Abstract
The brucellae are Gram-negative pathogens that cause brucellosis, a zoonosis of worldwide importance. The genus Brucella includes smooth and rough species that differ in that they carry smooth and rough lipopolysaccharides, respectively. Brucella abortus, B. melitensis, and B. suis are typical smooth species. However, these smooth brucellae dissociate into rough mutants devoid of the lipopolysaccharide O-polysaccharide, a major antigen and a virulence determinant encoded in regions wbo (included in genomic island-2) and wbk. We demonstrate here the occurrence of spontaneous recombination events in those three Brucella species leading to the deletion of a 5.5-kb fragment carrying the wbkA glycosyltranferase gene and to the appearance of rough mutants. Analysis of the recombination intermediates suggested homologous recombination between the ISBm1 insertion sequences flanking wbkA as the mechanism generating the deletion. Excision of wbkA was reduced but not abrogated in a recA-deficient mutant, showing the existence of both RecA-dependent and -independent processes. Although the involvement of the ISBm1 copies flanking wbkA suggested a transpositional event, the predicted transpositional joint could not be detected. This absence of detectable transposition was consistent with the presence of polymorphism in the inverted repeats of one of the ISBm1 copies. The spontaneous excision of wbkA represents a novel dissociation mechanism of smooth brucellae that adds to the previously described excision of genomic island-2. This ISBm1-mediated wbkA excision and the different %GC levels of the excised fragment and of other wbk genes suggest that the Brucella wbk locus is the result of at least two horizontal acquisition events.
INTRODUCTION
The brucellae are a group of Gram-negative pathogens that cause brucellosis, one of the most important bacterial zoonosis worldwide. Based on the appearance of the colony surface and the structure of the lipopolysaccharide (LPS), these bacteria are divided into rough (R) and smooth (S) species. The R species are represented by Brucella ovis and B. canis, produce matte, granular colonies, and carry an R-type LPS lacking the O-polysaccharide (O-PS). The S species, which are zoonotically more important, include B. melitensis, B. abortus, and B. suis, have an LPS with an N-formyl-perosamine O-PS, and produce glossy colonies. The S phenotype, however, is not stable, and S brucellae dissociate to generate R mutants closely similar to the R species in colony morphology and LPS structure. Under unfavorable growth conditions or after prolonged incubation, this dissociation is very noticeable. Thus, it soon caught the attention of brucellosis researchers, who described the colonial dissociation in great detail in the first half of the past century. This early work also established that R mutants are attenuated and not useful for serological diagnostic purposes (for a review of the early literature, see reference 37), two observations accounted for by the role of the O-PS as both a key virulence factor and a major diagnostic antigen in brucellosis serological tests (21, 27). Indeed, procedures that minimize the S-R dissociation and controls to exclude batches containing R bacteria are critical in brucellosis antigen and vaccine production (2).
The genetic basis underlying the S-R dissociation of S brucellae is only partially understood. Extensive analyses (for a review, see reference 16) have shown that the O-PS biosynthetic genes are located in two major regions, wbo and wbk, with different characteristics. The wbo region carries two putative glycosyltransferase genes (wboA and wboB) and is included in a large Brucella genomic island denominated GI-2 (32). The wbk region, which was probably acquired by horizontal transfer (15), carries the genes putatively necessary to synthesize N-formyl-perosamine and to prime bactoprenol for O-PS polymerization plus those of several glycosyltransferases and of the ABC transporters that translocate the O-PS. It is noteworthy that the wbk genes do not form a continuous unit since the region contains a number of insertion sequence (IS) open reading frames (ORFs) (Fig. 1A). Recently, we traced one mechanism of S-R dissociation to the spontaneous excision of GI-2 mediated by the recombination activity of a phage-related integrase present in this island (23). Stabilization of GI-2 by mutation of this integrase reduces but does not eliminate the S-R dissociation, suggesting additional mechanisms. Based on the characteristics of the wbk region, we hypothesized that spontaneous recombination between homologous sequences adjacent to the wbkA glycosyltransferase gene (Fig. 1A) could lead to its excision and generate S-R dissociation. We present here evidence showing that such an excision occurs spontaneously in vitro by homologous recombination and report that the deletion was found in a spontaneous R mutant directly isolated in a pure culture from goat milk.
Fig 1.
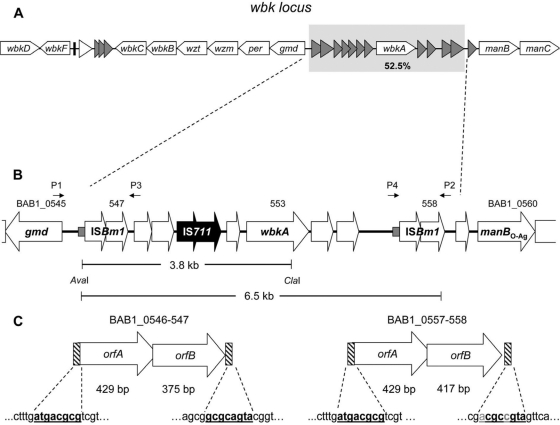
Genetic organization of the wbk locus of B. abortus 2308. (A) wbk locus (white arrows represent ORFs encoding LPS biosynthesis genes, gray triangles show transposase ORFs, a white triangle represents a gene encoding a putative regulatory protein, and the black bar indicates a gene encoding a tRNAGln). (B) wbkA region. Small gray boxes indicate the 60-nt repeat sequences located upstream of each ISBm1 (the sequences are AAAATTTTTTGGCATTTATCGGCCGCTATGATTCAAGGCTTCGAAATAGGGGAGCCTTTG and AAAATTTCTTGGCATTTATCGGCCGCTATGTTCAAGGCTTCGAAATAGGGAAGTCTTTG for left and right ISBm1 copies, respectively). The primer positions are depicted by small black arrows (P1, BAB1_0545Fb; P2, BAB1_0558R; P3, BAB1_0548R; P4, BAB1_0557F). The AvaI-ClaI segment indicates the location of the corresponding IS711-carrying 3.8-kb restriction fragment. The IS711 element is indicated in black. (C) Flanking ISBm1 with their corresponding inverted repeats (in boldface; polymorphism is indicated in gray).
MATERIALS AND METHODS
Bacterial strains and plasmids.
Table 1 lists the laboratory bacterial strains used in the present study. B. abortus 2308, B. melitensis 16M, and B. suis 1330 are strains commonly used in brucellosis studies that have been maintained as master stocks in either skimmed milk or Trypticase soy broth (Becton Dickinson, Sparks, MD)–5% sterile decomplemented calf serum–7% dimethyl sulfoxide at −85°C. Bacteria taken from these stocks were not passed more than once in vitro, and repeated controls showed the absence of R mutants detectable by the crystal violet dye exclusion test. In addition, a B. melitensis R mutant obtained in pure culture during routine bacteriological examination of goat milk was studied. The brucellae were grown on Brucella Trypticase soy agar (Becton Dickinson) plates (prepared at least 24 h before use and dried to remove syneresis water to avoid conditions known to favor S-R dissociation) at 37°C for 48 h. For cloning, E. coli was grown in Luria-Bertani broth supplemented with kanamycin (50 μg/ml) or chloramphenicol (20 μg/ml).
Table 1.
Bacterial strains used in this study
| Strain | Characteristics/relevant phenotypea | Source or referenceb |
|---|---|---|
| 2308 Nalr | B. abortus biovar 1 reference strain; S, Nalr | UNAV |
| R6 | Spontaneous B. abortus 2308 Nalr ΔwbkA mutant; R | This study |
| B10 | Chilean B. abortus field strain; S | 24 |
| R7 | Spontaneous ΔwbkA mutant, B. abortus B10 derivative; R | This study |
| R6/c-wbkA | R6 complemented with plasmid pMM14; S | This study |
| M939 | B. melitensis ΔwbkA field strain isolated from goat milk; R | CITA |
| BabΔISBm-L | B. abortus 2308 Nalr mutant carrying a deletion of 150 nt in BAB1_0546; S | This study |
| BabΔISBm-R | B. abortus 2308 Nalr mutant carrying a deletion of 150 nt in BAB1_0557; S | This study |
| BabΔISBm-LR | Double mutant in the wbkA-flanking ISBm1 copies; S | This study |
| BabΔrecA | B. abortus 2308 Nalr mutant with a recA(Δ80-283) gene deleted; S | This study |
| BabΔISBmLRΔrecA | recA mutant constructed on BabΔISBm-LR; S | This study |
Nalr, nalidixic acid resistance.
UNAV, Department of Microbiology and Parasitology, Universidad de Navarra, Navarra, Spain; CITA, Centro de Investigación yTecnología Agroalimentaria, Gobierno de Aragón, Spain.
Dissociation.
To promote dissociation, a protocol described previously was used (23). Bacteria taken directly from the frozen master stock (see above) were inoculated onto Trypticase soy agar plates, and a loopful of bacteria was transferred to a flask with 10 ml of brucella broth (Becton Dickinson) and grown at 37°C until reaching the stationary phase (typically within 48 h). An aliquot of this culture was adjusted to an A750 of 0.109 ± 0.005 using sterile broth, and 100 μl was inoculated in 10 ml of tryptic soy broth or the same broth supplemented with McIlvaine's buffer (pH 6.6) (3). After incubation for 10 days at 37°C, the CFU were counted, and the extent of S-R dissociation was determined by the crystal violet dye exclusion test (40). Two plates with approximately 1,500 colonies per plate (the limit that allows obtaining isolated colonies in 36 to 48 h) were examined, and three independent experiments performed.
Sequence analyses.
The complete sequence of B. abortus 2308 chromosome I was downloaded from GenBank (accession no. AM040264) (8), and similarity searches were performed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST). In silico restriction analysis of the wbk locus was performed with Vector NTI software (Invitrogen, Carlsbad, CA). The primers used for conventional PCR and real-time quantitative PCR (qPCR) assays (Table 2) were designed using the Primer3 web tool (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). DNA repeat sequences were predicted by use of the REPuter web tool (http://bibiserv.techfak.uni-bielefeld.de/reputer). DNA sequencing by the dideoxy method was performed using the Sequencing Unit of Centro de Investigación Médica Aplicada (CIMA; Universidad de Navarra). The sequence of the PCR product derived from the R6 strain was deposited in GenBank under accession no. JN982244.
Table 2.
Primers used in this study
| Primera | Sequence (5′-3′) | Purpose | Source or reference |
|---|---|---|---|
| BAB1_0553Fa | TGGTATATCCGGGTGTCTCG | wbkA detection | This study |
| BAB1_0553R | AAGCGTCCGTGCATTTCTAT | This study | |
| BAB1_0545F | CCGAATTCTTGGAATGGAGA | gmd detection | This study |
| BAB1_0545R | CAGTCGCGTAATGAGTCCAA | This study | |
| BAB1_0560F | CTTATGCAATGGCTCCCAAT | manBO-Ag detection | This study |
| BAB1_0560R | CATGTTCGGCGATAAATGTG | This study | |
| BAB1_0545Fb | TGCGACTTTCTTCACGATTG | Detection of wbkA deletion | This study |
| BAB1_0558R | GATCTTGGTATCGGCCTGTC | This study | |
| BAB1_0557F | CGCTTTAATATCTCGCGTTCC | Detection of excised circle | This study |
| BAB1_0548R | GGTCCCATCGGCATATCTT | This study | |
| recA_F1 | TGTTATCCCTGTCGCCAAAT | To construct recA deletion mutant | This study |
| recA_R2 | CTTTCCGGCCCATAGATTTC | This study | |
| recA_F3 | GAAATCTATGGGCCGGAAAGGCTGGTCGATCTTGGTGTC | This study | |
| recA_R4 | ACCGCTTTCCTGTCCAGATA | This study | |
| BAB1_0546_F1 | AGTCCACCTTGACATGCACA | To construct a 150-bp deletion ISBm1-left | This study |
| BAB1_0546_R2 | CCATAGCGTTCGGGAACTT | This study | |
| BAB1_0547_F3 | AAGTTCCCGAACGCTATGGAAAAAGGGGATGGAGACGAT | This study | |
| BAB1_0547_R4 | ACGATTGCAGCCTCTTCCAC | This study | |
| BAB1_0557_F1 | CGACGAAGGCGTTTTACAG | To construct a 150-bp deletion ISBm1-rightb | This study |
| BAB1_0558_R4 | GATCTTGGTATCGGCCTGTC | This study | |
| BAB1_0553F-gwc | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGCGAATTGGTGTCGAC | To complement the spontaneous ΔwbkA mutant | This study |
| BAB1_0553R-gw | GGGGACCACTTTGTACAAGAAAGCTGGGTCTTAATAGGTCATGAGCTTAGATT | This study | |
| 711u | CACAAGACTGCGTTGCCGACAGA | IS711 fingerprinting | 28 |
| 711d | CATATGATGGGACCAAACACCTAGGG | 28 | |
| scar_Fq | TTTGAAACGGTGCCCAAG | qPCR for chromosomal scar | This study |
| scar_Rq | ATAGACCCAGCGCGAAAAC | This study | |
| wbkA_Fq | TGCCGTCTCTCTACGAAGGT | qPCR for wbkA gene | This study |
| wbkA_Rq | TTCGGCTACGTTCAGAGGAT | This study |
F, forward; R, reverse.
These primers were used along with the primers BAB1_0546_R2 and BAB1_0547_F3 to construct the same deletion into ISBm1-right.
Sequences including att sites for BP clonase reaction are indicated in boldface.
DNA purification and PCR assays.
Genomic DNA (gDNA) was isolated from liquid cultures according to standard protocols (41), and PCR products were purified from agarose gels with a QIAEXII kit (Qiagen, Hilden, Germany). To determine the boundaries of spontaneous wbkA deletion, conventional PCR with primer pairs (Table 2) targeting the adjacent genes gmd (BAB1_0545) and manBO-Ag (BAB1_0560) and wbkA (BAB1_0553) was performed. The mixture included 10 ng of gDNA, 10 pmol of each primer, 0.2 mM deoxynucleoside triphosphates, 2 mM MgCl2, and 1 U of Immolase DNA polymerase (Bioline, London, United Kingdom) in a final volume of 25 μl. The amplification conditions were as follows: an initial step at 95°C for 5 min, followed by 30 cycles at 95°C for 20 s, annealing at 60°C for 25 s, and extension at 72°C for 30 s, with a final extension step of 5 min at 72°C. Recombination intermediates were amplified from 0.2-μg portions of gDNA samples using the primer pairs BAB1_0545Fb/BAB1_0558R (chromosomal scar) and BAB1_0557F/BAB1_0548R (excised circle) under the same conditions but for the extension steps that were 1 min. PCR assays were carried out in a GeneAmp PCR System 2700 (Applied Biosystems, Foster City, CA). Amplicons were resolved by electrophoresis in 1.0 to 2.0% TBE (45 mM Tris-borate, 1 mM EDTA [pH 8.0]) agarose gels.
qPCR assessment of wbkA excision.
To assess the extent of wbkA excision in each mutant, a qPCR assay was developed based on the relative quantification protocol described previously (5). The absolute quantification of chromosomal scars determined with the primers scar_Fq and scar_Rq was normalized to the amount of gDNA in each sample obtained by the absolute quantification of wbkA using the primers wbkA_Fq and wbkA_Rq (Table 2). Serial double dilutions of gDNA from B. abortus 2308 and R6 strains were used to generate calibration curves for the corresponding assay by plotting copy number versus log(CT) value. PCR fragments of 389 bp (scar) and 143 bp (wbkA gene) were obtained from a 20-μl reaction mixture containing 10 μl of 2× SYBR green PCR master mix (Applied Biosystems), the appropriate primers at a concentration of 0.5 μM, and 0.1 ng of gDNA for wbkA quantification or 100 ng for scar quantification. Amplification was carried out in a 7500 Real-Time PCR system (Applied Biosystems). The cycling conditions were as follows: 2 min at 50°C, 10 min at 95°C, and then 45 cycles of 15 s at 95°C and 1 min at 60°C. The amplification efficiency was obtained from the equation E = 10(−1/slope), using slope values determined from the standard curves. The results were analyzed with Applied Biosystems 7500 Fast System SDS software v1.3. For simplicity, the ratio Rscar obtained from three independent experiments was expressed as a percentage (i.e., the number of scars in 100 genomes). A fragment of 853 bp from circular intermediates was quantified using 100 ng of gDNA and the same amplification conditions with the primers BAB1_0557F and BAB1_0548R (Table 2) and dilutions of plasmid pMM99 as a standard. The efficiencies of the amplifications of the scar, wbkA, and the circular intermediate were 1.82, 1.98, and 1.60, respectively.
Mutagenesis.
To construct a ΔrecA mutant of B. abortus 2308, PCR overlap was used to generate an in-frame deleted allele that was then introduced into the genome by allelic exchange (7). Briefly, the two fragments obtained with the primer pairs recA-F1/recA-R2 and recA-F3/recA-R4 (Table 2) were ligated by overlap PCR, and the resulting fragment (containing the ΔrecA allele lacking the nucleotides corresponding to amino acids 80 to 283) was cloned into pCR2.1 TOPO (Invitrogen) to produce plasmid pMM62 (Table 3). A BamHI-XbaI fragment from this plasmid was subcloned into pJQK (31), and the resulting plasmid (pMM63) was introduced into B. abortus by mating using the E. coli S17-1λpir strain (35). Single-crossover transconjugants were selected on medium containing nalidixic acid (25 μg/ml) and kanamycin (50 μg/ml). After 24 h in broth without antibiotics, double-crossover mutants were selected on nalidixic acid–5% sucrose plates and confirmed by PCR. Mutants were characterized for sensitivity to methyl methanesulfonate and H2O2 by using the Kirby-Bauer disk diffusion assay (34). The same mutational strategy was utilized to produce ISBm1 mutants by making 150-nucleotide (nt) deletions of each orfA, which introduced several stop codons affecting the transposase catalytic domain. The BAB1_0557_F1, BAB1_0546_R2, BAB1_0547_F3, and BAB1_0558_R4 primers were used to construct the mutator plasmid pMM73, which was then introduced into B. abortus 2308 to obtain the BabΔISBm-R mutant. Similarly, the primers pairs BAB1_0546_F1/BAB1_0546_R2 and BAB1_0547_F3/BAB1_0547_R4 were used to generate the mutator plasmid pMM78 for the construction of a BabΔISBm-L mutant. Next, a double BabΔISBm-LR mutant was generated by the introduction of plasmid pMM73 in BabΔISBm-R. For the complementation of ΔwbkA (BabΔwbkA) spontaneous mutants, a copy of wbkA generated by PCR with the primers BAB1_0553F-gw and BAB1_0553R-gw (Table 2) was introduced into the pDONR221 donor vector using clonase BP (Gateway Technology; Invitrogen). The complementation plasmid pMM14 was obtained by clonase LR reaction between the pMR10 destination vector (19), and the donor plasmid pMM13 was cloned into the E. coli OmniMAX 2-t1 strain. After this, plasmid pMM14 was transferred to the E. coli S17-1λpir strain, which was then used to introduce pMM14 into B. abortus ΔwbkA mutants. Restoration of the S phenotype was assessed by using the crystal violet dye exclusion test, the agglutination of whole bacteria, and Western blotting of sodium dodecyl sulfate-proteinase K bacterial extracts with the serum of a rabbit hyperimmunized with S B. abortus and purified S and R-LPS as controls (16).
Table 3.
Plasmids and E. coli strains used in this study
| Plasmid or E. coli strain | Characteristicsa | Source or reference |
|---|---|---|
| Plasmids | ||
| pCR2.1 TOPO | Cloning plasmid; Kmr | Invitrogen |
| pJQK | Suicide vector for mutagenesis by gene replacement; Kmr | 31 |
| pDONR221 | Cloning plasmid (Gateway Technology); Kmr | This study |
| pMR10 | Destination vector (Gateway Technology); Kmr Cmr | 19 |
| pMM13 | Donor vector, pDONR221 derivative containing a copy of the B. abortuswbkA gene (BAB1_0553); Kmr | This study |
| wbkA-pMM14 | Complementation vector, pMR10 derivative carrying the att fragment from pMM13, Kmr Cmr | This study |
| pMM62 | pCR2.1 TOPO derivative containing a mutant allele of recA(Δ80-283) (BAB1_1224) | This study |
| pMM63 | Mutator plasmid, pJQK derivative containing the BamHI-XbaI fragment of pMM62 | This study |
| pMM72 | pCR2.1 derivative containing F1R4ΔBAB1_0557 fragment generated by PCR overlap | This study |
| pMM73 | Mutator plasmid, pJQK derivative containing the BamHI-XbaI fragment of pMM72 | This study |
| pMM77 | pCR2.1 derivative containing F1R4ΔBAB1_0546 fragment generated by PCR overlap | This study |
| pMM78 | Mutator plasmid, pJQK derivative containing the BamHI-XbaI fragment of pMM77 | This study |
| pMM99 | pCR2.1 derivative containing a fragment of 853 bp from a wbkA deletion intermediate | This study |
| E. coli strains | ||
| TOP10F | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| OmniMAX 2-t1 | F′ [proAB+lacIqlacZΔM15 Tn10(TetR) Δ(ccdAB)] mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80(lacZ)ΔM15 Δ(lacZYA-argF)U169endA1recA1supE44 thi-1 gyrA96relA1tonA panD | Invitrogen |
| S17-1λpir | Mating strain with plasmid RP4 inserted into the chromosome | 35 |
Cmr, chloramphenicol resistance; Strr, streptomycin resistance; Kmr, kanamycin resistance.
IS711-fingerprinting.
The IS711 sequence was detected by Southern blotting with 1 to 2 μg of AvaI-ClaI double-digested B. abortus gDNA resolved in 1% agarose in TBE buffer at 25 mA for 10 h. The IS711 probe was generated by PCR with the primers 711u and 711d (Table 2) (28) and biotinylated by direct labeling (GE Healthcare, Buckinghamshire, United Kingdom). Chemiluminescent detection of the hybridized DNA was done using a commercial kit (Amersham/GE Healthcare), and films were developed by conventional photographic methods.
RESULTS
Excision of wbkA occurs spontaneously without affecting other wbk genes.
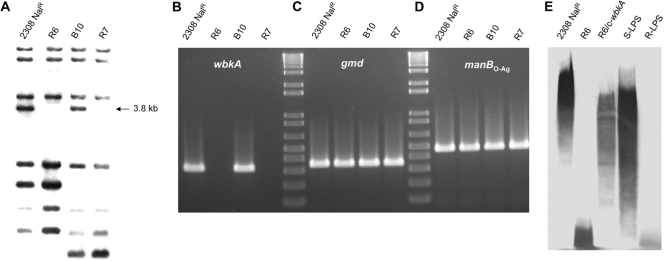
Previous works have shown that all wbk O-PS genes except wbkA are concatenated in sections of 2 to 6 ORFs and that wbkA is flanked by a high number of transposase ORFs (Fig. 1A) (6, 15, 16). Although the presence of transposases and inactivated remnants results in various sets of repeated sequences that make uncertain an accurate prediction of boundaries, the size of the fragment bearing wbkA could be estimated in 6.5 kb. This fragment (referred to here as the wbkA region) contains one IS711 plus two nearly identical ISBm1 copies at each end with a 60-nt imperfect repeat sequence upstream of each orfA (Fig. 1B). A closer analysis of the ISBm1 copies revealed the existence of inverted repeats susceptible to transpositional recombination (Fig. 1C). These features are consistent with the hypothesis that wbkA could be excised through events involving the flanking sequences but no adjacent wbk genes. To address this possibility, 12 spontaneous R mutants obtained in vitro from different B. abortus cultures were screened for wbkA deletions by IS711 fingerprinting, taking advantage of the existence (predicted by in silico restriction analysis) of a 3.8-kb AvaI-ClaI fragment carrying IS711 (Fig. 1B). Two mutants (named R6 and R7) lacked the 3.8-kb fragment (Fig. 2A). Additional analyses by PCR with primers targeting wbkA, gmd, and manBO-Ag (Table 2) showed that mutants R6 and R7 carried a deletion encompassing wbkA but not the two adjacent LPS genes (Fig. 2B, C, and D), suggesting that the deletion affected only wbkA. This was confirmed by complementation of R6, R7, and four additional B. abortus 2308 spontaneous R ΔwbkA mutants with the plasmid wbkA-pMM14, which restored the S phenotype as judged by the crystal violet exclusion test, agglutination, and Western blotting with antibodies to S B. abortus (Fig. 2E). The rate of spontaneous ΔwbkA mutation was calculated by probing R colonies (i.e., crystal violet positive) obtained in dissociation experiments. Under the conditions used (prolonged incubation in static, oxygen-limiting cultures), BaΔwbkA mutants represented approximately 8 to 10% of the R mutants, a value lower than that reported for GI-2 deletion (23).
Fig 2.
The spontaneous deletion of wbkA is linked to the emergence of R mutants. (A) IS711 fingerprinting characterization of R mutants (the arrow indicates the 3.8-kb fragment absent in R mutants). (B) PCR detection of wbkA gene (BAB1_0553). (C) PCR detection of gmd gene (BAB1_0545). (D) PCR detection of manBO-Ag gene (BAB1_0560). (E) Western blot analysis with LPS antibodies. Strains 2308 Nalr and B10 are the B. abortus parental S strains of R mutants R6 and R7, respectively, and strain R6/c-wbkA is plasmid wbkA-pMM14 complemented R6 mutant (Table 1). S-LPS and R-LPS are the purified products used as controls.
The excision of wbkA is generated by a recombination event involving the flanking ISBm1 copies.
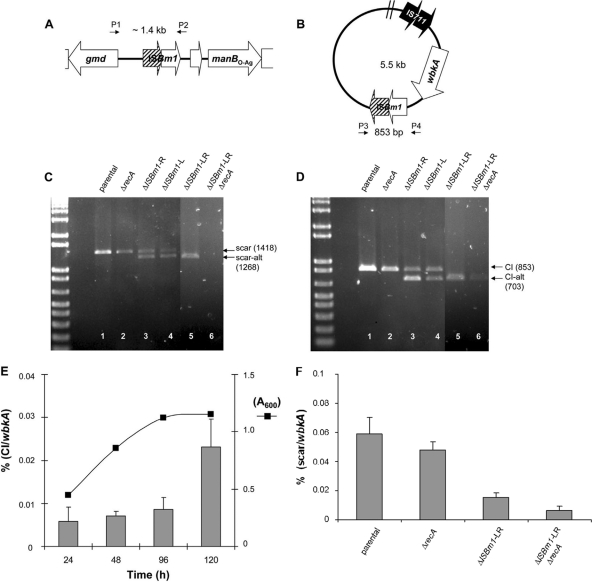
Two possible wbkA excision mechanisms are deletion by homologous recombination and IS-mediated transposition. The former involves RecA activity, and the latter involves transposition or cooperatively acting IS-encoded proteins (13, 17, 18, 22). Concerning the first possibility, RecA can recognize nearly identical IS pairs as substrates of excisive recombination if they are oriented as direct repeats (13, 22). Accordingly, an intrachromosomal exchange between ISBm1 copies should result in the products illustrated in Fig. 3A and B. PCR assays with the primers pairs BAB1_0545Fb/BAB1_0558R and BAB1_0557F/BAB1_0548R showed that B. abortus 2308 (Fig. 3C and D, lane 1), as well as B. melitensis 16M and B. suis 1330 (data not shown), yielded both predicted products. Moreover, the circular intermediate that should result from active recombination was also demonstrated by qPCR, a method that also revealed a progressive increase in this product as the bacteria entered the stationary phase of growth (Fig. 3E). A spontaneous B. melitensis R mutant isolated in a pure culture from goat milk also showed the chromosomal scar corresponding to the wbkA deletion (not shown). Consistent with the homologous recombination mechanism, sequencing revealed a crossover between ISBm1 ORFs. In order to study the involvement of RecA, a recA-deficient mutant (BabΔrecA) was constructed by allelic exchange. The BabΔrecA mutant failed to grow in the presence of methyl methanesulfonate, a potent DNA damage inductor (38), and was refractory to the integration of plasmids successfully used in several allelic exchange experiments in our laboratory (not shown), confirming that it was defective in RecA-dependent homologous recombination. Although with less intensity, BabΔrecA yielded the same products as the wild-type strain (Fig. 3C and D, lane 2), strongly suggesting some degree of RecA-independent homologous recombination.
Fig 3.
IS-mediated recombination is responsible for the loss of wbkA. (A) Genetic organization of the chromosomal scar left behind the excision. (B) Genetic organization of the closed-circular intermediate or CI carrying the copy of IS711 (in both panels A and B, hatched arrows indicate the crossover between the ISBm1 ORFs, and the primer [see the legend for Fig. 1] positions are depicted by small black arrows). (C and D) PCR detection of recombination intermediates in B. abortus recA and ISBm1-related mutants (note that scar-alt and CI-alt correspond to alternative recombination products). (E) PCR quantification of circular intermediates during growth (results are the average ± the standard error of triplicate measurements). (F) PCR quantification of chromosomal scars (results are the average ± the standard error of duplicate measurement from three independent experiments).
To test the existence of IS-mediated transposition, ISBm1 mutants were constructed by removing 150 nt from identical sites of one (mutants BabΔISBm-L and BabΔISBm-R) or both (BabΔISBm-LR) flanking transposases (Table 1). PCR analyses demonstrated products of the same size as those produced by the wbkA in the parental strain. In addition, these analyses showed the alternative 150-bp-lacking recombination species (Fig. 3C and D, lanes 3, 4, and 5) that result from the resolution of the Holliday junction produced by the strand exchange between asymmetric repeats. Since a chromosomal joint with no traces of ISBm1 sequences is expected from a transpositional deletion (10), these results were consistent with homologous recombination but not with transposition. To test whether a RecA-independent recombination takes place in the ISBm1 mutants, the mutation used to construct BabΔrecA was introduced in BabΔISBm-LR by allelic exchange to generate BabΔISBm-LRΔrecA. The presence of the scar was then investigated by conventional PCR. The results showed that the scar was still detectable, thus demonstrating that the recombination was not only RecA dependent. Moreover, although conventional PCR did not show the products resulting from the scar (1,268 bp; Fig. 3C, lane 6), traces of the expected circular intermediate (703 bp; Fig. 3D, lane 6) were detected in the triple mutant. To confirm the existence of a residual RecA-independent recombination, quantitative measurements of chromosomal scars were performed by qPCR. This method revealed that, although with a nearly 10-fold reduction with respect to the parental strain, the triple mutant displayed the wbkA excision scar (Fig. 3F). These same experiments also showed that the deletions created in the IS repeats impaired the excision rate, as expected from homologous recombination. In summary, wbkA is undergoing a spontaneous excision by both RecA-dependent and RecA-independent pathways.
DISCUSSION
S-R dissociation has a deep impact on the antigenic structure and virulence of the major zoonotic brucellae and on the stability of S vaccines (2, 37). Early work showed that unfavorable growth conditions, oxygen limitation in particular, facilitate the establishment of R variants (20) and that the effect of low oxygen tension can be abrogated by supplying alternative proton/electron acceptors such as nitrate, resazurine, or methylene blue (14). Two complementary hypotheses that could explain these observations are (i) that mutants not synthesizing the costly O-PS are more competitive when energy shortage and no selective pressure to maintain the O-PS concur and (ii) that S brucellae have genetic features introducing instability in O-PS genes. Concerning the second hypothesis, we have shown previously the role of the phage-related integrase in GI-2 excision and S-R dissociation (23), and we present here evidence for an additional mechanism involving the deletion of wbkA by homologous recombination. Clearly, these two excision pathways belong to a set of mechanisms affecting LPS biosynthesis genes in Gram-negative bacteria, including site-specific recombination (23), complex genome rearrangements not dependent on homology at boundaries (39), insertion-deletions, and point mutations (1, 39). Moreover, in the case of the S-R dissociation of brucellae, the instability may be connected to the origin of the O-PS genes. GI-2 bears traits that indicate horizontal acquisition, and this may be also true for wbk and, within this region, for wbkA. The %GC of the 6.5-kb fragment is 52.5, higher than the 50% of the entire wbk region, and both are different from the average %GC of the Brucella genome (56 to 58%). These percentages and the peculiarities of wbkA and its flanking sections are compatible with the acquisition of wbkA by a preexisting region comprising the remaining wbk genes. Horizontal acquisition of the complete wbk has been proposed before (6, 15), an event that may have happened before the brucellae became intracellular parasites. Noteworthy, the wbkA region is absent from the O-PS encoding region of Yersinia enterocolitica O:9, a bacterium that also makes an N-formylperosamine homopolymers, albeit with some structural differences with that of S brucellae (30, 36). The analysis of the structure of the ISBm1 copies flanking wbkA showed that, in spite of the absence of any evident duplicated insertion site, they carry canonical terminal inverted repeat of 8 bp which might be involved in the transpositional release of the 6.5-kb fragment. In addition, the organization of this region resembles that of composite transposons (22), and there is experimental evidence for the expression of ISBm1-related ORFs at the beginning of the host-pathogen interaction (33). Thus, an excision mechanism mediated by transposition seemed possible, which would be in accordance with a foreign origin of this locus in wbk. Nevertheless, the predicted transpositional joint was not detected, suggesting that the ability to transpose has been lost. In fact, the single nucleotide polymorphisms (SNPs) present in inverted repeats in one of the ISBm1 copies (Fig. 1C) could prevent transposition, so that only excision by homologous recombination remains.
Homologous recombination is one of the mechanisms responsible for large rearrangements in bacterial genomes. These changes are part of the plasticity showed by chromosomes and are often involved in intra- or interspecies polymorphism (9, 22, 29). There are examples in the literature in which ISs appear as rearrangement-promoting elements, playing a role in the deletion of fragments encoding virulence factors in pathogens such as Y. pestis and uropathogenic E. coli (12, 26). In bacteria belonging to the Mycobacterium tuberculosis complex, comparative analyses suggest that IS6110-mediated deletions have shaped extensively their genomes (4, 11). In a way similar to the wbkA region, the homologous recombination between IS1610 copies participates in the formation of R variants in M. avium (10). In all of these cases, RecA occupies a central role prompting these genome deletions. Our results also show that, after inactivation of recA, homologous recombination still operates at an extent that is low but enough to produce wbkA deletions in Brucella. This observation strongly suggests that recombination-promoting genes other than recA participate in the excision of the wbkA region. The mechanism of homologous recombination and the genes involved have not been studied in detail in brucellae. In the closely related Rhizobiaceae, the products of the genes ruvB and radA migrate to the Holliday intermediary and, although modestly, seem to contribute to homologous recombination (25, 29). In Brucella, a previous work has shown the presence of a putative recA homolog (radA, BAB1_0474), which does not perform a protective DNA repair function and is devoid of physiological importance (34). Preliminary results indicate that the residual homologous recombination observed in the wbkA excision is not radA dependent.
In regard to the frequency of dissociation, the data from this and previous works (23, 39) show that deletion of wbkA and of GI-2, plus mutations in the mannose genes involved in LPS core oligosaccharide synthesis, is responsible for the majority of R mutants obtained in the laboratory. In this context, it is worth commenting the stochasticity of mutations involved in S-R dissociation. Mutations affecting mannose synthesis genes outside wbk correspond to point mutations, indels, and extensive deletions and, indeed, it seems possible that stochastic mutations affect other LPS genes. In contrast, wbkA and GI-2 deletions are generated through well-defined paths and, therefore, the corresponding mutants are not randomly generated. Accordingly, the identification of S-R dissociation mechanisms such as those dependent on GI-2 and wbkA entail the possibility of controlling these excisions to obtain more stable strains for antigen and vaccine production. Moreover, it has been recently hypothesized that dissociation may be part of the natural course of brucellosis infection (39). In this context, the isolation of a ΔwbkA B. melitensis R mutant strain from goat milk that lacked detectable S colonies is interesting. Isolation of R mutants in pure culture from goat milk but not from other infected samples has been occasionally observed in the laboratory of one of the authors, suggesting that these animals carry R mutants in the mammary glands. The biological significance of the loss of O-PS expression in vivo is currently being investigated.
ACKNOWLEDGMENTS
We thank A. Delgado for skilful technical assistance.
This study was funded by the Ministerio de Ciencia y Tecnología of Spain (AGL2008-04514) and the Universidad Austral de Chile (DID S-2009-33).
Footnotes
Published ahead of print 10 February 2012
REFERENCES
- 1. Adone R, Francia M, Ciuchini F. 2008. Evaluation of Brucella melitensis B115 as rough-phenotype vaccine against B. melitensis and B. ovis infections. Vaccine 26: 4913– 4917 [DOI] [PubMed] [Google Scholar]
- 2. Alton G, Jones L, Angus R, Verger JM. 1988. The production of Brucella vaccines: techniques for the brucellosis laboratory. INRA, Paris, France [Google Scholar]
- 3. Braun W. 1946. Dissociation in Brucella abortus: a demonstration of the role of inherent and environmental factors in bacterial variation. J. Bacteriol. 51: 327– 349 [DOI] [PubMed] [Google Scholar]
- 4. Brosch R, et al. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. U. S. A. 99: 3684– 3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burrus V, Waldor MK. 2003. Control of SXT integration and excision. J. Bacteriol. 185: 5045– 5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cloeckaert A, Grayon M, Verger JM, Letesson JJ, Godfroid F. 2000. Conservation of seven genes involved in the biosynthesis of the lipopolysaccharide O-side chain in Brucella spp. Res. Microbiol. 151: 209– 216 [DOI] [PubMed] [Google Scholar]
- 7. Conde-Alvarez R, et al. 2006. Synthesis of phosphatidylcholine, a typical eukaryotic phospholipid, is necessary for full virulence of the intracellular bacterial parasite Brucella abortus. Cell. Microbiol. 8: 1322– 1335 [DOI] [PubMed] [Google Scholar]
- 8. Chain PS, et al. 2005. Whole-genome analyses of speciation events in pathogenic brucellae. Infect. Immun. 73: 8353– 8361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dobrindt U, Hacker J. 2001. Whole genome plasticity in pathogenic bacteria. Curr. Opin. Microbiol. 4: 550– 557 [DOI] [PubMed] [Google Scholar]
- 10. Eckstein TM, Inamine JM, Lambert ML, Belisle JT. 2000. A genetic mechanism for deletion of the ser2 gene cluster and formation of rough morphological variants of Mycobacterium avium. J. Bacteriol. 182: 6177– 6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fang Z, et al. 1999. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J. Bacteriol. 181: 1014– 1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fetherston JD, Schuetze P, Perry RD. 1992. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol. Microbiol. 6: 2693– 2704 [DOI] [PubMed] [Google Scholar]
- 13. Galas D, Chandler M. 1989. Bacterial insertion sequences, p 109– 162 In Berg D, Howe M. (ed), Mobile DNA. ASM Press, Washington, DC [Google Scholar]
- 14. Gerhardt P. 1958. The nutrition of brucellae. Bacteriol. Rev. 22: 81– 98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Godfroid F, et al. 2000. Genetic organisation of the lipopolysaccharide O-antigen biosynthesis region of Brucella melitensis 16M (wbk). Res. Microbiol. 151: 655– 668 [DOI] [PubMed] [Google Scholar]
- 16. Gonzalez D, et al. 2008. Brucellosis vaccines: assessment of Brucella melitensis lipopolysaccharide rough mutants defective in core and O-polysaccharide synthesis and export. PLoS One 3: e2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gray YH. 2000. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 16: 461– 468 [DOI] [PubMed] [Google Scholar]
- 18. Gueguen E, Rousseau P, Duval-Valentin G, Chandler M. 2005. The transpososome: control of transposition at the level of catalysis. Trends Microbiol. 13: 543– 549 [DOI] [PubMed] [Google Scholar]
- 19. Hallez R, Letesson JJ, Vandenhaute J, De Bolle X. 2007. Gateway-based destination vectors for functional analyses of bacterial ORFeomes: application to the Min system in Brucella abortus. Appl. Environ. Microbiol. 73: 1375– 1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Henry B. 1933. Dissociation in the genus Brucella. J. Infect. Dis. 52: 374– 402 [Google Scholar]
- 21. Lapaque N, Moriyon I, Moreno E, Gorvel JP. 2005. Brucella lipopolysaccharide acts as a virulence factor. Curr. Opin. Microbiol. 8: 60– 66 [DOI] [PubMed] [Google Scholar]
- 22. Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62: 725– 774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mancilla M, López-Goñi I, Moriyón I, Zárraga AM. 2010. Genomic island-2 is an unstable genetic element contributing to Brucella lipopolysaccharide spontaneous smooth to rough dissociation. J. Bacteriol. 192: 6346– 6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mancilla M, Villarroel M, Saldías ME, Soto J, Zárraga AM. 2008. Genotipos de aislados de campo de Brucella abortus de distintas regiones geográficas de Chile. Arch. Med. Vet. 40: 187– 192 [Google Scholar]
- 25. Martinez-Salazar JM, Zuniga-Castillo J, Romero D. 2009. Differential roles of proteins involved in migration of Holliday junctions on recombination and tolerance to DNA damaging agents in Rhizobium etli. Gene 432: 26– 32 [DOI] [PubMed] [Google Scholar]
- 26. Middendorf B, et al. 2004. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J. Bacteriol. 186: 3086– 3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moriyon I, et al. 2004. Rough vaccines in animal brucellosis: structural and genetic basis and present status. Vet. Res. 35: 1– 38 [DOI] [PubMed] [Google Scholar]
- 28. Ocampo-Sosa AA, Aguero-Balbin J, Garcia-Lobo JM. 2005. Development of a new PCR assay to identify Brucella abortus biovars 5, 6, and 9 and the new subgroup 3b of biovar 3. Vet. Microbiol. 110: 41– 51 [DOI] [PubMed] [Google Scholar]
- 29. Orozco-Mosqueda Mdel C, Altamirano-Hernandez J, Farias-Rodriguez R, Valencia-Cantero E, Santoyo G. 2009. Homologous recombination and dynamics of rhizobial genomes. Res. Microbiol. 160: 733– 741 [DOI] [PubMed] [Google Scholar]
- 30. Perry RD, Bundle DR. 1990. Lipopolysaccharide antigens and carbohydrates of Brucella, p 76– 88 In Adams LG. (ed), Advances in brucellosis research. Texas A&M University Press, College Station, TX [Google Scholar]
- 31. Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127: 15– 21 [DOI] [PubMed] [Google Scholar]
- 32. Rajashekara G, Covert J, Petersen E, Eskra L, Splitter G. 2008. Genomic island 2 of Brucella melitensis is a major virulence determinant: functional analyses of genomic islands. J. Bacteriol. 190: 6243– 6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rossetti CA, Galindo CL, Lawhon SD, Garner HR, Adams LG. 2009. Brucella melitensis global gene expression study provides novel information on growth phase-specific gene regulation with potential insights for understanding Brucella-host initial interactions. BMC Microbiol. 9: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roux CM, et al. 2006. RecA and RadA proteins of Brucella abortus do not perform overlapping protective DNA repair functions following oxidative burst. J. Bacteriol. 188: 5187– 5195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Simon R, Priefer U, Pehle A. 1983. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1: 784– 890 [Google Scholar]
- 36. Skurnik M, et al. 2007. Characterization and biological role of the O-polysaccharide gene cluster of Yersinia enterocolitica serotype O:9. J. Bacteriol. 189: 7244– 7253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Spink W. 1956. The nature of brucellosis. Lund Press, Inc., Minneapolis, MN [Google Scholar]
- 38. Tatum FM, Morfitt DC, Halling SM. 1993. Construction of a Brucella abortus RecA mutant and its survival in mice. Microb. Pathog. 14: 177– 185 [DOI] [PubMed] [Google Scholar]
- 39. Turse JE, Pei J, Ficht TA. 2011. Lipopolysaccharide-deficient Brucella variants arise spontaneously during infection. Front. Microbiol. 2: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. White PG, Wilson JB. 1951. Differentiation of smooth and nonsmooth colonies of brucellae. J. Bacteriol. 61: 239– 240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilson K. 2001. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. Chapter 2: Unit 2.4. [DOI] [PubMed] [Google Scholar]