50 in FMC defined medium (51) containing 10 mM glucose and grown until the OD600 reached 0.2. Cells were then incubated with [32P]orthophosphate for an additional hour, with or without 0.003% hydrogen peroxide. Cells were harvested, and nucleotides were extracted using ice-cold 13 M formic acid. Acid extracts were spotted onto a polyethyleneimine (PEI)-cellulose plate (Selecto Scientific, Inc., Suwanee, GA) for separation of the phosphorylated nucleotides by thin-layer chromatography (TLC). The plates were chromatographed with 1.5 M KH2PO4 (pH 3.5) and then exposed to X-ray film (Kodak) at −70°C.
RESULTS
Organization of the relQ operon.
Based on the genome sequence of S. mutans UA159, four genes appear to be part of the relQ operon: relQ, ppnK (NAD+ kinase), rluE (pseudouridine synthase), and pta (phosphotransacetylase) (Fig. 1) (18, 30). Using reverse transcriptase PCR (RT-PCR), we observed that relQ could be cotranscribed with ppnK, rluE, and pta (data not shown). The relQ gene (GenBank accession no. SMU.1046c) has been shown experimentally to be a small (p)ppGpp synthetase (30), in that purified recombinant RelQ could produce (p)ppGpp from GTP and ATP and relQ could complement a relA/spoT mutant strain of E. coli. The ppnK gene, alternatively referred to as nadK, encodes a predicted NAD+ kinase that utilizes ATP to catalyze the conversion of NAD+ to NADP+ (15). NADP+ is an essential cofactor in energy metabolism and reductive biosynthetic reactions, including fatty acid biosynthesis (14). The rluE gene encodes a pseudouridine synthase that is predicted to catalyze the formation of pseudouridine (Ψ) in the 23S rRNA of the large ribosomal subunit. The isomerization of uridine to pseudouridine is the most abundant posttranscriptional modification of RNA in all living organisms (42). The pta gene, encoding the only predicted phosphotransacetylase enzyme in S. mutans, is involved in acetate metabolism in many bacteria. Pta catalyzes the following reaction: acetyl-CoA + phosphate ↔ acetyl phosphate + HS-CoA (22, 49). In S. mutans, the acetyl phosphate can be utilized to generate an additional molecule of ATP compared with the homofermentative production of lactate.
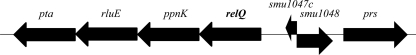
Fig 1.
Schematic representation of the relQ operon. RelQ has been demonstrated to be a small (p)ppGpp synthetase. The ppnK is predicted to encode an NAD+ kinase, rluE codes for a pseudouridine synthase, and pta codes for a phosphotransacetylase. SMU.1047c and SMU.1048 (GenBank designations) encode a hypothetical small protein and a protein with similarity to adenylate cyclases, respectively. The prs gene encodes a predicted phosphoribosyl pyrophosphate synthetase. Data were retrieved from the Oral Pathogen Sequence Database (http://www.oralgen.lanl.gov).
Transcriptional organization of the relQ operon.
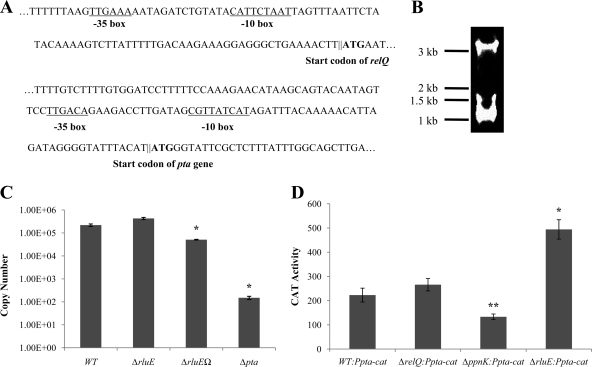
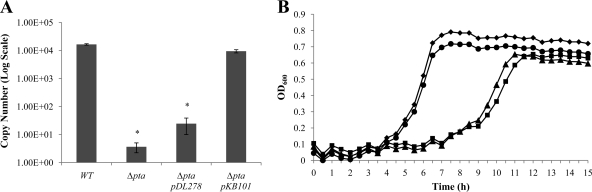
Two potential promoters were identified 56 bp and 35 bp upstream of the relQ and pta start sites, respectively, using BPROM bacterial promoter identification software (Softberry, Inc., NY) (Fig. 2A). Northern blotting with a biotin-labeled probe specific for pta transcripts was conducted and, consistent with our RT-PCR results, two transcripts of approximately 3.4 kb and 1.0 kb were observed that included the entire relQ operon and a monocistronic pta transcript, respectively (Fig. 2B). We next constructed three deletion mutants (ΔrluE [nonpolar], ΔrluEΩ [polar], and Δpta [nonpolar]) in a wild-type S. mutans background by allelic exchange (1, 9) and quantified pta-containing transcripts by the use of real-time RT-PCR (Fig. 2C). Copy numbers of the pta transcript in the wild-type and ΔrluE strains were 2.19 × 105 and 4.28 × 105 (μg of input RNA)−1, respectively. The amount of pta mRNA in the Δpta mutant was at the lower limit of detection, but 5.10 × 104 copies of pta mRNA (μg input RNA)−1 were detected in the ΔrluEΩ (polar) strain (Fig. 2C), suggesting that the pta gene could be transcribed from its own promoter.
Fig 2.
(A) Promoters predicted by BPROM (see text) to drive transcription of the relQ operon (top two lines) and pta (bottom three lines). Underlined letters indicate −35 and −10 regions for the putative promoters. Bold letters indicate the predicted start codons (ATG) of relQ and pta. (B) Northern blot analysis with a biotin-labeled probe specific for the pta transcript. (C) Real-time PCR results measuring the copy numbers of pta transcripts in the wild type and relQ operon mutations. (D) Promoter-cat fusion activity from the pta promoter located within the rluE gene. Cells were grown to mid-exponential phase in BHI broth at 37°C in a 5% CO2 atmosphere, and CAT activity was measured as described in Materials and Methods. *, data differ from the wild-type genetic background data at P < 0.001; **, data differ from the wild-type genetic background data at P < 0.01 (Student's t test). WT, wild type.
To determine whether a functional promoter was located 5′ to the pta gene, a 291-bp fragment directly upstream of the ATG start site of pta was cloned behind a promoterless chloramphenicol acetyltransferase (cat) gene in pJL105 integration vector (57). The construct was used to transform S. mutans strain UA159 and strains carrying mutations in the relQ operon to create a total of four cat fusion strains (WT::Ppta-cat, ΔrelQ::Ppta-cat, ΔppnK::Ppta-cat, and ΔrluE::Ppta-cat). When grown in BHI medium, the wild-type and ΔrelQ strains expressed 223 ± 28.6 and 266 ± 25.7 units of CAT activity, respectively (Fig. 2D). Interestingly, compared to that measured in the wild-type genetic background, CAT activity in the ΔppnK strain was decreased to 133 ± 11.4 units (P < 0.01) but was increased in the ΔrluE background (494 ± 40 units; P < 0.001). Collectively, these results support the idea that transcription of the genes in the relQ operon arises from a promoter 5′ to relQ and from an active promoter within the rluE gene and that expression from the internal promoter can be influenced in opposite ways by loss of PpnK or RluE.
Inactivation of the pta gene affects growth and biofilm formation of S. mutans.
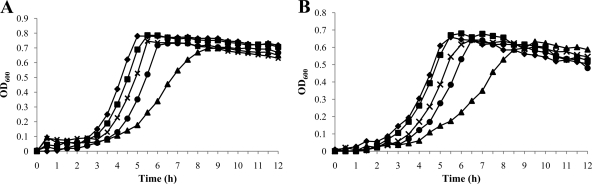
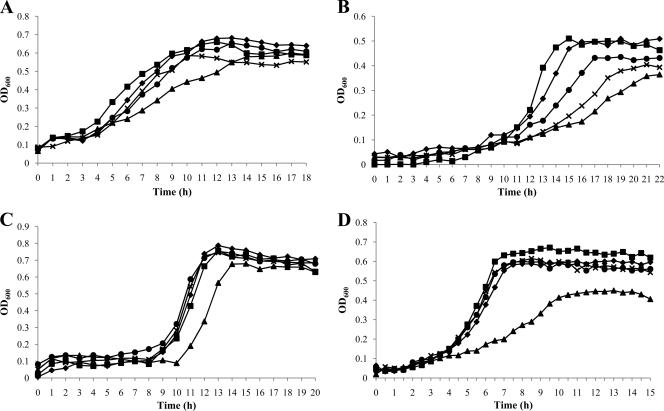
Growth of the wild-type and mutant strains was monitored in a Bioscreen C Lab system using BHI broth with a mineral oil overlay to create comparatively anaerobic conditions (Fig. 3A) or without a mineral oil overlay for comparatively aerobic conditions (Fig. 3B). Under anaerobic conditions, the wild-type strain and strains carrying the nonpolar ΔrelQ, ΔppnK, or ΔrluE mutation had exponential growth rates that were not significantly different. However, the strain harboring only the pta mutation had a doubling time of 75 min ± 9.4 min, which was significantly lower than that of the parental strain (Fig. 3A). Under aerobic conditions, the wild-type, ΔrelQ, ΔppnK, and ΔrluE strains again did not differ in their growth rates, but the single pta mutant did again grow more slowly than the wild-type strain (Fig. 3B). Thus, deletion of pta alone had the most profound impact on growth. Importantly, though, introduction of the rluE mutation into a strain carrying an inactivated pta gene (either the ΔrluE ΔptaE double mutant or the polar ΔrluE mutant [ΔrluEΩ] strain) reversed the effects of the pta mutation, restoring growth rates to wild-type levels (Fig. 3).
Fig 3.
Growth curves of wild-type and mutant strains obtained during growth in BHI medium. The wild-type (◆), ΔrelQ (■), Δpta (▲), ΔrluEΩ (×), and ΔrluE ΔptaE (●) strains were grown at 37°C with a mineral oil overlay (A) or without a mineral oil overlay (B). Optical density at 600 nm was monitored every 30 min at 37°C using the Bioscreen C Lab system. The results represent the mean values of triplicate growth curves for three independent isolates.
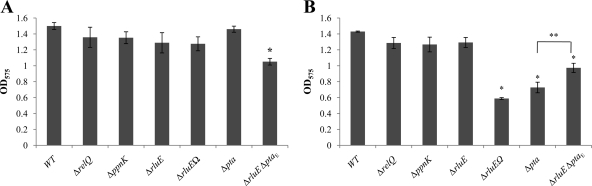
The ability of the various relQ operon mutants to form biofilms was compared with that of the wild-type strain in BM broth supplemented with either 10 mM sucrose (Fig. 4A) or 20 mM glucose (Fig. 4B) such that the media contained equal amounts of carbohydrate on a weight-to-volume basis. The Δpta mutant formed a smaller amount of biofilm (P < 0.001) when grown in BM supplemented with 20 mM glucose than the parental strain, but the biofilms formed by these two strains in sucrose were equivalent. Notably, though, combining of the rluE and pta mutations in a strain resulted in significantly enhanced biofilm formation in glucose compared to that seen with the strain carrying only the pta mutation (Fig. 4B), albeit the level did not reach that seen with the wild-type strain.
Fig 4.
Biofilm formation. S. mutans strains were grown to an OD600 of 0.5 in BHI broth, diluted 1:50 in BM semidefined medium supplemented with 10 mM sucrose (A) or with 20 mM glucose (B) in a 96-well microtiter plate, and incubated at 37°C with 5% CO2 for 48 h. To quantify biofilm formation, the plates were washed twice, stained with 0.1% crystal violet, and resuspended with an ethanol-acetone (8:2 [vol/vol]) mixture. The optical density of the stained biofilm was measured at OD575. Data represent the means and standard deviations (error bars) of the results obtained with three separate isolates assayed in triplicate. *, data differ from the wild-type genetic background data at P ≤ 0.001 (Student's t test); **, data differ from the Δpta strain data at P < 0.005. WT, wild type.
To confirm that the phenotypic changes described above resulted directly from loss of the pta gene, a plasmid-borne copy of the pta gene driven from its own promoter (pKB101) was introduced into the pta mutant strains. The transcript levels of pta were measured in the complemented strains by real-time RT-PCR and were found not to differ from that of the wild-type strain (Fig. 5A). When the pta deletion mutant carried the wild-type copy of pta on pKB101, growth and biofilm formation were restored to the levels observed for S. mutans UA159 (Fig. 5B).
Fig 5.
Complementation analysis of the S. mutans Δpta mutant. (A) The copy numbers of the pta transcripts were determined using qRT-PCRs. 16S rRNA was used as a reference transcript for normalization of the data. Values shown represent the means ± standard deviations for RNA data from three separate cultures. *, data differ from the wild-type genetic background data at P < 0.001 (Student's t test). (B) The wild-type strain (◆), Δpta strain (▲), pta strain harboring pDL278 (■), and complemented Δpta strain harboring pBK101 (●) were grown to mid-exponential phase in BHI medium and diluted 1:100 into fresh BHI medium with a mineral oil overlay. Optical density at 600 nm was monitored every 30 min at 37°C using the Bioscreen C Lab system. The results are expressed as means of the results determined with triplicate experiments performed with three independent isolates.
Contribution of the S. mutans relQ operon to stress tolerance.
Acid tolerance is recognized as an essential characteristic of cariogenic bacteria (28). To assess whether the mutant strains had altered resistance to low pH, we compared the growth of the wild-type strain to that of the mutants under anaerobic conditions in BHI broth that had been acidified to pH 5.5 (Fig. 6A). Growth of all strains at pH 5.5 was slower than at neutral pH in plain BHI broth (Fig. 3). Interestingly, the ΔrelQ mutant exhibited faster growth at pH 5.5 (145 ± 12.6 min) than the wild-type strain (185 ± 13 min) (P < 0.01). In contrast, the Δpta mutant grew much slower (279 ± 16.8 min) than all other strains (P < 0.001). Again, introduction of the rluE mutation into the pta mutant restored the ability of the strain to grow at wild-type rates at low pH (Fig. 6A).
Fig 6.
Growth under various stress conditions. The wild-type (◆), ΔrelQ (■), Δpta (▲), ΔrluEΩ (×), and ΔrluE ΔptaE (●) strains were grown in triplicate to mid-exponential phase in BHI medium and diluted to 1:100 into fresh BHI medium supplemented with HCl to lower the pH 5.5 under anaerobic conditions (A), with 25 mM paraquat under anaerobic conditions (B), with 0.003% hydrogen peroxide (H2O2) under anaerobic conditions (C), or with 50 mM sodium acetate under aerobic conditions (D). Optical density at 600 nm was monitored every 30 min at 37°C using the Bioscreen C Lab system. Data are expressed as means of the results determined with triplicate wells for three independent isolates.
Hydrogen peroxide confers a type of oxidative stress to cells different from paraquat (3), with paraquat forming superoxide anion (O2−) and H2O2 generating primarily hydroxyl-radical species (47). When growing in the presence of paraquat (Fig. 6B), all strains exhibited slower growth than in the absence of the stress agent. However, the pta deletion mutant again displayed a growth defect, achieving a peak OD600 of only 0.25 with a minimum doubling time of 193 ± 3.9 min compared to the final OD600 of 0.6 and minimum doubling time of 89 ± 7.7 min achieved by the parental strain. When growth phenotypes were compared in BHI broth supplemented with 0.003% H2O2 with a mineral oil overlay to limit additional oxidative stress (Fig. 6C), only the pta deletion mutant exhibited significantly impaired growth. Also, as noted for other stresses, the strains carrying both the rluE and pta mutations grew faster and achieved higher final yields than strains carrying only the pta mutation (Fig. 6B and C).
Effects of exogenous acetate on growth of S. mutans.
Pta is known to be important in both the formation of acetate and the assimilation of exogenous acetate (12, 54). To see if the presence of excess exogenous acetate could alter the growth phenotypes of the various S. mutans strains used in this study, growth was monitored in plain BHI broth and BHI broth supplemented with 50 mM sodium acetate (both media had an initial pH value of ∼7.3). Interestingly, the growth of the Δpta mutant was strongly inhibited in cultures containing 50 mM acetate compared to that of the wild-type strain and other mutants (Fig. 6D). Again, introduction of an rluE deletion into the pta mutant background resulted in restoration of growth to the level of the wild-type strain in acetate (Fig. 6D).
(p)ppGpp accumulation under selected stress conditions.
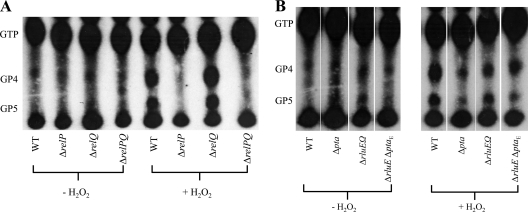
Changes in the cellular concentrations of (p)ppGpp mediate adjustments in stable RNA synthesis (20, 29, 31, 36). Since the relQ gene product possesses efficient (p)ppGpp synthetic activity (1, 27), we hypothesized that the H2O2-sensitive phenotypes of the mutant strain (Fig. 6C) could be associated with enhanced (p)ppGpp production. Accumulation of (p)ppGpp in the ΔrelQ strain (30) was relatively more abundant than that seen with the wild-type strain, whereas the ΔrelP and ΔrelPQ strains failed to accumulate (p)ppGpp during exponential-phase growth, suggesting that RelP synthesizes (p)ppGpp in response to H2O2-stressed conditions (Fig. 7A). Of particular interest, the pta strain exhibited lower (p)ppGpp accumulation (Fig. 7B) than the wild-type strain. Further, accumulation of (p)ppGpp in the ΔrluEΩ (polar) mutant strain and the ΔrluE ΔptaE double mutant strain was less than in the wild-type strain. Collectively, these results indicate that gene products in the relQ operon influenced (p)ppGpp accumulation under the particular environmental conditions.
Fig 7.
Accumulation of (p)ppGpp in S. mutans strains in the presence or absence of hydrogen peroxide. (p)ppGpp accumulation of the wild-type strain compared (A) with that of the ΔrelP, ΔrelQ, and ΔrelPQ mutants and (B) with that of the Δpta, ΔrluEΩ, and ΔrluE ΔptaE mutants. Cells growing exponentially in FMC medium were labeled with [32P]orthophosphate and subjected to the stress conditions for 1 h. Extracts derived from treatment of the cells with 13 M formic acid were normalized to counts per minute (CPM) and spotted onto PEI-cellulose plates for TLC using 1.5 M KH2PO4 buffer.
DISCUSSION
The critical roles of (p)ppGpp metabolism in homeostasis and stress tolerance, and the key role of RSH enzymes in these processes, have been studied in detail in a variety of diverse bacteria (1, 27, 30, 40, 43, 56). The relatively recent identification of the small (p)ppGpp synthetases RelP and RelQ, which share limited homology with RelA- and SpoT-like enzymes (30), has raised new questions about how bacteria may utilize these enzymes to fine-tune gene expression and physiologic responses to environmental stimuli (11). The RelP enzyme of S. mutans is responsible for (p)ppGpp production in exponentially growing cells, and optimal production of RelP requires activation by the RelRS two-component system (30, 46) in response to some as-yet-unidentified signal. Notably, cells lacking RelP or RelRS grow more rapidly than the parental strain when exposed to oxidative stress, indicating that RelP-dependent (p)ppGpp production may slow growth to minimize damage from oxygen radicals produced by metabolism. On the other hand, the contributions of RelQ to (p)ppGpp metabolism and the conditions that trigger RelQ-dependent alarmone synthesis remain undefined. This report provides novel and significant insights into how control of carbohydrate metabolism, a central component of S. mutans virulence, may be integrated with growth and survival decisions and stress tolerance in this caries pathogen by linking acetate metabolism with (p)ppGpp metabolism, NADP production, and rRNA modification.
The transcriptional organization and coordinate regulation of the four genes in the relQ operon of S. mutans UA159 imply a functional interrelationship between the gene products. S. mutans generates energy almost exclusively through glycolysis, using hexoses to form two molecules of pyruvate, two ATP molecules, and two molecules of NADH, which correspond to four reducing equivalents (54). To maintain glycolytic flux, the organism must reoxidize NADH to NAD+. NAD+ molecules can be regenerated by placing the reducing equivalents onto partially oxidized metabolic intermediates (d-lactate, succinate, formate, and ethanol) (15), which can be excreted into the environment (54) and can damage the tissues of the oral environment. Alternatively, generation of acetate affords the organisms an additional ATP (34, 50, 52, 54), as the acetyl-CoA derived from pyruvate by PDH is acted on by the Pta enzyme to create acetyl phosphate, which in turn is converted to acetate and ATP by acetate kinase (Ack) in the presence of ADP. NAD can also be generated from NADH by oxygen-consuming NADH oxidases, which are believed to be a primary defense of S. mutans against ROS damage. The primary route for NADH generation by S. mutans involves shunting pyruvate into a partial TCA cycle (2, 5), and the genes for these TCA enzymes are upregulated in cells growing in the presence of air compared to anaerobically cultured cells (3). NAD can also be phosphorylated to NADP(H) by NAD+ kinase and ATP, and S. mutans has a small number of NADP-dependent enzymes that function in redox reactions and carbohydrate or amino acid metabolism. Thus, three gene products encoded by the relQ operon play critical roles in carbon flow and energy generation (Pta), oxidation-reduction reactions (NadK), and growth rate regulation and stress tolerance by virtue of the ability to produce (p)ppGpp (RelQ).
The rluE gene is also transcribed as part of the relQ operon and encodes a pseudouridine synthase that is predicted to catalyze the formation of pseudouridine (Ψ) in the 23S rRNA (5, 13, 42). Relatively little is known about the specific roles of RluE in the physiology of bacteria, but the major role of Ψ appears to be to stabilize the RNA tertiary structure that is important for mRNA translation (42). Deletion of genes involved in incorporation of Ψ into rRNA affects translational efficiency and fidelity, and mutants of E. coli that lack key Ψ residues in the 23S RNA display slower growth and compete less efficiently with the parental organism (41, 45). Therefore, like the other gene products encoded by the relQ operon, RluE is intimately tied to physiologic homeostasis and growth.
Similar to those of other bacteria, the S. mutans genome contains multiple pseudouridine synthases that are predicted, based on very high degrees of sequence conservation with gene products of known function, to participate in the posttranscriptional modification of functional RNAs with pseudouridine, including tRNAs (e.g., truA), the small ribosomal subunit RNA (e.g., rsuA), and the large ribosomal subunit RNA (rluB, -D, and -E). Interestingly, the four-gene relQ operon, including the rluE homologue, is conserved in multiple streptococcal species and some other lactic acid bacteria, including Streptococcus gordonii, Streptococcus mitis, Streptococcus sanguinis, Streptococcus agalactiae, Streptococcus thermophilus, Streptococcus pyogenes, and some Lactobacillus species, but not in Enterococcus spp. and a number of other related Gram-positive species. We have also observed that the relQ operon is conserved in 57 strains of S. mutans for which we have recently obtained high-coverage draft genomes (unpublished data). It should also be mentioned that annotation of this gene in various databases is inconsistent; for example, the apparent homologue of rluE of S. mutans is annotated as rluD in certain pneumococci. Unfortunately, functional studies are lacking at this time to determine which residues in the 23S RNA are impacted by deletion of this gene in streptococci. Nonetheless, the high degree of conservation of the relQ operon in this group of metabolically similar organisms, coupled with the studies presented here that show coordinate regulation of these genes and interactions of the pathways in which these gene products participate, provides compelling evidence that the relQ operon has critical functions in organisms with physiology or ecological niches that are similar to those of S. mutans.
Phenotypic analysis of the various mutants revealed that only strains carrying a pta deletion displayed growth impairment and elevated sensitivities to selected stressors. In E. coli, pta mutants impact the expression profile of the genes for the TCA cycle and grow more slowly than the corresponding wild-type strains under aerobic conditions in defined minimal media supplemented with pyruvate (34, 35, 44, 54). More recently, studies in S. pneumoniae have revealed important roles for acetate kinase and acetyl phosphate in homeostasis and gene regulation (44). It is known that growth of S. mutans in the presence of oxygen causes an upregulation of the genes for the PDH complex and for the partial TCA cycle of S. mutans, presumably to allow additional energy production in the form of ATP (12) via the acetate kinase pathway, as well as to generate NADH that is needed for oxidative stress tolerance via the NADH oxidase enzymes (2, 3, 5). Given that the pyruvate formate lyase complex is inactivated by oxygen, then one would predict that Pta and Ack are critical for S. mutans in controlling carbon flow and ATP generation under aerobic conditions. Thus, as part of our working model, we propose that mutations in the pta gene alter steady-state levels of pyruvate and acetyl-CoA (17), impeding the ability of cells to generate ATP and adversely affecting NAD/NADH balances that are essential for optimal growth of S. mutans, particularly under aerobic conditions or in the presence of oxidative stress. The acid-sensitive phenotype of the pta mutant could be directly associated with diminished ATP generation, since the F1FO-ATPase requires ATP for proton extrusion or, more likely, arises from additional alterations in metabolic pathways (e.g., membrane biogenesis), signaling, and gene regulation. It should also be noted here that the relQ mutant grew more rapidly in acid than the parental strain. We are therefore investigating the possibility that RelQ may play a role in slowing the growth of S. mutans under strongly acidic conditions to protect the cells from acid damage, similar to the proposed role for RelPRS in cells experiencing oxidative stress.
While deletion of rluE alone did not discernibly impact growth or stress tolerance in S. mutans under the conditions tested, the phenotype of strains carrying deletions of both rluE and pta was striking. In particular, introduction of the rluE knockout into a strain carrying a pta deletion consistently reversed the slow growth and poor stress tolerance phenotypes of the pta mutant and partially restored the ability of the bacteria to form biofilms in the presence of glucose. Moreover, deletion of either rluE or pta impacted (p)ppGpp accumulation in the presence of H2O2. These important findings provide strong evidence for coordination and integration of (p)ppGpp and acetate metabolism, which are closely linked to the energy state and redox balance of the cells, with translational efficiency. Although significant challenges to dissection of the underlying basis for these connections remain, one can postulate that ATP and acetyl phosphate levels are integrated with sensing of the redox environment and NAD/NADP and NAD/NADH balances to regulate growth through the production of (p)ppGpp and translational efficiency. One also cannot exclude the possibility that the modifications of rRNA by RluE impact the way in which (p)ppGpp influences gene expression via interactions with ribosomes.
Production of the molecular alarmone (p)ppGpp is crucial under various conditions that require transitioning of bacteria between growth and survival modes. Multiple studies have demonstrated that perturbations in the normal pathways for accumulation or turnover of (p)ppGpp elicit a wide range of altered phenotypes, including alterations in susceptibility to stress, requirements for amino acids, and changes in cellular morphology (32, 43). In S. mutans, a classical RSH enzyme (RelA) regulates the stringent response induced by exposure to mupirocin, which is a relatively severe starvation stress. The predicted roles for RelP and RelQ, which lack any apparent domains for allosteric regulation, appear more subtle and to involve maintaining basal levels of (p)ppGpp to optimize growth and allow more modest adaptations on the genetic and physiologic levels to the rapidly and often changing environments in the human oral cavity (30).
We have also reported that the RelPRS system can be activated by (p)ppGpp and is able to moderate growth of the organisms in response to oxidative stressors (46). The present data indicated that ΔrelP and ΔrelPQ mutant strains had a (p)ppGpp0(null) phenotype in the presence of hydrogen peroxide, suggesting that RelP, and not RelA, is the principal source of alarmone in response to hydrogen peroxide stress. Importantly, hydrogen peroxide production by commensal streptococci appears to be antagonistic to the growth of S. mutans (30, 46) and is therefore an important ecological factor impacting dental biofilm ecology. In contrast, the contribution of RelQ to (p)ppGpp metabolism appears minimal, but the finding that a relQ deletion mutant produced elevated levels of alarmone in response to oxidative stress is potentially significant (Fig. 7A). The basis for this finding is not clear at this time, but it does imply that there is some crosstalk or cross-regulation between the relQ and relPRS operons and systems. This idea is further supported by the finding that inactivation of pta caused a decrease in (p)ppGpp accumulation, presumably mediated by RelP, in response to peroxide stress. However, we cannot exclude the possibility that the slower growth of this mutant, particularly in the presence of H2O2, may have impacted assimilation of the [32P]orthophosphate or incorporation of the label into nucleotides. There is also the issue of the lack of knowledge about the factors that influence (p)ppGpp hydrolase activity by the RelA enzyme of S. mutans, so it is not known how RelA-dependent (p)ppGpp hydrolysis may have contributed to the net levels of alarmone.
Of particular interest, the Δpta strains exhibited a lower growth rate and impairment of biofilm formation. The mutant also showed a decrease in RelP-dependent production of (p)ppGpp under oxidative stress conditions (46) but no significant changes in (p)ppGpp levels under nonstressed conditions. Thus, the slower growth phenotype of the pta strain cannot be attributed in any major way to accumulation of alarmone under either stressed or nonstressed conditions. Rather, as noted above, decreases in the acetyl phosphate pools (data not shown), perturbations in the steady-state levels of pyruvate, acetyl-CoA, or TCA intermediates, and disruption of NAD/NADH balances may all contribute to the observed growth changes. Notably, decreases in the acetyl phosphate pools may lower (p)ppGpp levels by diminished activation of the relP operon by the RelRS two-component system, since acetyl phosphate may be a cognate substrate for the RelS sensor kinase. We are currently investigating how acetyl phosphate levels may integrate relQ and relP operon expression with the energy state of the cell and with alterations in carbon utilization associated with changes in redox environment.
The most enigmatic data of this study deal with the interrelationship of the rluE and pta genes. As observed, the addition of an rluE deletion restored growth phenotypes of a pta-deleted mutant under the stress conditions tested. A critical issue is how a lack of pseudouridine in the ribosomes restores growth of a strain lacking Pta across a wide range of environmental conditions (low pH, oxidative stress, biofilms). In E. coli, pseudouridine synthases, encoded by rluA, rluB, rluD, rluE, or rluF, are responsible for the production of pseudouridine at different positions in23S rRNA (13, 41, 45). The strains deficient in these synthases had no effect on normal growth (13), except for the rluD mutant, which displayed growth defects (45). Thus, in E. coli and S. mutans, loss of RluE does not have a discernible effect on growth, but the effects of loss of RluE in a pta mutant background have been tested only in this study. To begin to discern the basis for this important and unusual observation, we examined whether cross-regulation of expression of the rluE and pta genes could occur in S. mutans. The results indicated that expression from the proximal pta promoter, as measured from a cat fusion integrated at a remote site in the chromosome, was significantly upregulated when a nonpolar rluE mutation was present (Fig. 2D). Notably, the levels of pta mRNA were not diminished in the nonpolar rluE mutant (data not shown), so the increase in pta promoter activity probably did not arise from compensatory expression associated with low Pta or acetyl phosphate levels. Collectively, then, the simplest interpretation of the finding that loss of RluE reverses the effects of loss of Pta is that fine-tuning of growth and gene expression in response to shifts in acetate metabolism require modifications to rRNA that could affect translation. One possible target for alterations in translation could involve the amino acid or lipid biosynthetic pathways, many of which are fueled by acetyl-CoA. Also noteworthy is that S. mutans encodes an NADP-dependent nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase that directly produces 3-phosphoglycerate (GapN), which is a precursor for serine metabolism (7). Future studies aimed at examining overlaps in the transcriptome in mutants lacking individual genes and combinations of genes in the relQ operon should shed more light on the genetic networks or metabolic pathways that allow an rluE mutation to suppress phenotypes associated with loss of Pta.
The importance of considering acetyl phosphate levels in interpretation of these findings cannot be overstated. Previous research has demonstrated that acetyl phosphate acts as a global signal governing gene expression in many bacteria, in part through its ability to serve as a substrate for certain two-component signal transduction systems (TCSs) (16, 35, 54). Most of these TCSs consist of a sensor kinase and a response regulator (RR). Although autophosphorylation of RRs in vivo by acetyl phosphate has not been proven, many RRs autophophorylate using a phosphoryl group from acetyl phosphate in vitro, and many of these TCS influence cellular processes, such as biofilm formation and nutrient metabolism (34, 35, 55). Studies are under way to dissect the contributions of acetyl phosphate to S. mutans physiology and stress tolerance to fully appreciate how the organism links (p)ppGpp metabolism, growth rate control, and stress tolerance.
ACKNOWLEDGMENTS
We thank members of our laboratory for their assistance and helpful discussions.
This work was supported by DE13239 from the National Institute of Dental and Craniofacial Research.
Footnotes
Published ahead of print 17 February 2012
REFERENCES
- 1. Abranches J, et al. 2009. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J. Bacteriol. 191: 2248– 2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahn SJ, Browngardt CM, Burne RA. 2009. Changes in biochemical and phenotypic properties of Streptococcus mutans during growth with aeration. Appl. Environ. Microbiol. 75: 2517– 2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahn SJ, Wen ZT, Burne RA. 2007. Effects of oxygen on virulence traits of Streptococcus mutans. J. Bacteriol. 189: 8519– 8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahn SJ, Wen ZT, Burne RA. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 74: 1631– 1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ajdic D, et al. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99: 14434– 14439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Banas JA. 2004. Virulence properties of Streptococcus mutans. Front. Biosci. 9: 1267– 1277 [DOI] [PubMed] [Google Scholar]
- 7. Boyd DA, Cvitkovitch DG, Hamilton IR. 1995. Sequence, expression, and function of the gene for the nonphosphorylating, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase of Streptococcus mutans. J. Bacteriol. 177: 2622– 2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown AT, Wittenberger CL. 1972. Fructose-1,6-diphosphate-dependent lactate dehydrogenase from a cariogenic streptococcus: purification and regulatory properties. J. Bacteriol. 110: 604– 615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burne RA, Chen YY, Wexler DL, Kuramitsu H, Bowen WH. 1996. Cariogenicity of Streptococcus mutans strains with defects in fructan metabolism assessed in a program-fed specific-pathogen-free rat model. J. Dent. Res. 75: 1572– 1577 [DOI] [PubMed] [Google Scholar]
- 10. Carlsson J, Kujala U, Edlund MB. 1985. Pyruvate dehydrogenase activity in Streptococcus mutans. Infect. Immun. 49: 674– 678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cashel M, Gentry DR, Hernandez VJ, Vinella D. 1996. The stringent response, p 1458– 1496 In Neidhardt R. C. I. F. C., Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Reznikoff W., Riley M., Shaechter M., Umbarger A. E. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 12. Chang DE, Shin S, Rhee JS, Pan JG. 1999. Acetate metabolism in a pta mutant of Escherichia coli W3110: importance of maintaining acetyl coenzyme A flux for growth and survival. J. Bacteriol. 181: 6656– 6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Del Campo M, Kaya Y, Ofengand J. 2001. Identification and site of action of the remaining four putative pseudouridine synthases in Escherichia coli. RNA. 7: 1603– 1615 [PMC free article] [PubMed] [Google Scholar]
- 14. Garavaglia S, Galizzi A, Rizzi M. 2003. Allosteric regulation of Bacillus subtilis NAD kinase by quinolinic acid. J. Bacteriol. 185: 4844– 4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grose JH, Joss L, Velick SF, Roth JR. 2006. Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 103: 7601– 7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grundy FJ, Waters DA, Allen SH, Henkin TM. 1993. Regulation of the Bacillus subtilis acetate kinase gene by CcpA. J. Bacteriol. 175: 7348– 7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gupta S, Clark DP. 1989. Escherichia coli derivatives lacking both alcohol dehydrogenase and phosphotransacetylase grow anaerobically by lactate fermentation. J. Bacteriol. 171: 3650– 3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Higuchi M, et al. 1993. Identification of two distinct NADH oxidases corresponding to H2O2-forming oxidase and H2O-forming oxidase induced in Streptococcus mutans. J. Gen. Microbiol. 139: 2343– 2351 [DOI] [PubMed] [Google Scholar]
- 19. Higuchi M, et al. 1999. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J. Bacteriol. 181: 5940– 5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. 2004. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected]. Cell 117: 57– 68 [DOI] [PubMed] [Google Scholar]
- 21. Kari C, Torok I, Travers A. 1977. ppGpp cycle in Escherichia coli. Mol. Gen. Genet. 150: 249– 255 [DOI] [PubMed] [Google Scholar]
- 22. Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. 2007. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J. Bacteriol. 189: 5574– 5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kodama T, et al. 1987. Effects of oxygen on glucose-limited growth of Streptococcus mutans. Infect. Immun. 55: 169– 173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kolenbrander PE, et al. 2006. Bacterial interactions and successions during plaque development. Periodontol. 2000 42: 47– 79 [DOI] [PubMed] [Google Scholar]
- 25. Kremer BH, et al. 2001. Characterization of the sat operon in Streptococcus mutans: evidence for a role of Ffh in acid tolerance. J. Bacteriol. 183: 2543– 2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lemos JA, Abranches J, Burne RA. 2005. Responses of cariogenic streptococci to environmental stresses. Curr. Issues Mol. Biol. 7: 95– 107 [PubMed] [Google Scholar]
- 27. Lemos JA, Brown TA, Jr, Burne RA. 2004. Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect. Immun. 72: 1431– 1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lemos JA, Burne RA. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154: 3247– 3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lemos JA, Chen YY, Burne RA. 2001. Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 183: 6074– 6084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lemos JA, Lin VK, Nascimento MM, Abranches J, Burne RA. 2007. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol. Microbiol. 65: 1568– 1581 [DOI] [PubMed] [Google Scholar]
- 31. Magnusson LU, Farewell A, Nystrom T. 2005. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13: 236– 242 [DOI] [PubMed] [Google Scholar]
- 32. Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nystrom T. 2007. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J. Bacteriol. 189: 5193– 5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marquis RE. 1995. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J. Ind. Microbiol. 15: 198– 207 [DOI] [PubMed] [Google Scholar]
- 34. McCleary WR, Stock JB. 1994. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem. 269: 31567– 31572 [PubMed] [Google Scholar]
- 35. McCleary WR, Stock JB, Ninfa AJ. 1993. Is acetyl phosphate a global signal in Escherichia coli? J. Bacteriol. 175: 2793– 2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mechold U, Cashel M, Steiner K, Gentry D, Malke H. 1996. Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. J. Bacteriol. 178: 1401– 1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mechold U, Murphy H, Brown L, Cashel M. 2002. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of RelSeq, the Rel/Spo enzyme from Streptococcus equisimilis. J. Bacteriol. 184: 2878– 2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nanamiya H, et al. 2008. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 67: 291– 304 [DOI] [PubMed] [Google Scholar]
- 39. Nascimento MM, Lemos JA, Abranches J, Lin VK, Burne RA. 2008. Role of RelA of Streptococcus mutans in global control of gene expression. J. Bacteriol. 190: 28– 36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nguyen D, et al. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334: 982– 986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ofengand J, Campo MD. 2004. Chapter 4.6.1, Modified nucleosides in Escherichia coli ribosomal RNA. In Curtiss R, III, et al. (ed), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC [Google Scholar]
- 42. Ofengand J, Del Campo M, Kaya Y. 2001. Mapping pseudouridines in RNA molecules. Methods 25: 365– 373 [DOI] [PubMed] [Google Scholar]
- 43. Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62: 35– 51 [DOI] [PubMed] [Google Scholar]
- 44. Ramos-Montañez S, Kazmierczak KM, Hentchel KL, Winkler ME. 2010. Instability of ackA (acetate kinase) mutations and their effects on acetyl phosphate and ATP amounts in Streptococcus pneumoniae D39. J. Bacteriol. 192: 6390– 6400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Raychaudhuri S, Conrad J, Hall BG, Ofengand J. 1998. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli RNA. 4: 1407– 1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Seaton K, Ahn SJ, Sagstetter AM, Burne RA. 2011. A transcriptional regulator and ABC transporters link stress tolerance, (p)ppGpp, and genetic competence in Streptococcus mutans. J. Bacteriol. 193: 862– 874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Senadheera MD, et al. 2007. The Streptococcus mutans vicX gene product modulates gtfB/C expression, biofilm formation, genetic competence, and oxidative stress tolerance. J. Bacteriol. 189: 1451– 1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shubeita HE, Sambrook JF, McCormick AM. 1987. Molecular cloning and analysis of functional cDNA and genomic clones encoding bovine cellular retinoic acid-binding protein. Proc. Natl. Acad. Sci. U. S. A. 84: 5645– 5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Speckman RA, Collins EB. 1973. Incorporation of radioactive acetate into diacetyl by Streptococcus diacetilactis. Appl. Microbiol. 26: 744– 746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Takahashi S, Abbe K, Yamada T. 1982. Purification of pyruvate formate-lyase from Streptococcus mutans and its regulatory properties. J. Bacteriol. 149: 1034– 1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Terleckyj B, Willett NP, Shockman GD. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect. Immun. 11: 649– 655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wanner BL. 1993. Gene regulation by phosphate in enteric bacteria. J. Cell. Biochem. 51: 47– 54 [DOI] [PubMed] [Google Scholar]
- 53. Wen ZT, Burne RA. 2004. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 186: 2682– 2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wolfe AJ. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69: 12– 50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wolfe AJ, et al. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol. Microbiol. 48: 977– 988 [DOI] [PubMed] [Google Scholar]
- 56. Wu J, Xie J. 2009. Magic spot: (p) ppGpp. J. Cell. Physiol. 220: 297– 302 [DOI] [PubMed] [Google Scholar]
- 57. Zeng L, Wen ZT, Burne RA. 2006. A novel signal transduction system and feedback loop regulate fructan hydrolase gene expression in Streptococcus mutans. Mol. Microbiol. 62: 187– 200 [DOI] [PubMed] [Google Scholar]