Abstract
Oral bacterial biofilms are highly complex microbial communities with up to 700 different bacterial taxa. We report here the use of metatranscriptomic analysis to study patterns of community gene expression in a multispecies biofilm model composed of species found in healthy oral biofilms (Actinomyces naeslundii, Lactobacillus casei, Streptococcus mitis, Veillonella parvula, and Fusobacterium nucleatum) and the same biofilm plus the periodontopathogens Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans. The presence of the periodontopathogens altered patterns in gene expression, and data indicate that transcription of protein-encoding genes and small noncoding RNAs is stimulated. In the healthy biofilm hypothetical proteins, transporters and transcriptional regulators were upregulated while chaperones and cell division proteins were downregulated. However, when the pathogens were present, chaperones were highly upregulated, probably due to increased levels of stress. We also observed a significant upregulation of ABC transport systems and putative transposases. Changes in Clusters of Orthologous Groups functional categories as well as gene set enrichment analysis based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways showed that in the absence of pathogens, only sets of proteins related to transport and secondary metabolism were upregulated, while in the presence of pathogens, proteins related to growth and division as well as a large portion of transcription factors were upregulated. Finally, we identified several small noncoding RNAs whose predicted targets were genes differentially expressed in the open reading frame libraries. These results show the importance of pathogens controlling gene expression of a healthy oral community and the usefulness of metatranscriptomic techniques to study gene expression profiles in complex microbial community models.
INTRODUCTION
Biofilms growing in the oral cavity are among the most complex microbial communities in the human body, with more than 700 bacterial taxa identified so far (59). Many of the species are noncultivable organisms whose only information we possess derives from their 16S rRNA phylogenetic affiliation (58, 59). Oral biofilms show all of the characteristic features of biofilm architecture, yet are distinct in that they consist of a high density of cells and the array of species have specific cell-cell interactions that lead to the development of these communities (36, 48). Moreover, dental biofilms form in a well-defined pattern with a highly complex spatial organization (36, 83). The first step in dental plaque formation builds on the abiotic coating of the tooth surface by a pellicle of salivary proteins that precondition the enamel to be colonized by a first group of bacteria which are primarily Gram-positive cocci (streptococcal species). During the next stage of development, Fusobacterium species attach to the streptococci and serve as a bridge to the third set of organisms, which are predominantly Gram-negative anaerobes (37). However, in situ studies of initial enamel colonization present a more complex scenario. Although there are variations from individual to individual, in general, between 4 and 8 h from the initiation of colonization, communities are dominated by Streptococcus spp. (11, 16). Other frequently observed genera were Actinomyces and Veillonella (1, 16).
The nature of these interactions is partially driven by coaggregation, cell-to-cell specific recognition of genetically distinct cell types, and these specific interactions play a critical role in biofilm architecture (18, 23, 36). The stability of this biofilm depends on changes in environmental factors, as well as on interspecies communication (e.g., quorum-sensing signaling) that modulates the stability and composition of the biofilm (50, 63, 73). Microbial cell-cell interactions in the oral flora are believed to play an integral role in the development of dental plaque and, ultimately, its pathogenicity (41).
Although mature, healthy biofilms are resilient structures, due to factors not completely understood, they can progress toward polymicrobial infections, with a coordinate action of the biofilm leading to pathogenesis. The pathogenic biofilm causes a progressive loss of bone surrounding the teeth, which, if left untreated, results in loosening and eventual loss of the teeth (2, 8). The oral biofilm undergoes a change in composition from healthy to the most severe forms of periodontitis. Thus, periodontal health is the result of a dynamic equilibrium between the microbial flora and the host, characterized by minimal inflammatory episodes. Microbial interactions play a critical role in the evolution of the disease. Besides coaggregation, competition for nutrients and synergistic interactions are crucial in the development of the oral biofilm (26, 38). For example, Porphyromonas gingivalis, a major periodontal pathogen, metabolizes succinate produced by Treponema denticola, while T. denticola uses isobutyric acid secreted by P. gingivalis (21).
High complexity of dental plaque and variability among individuals make reproducing and interpreting in vivo studies of oral biofilms difficult. To overcome problems associated with in vivo studies, a large number of different laboratory model systems, which are more controllable, have been developed (18, 22, 70, 74). Most model systems are based on flow cells (18) or chemostats with removable inserts, providing a surface for biofilm formation (34). However, the use of these devices with a multispecies biofilm model is difficult to maintain during long periods of time and is cumbersome to construct (22). Hence, some authors have opted for using static systems that simplify the setup and manipulation of the oral biofilm model (22, 39, 64).
Techniques like quantitative PCR (qPCR) can be used to quantify gene expression in natural samples, although this technique is typically limited to analyzing a small number of known genes. Other techniques, such as microarray analysis (78) or proteomic analysis (80), have been restricted to the analysis of expression profiles of one organism. In general, most expression analysis studies have been performed on monospecies biofilms under different laboratory conditions trying to mimic environmental conditions (45, 52, 80).
Probably the most important limiting factor to study gene expression in complex microbial communities is the small amount of biomass either from plaque samples or in vitro biofilms. Moreover, the half-life of prokaryotic mRNA is short (3, 65), and mRNA in bacteria and archaea usually comprises only a small fraction of total RNA. To overcome these challenges, several approaches have been recently developed. rRNA subtraction has been used in combination with randomly primed reverse transcription-PCR (RT-PCR) to generate microbial community cDNA for cloning and downstream sequence analysis (61). We have applied a method based on linear amplification of the mRNA that allows for evaluation of gene expression in the whole microbial community from small amounts of RNA in environmental samples (19). The use of next-generation sequencing has substantially widened the scope of metagenomic analysis of environmentally derived samples and, in our case, has facilitated the study of transcriptome analysis of complex microbial communities.
Here we report the use of an oral biofilm model growing on hydroxyapatite disks to study gene expression patterns of the whole microbial community in situ and the effect of the presence of periodontal pathogens on the healthy community. Two different multispecies biofilms based on McKee et al. (49) were studied, one with 5 of the most abundant and frequently found species in dental plaque from healthy individuals (Actinomyces naeslundii, Lactobacillus casei, Streptococcus mitis, Veillonella parvula, and Fusobacterium nucleatum) (6, 58, 81) and the other with those same species plus two of the most important periodontopathogens: Porphyromonas gingivalis and Aggregatibacter (formerly Actinobacillus) actinomycetemcomitans. We wanted to asses whether the presence of periodontopathogens has any effect on a healthy, already established biofilm. The use of next-generation sequencing technologies was invaluable to obtain a deeper coverage and understanding of the complex metatranscriptome resulting from the multispecies community.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The species used in the study were A. naeslundii (MG1), L. casei (ATCC 334), S. mitis (ATCC 49456), V. parvula (ATCC 17745), and F. nucleatum (ATCC 10953). In the pathogenic biofilm, we used these same previously cited species plus the periodontopathogens P. gingivalis (ATCC 33277) and A. actinomycetemcomitans (ATCC 33384). Strains were grown under anaerobic conditions at 37°C for 72 h in a Coy anaerobic chamber on brain heart infusion agar (Difco) plates supplemented with 5% horse blood (Northeastern Laboratory, ME), 1 μg/ml hemin, and 1 μg/ml of vitamin K.
Biofilm growth.
Biofilms were grown on sterile hydroxyapatite disks of 7 mm by 1.8 mm (Clarkson Inc.) placed into each well of a 24-well cell culture plate (Nalgene Nunc International, Denmark). Wells were filled with 1 ml of the mucin growth medium (MGM), used by Kinniment et al. (34), which presents a high concentration of proteins, and supplemented with 4 ml of resazurin from a 25-mg/100-ml solution, 1 μg/ml hemin, and 1 μg/ml of vitamin K. pH was adjusted to 7.4 prior to autoclaving. In order to form the acquired pellicle, the disks were allowed to stay at least 60 h in contact with the medium under anaerobic conditions prior to inoculation. This period of preincubation allowed us to check for possible contamination as well. Each of the strains grown on agar were resuspended in MGM until the suspension reached a turbidity of a McFarland standard of 3 (approximately 108 CFU/ml). Finally, 1 ml of each bacterial suspension was mixed and 50 μl of this suspension was added into each hydroxyapatite disk-containing well that contained 3 ml of MGM. Plates were incubated under anaerobic conditions at 37°C. After 24 h, disks were used for analysis. For each condition (healthy and pathogenic biofilms), we used 6 hydroxyapatite disks with their corresponding planktonic phase. All of them were combined before nucleic acid extraction.
For the pathogenic biofilm, the periodontopathogens P. gingivalis (ATCC 33277) and A. actinomycetemcomitans (ATCC 33384) were added to the biofilm after 24 h and incubated for another 6 h. The inocula were obtained as described above, and each of the strains grown on agar was resuspended in MGM until reaching a turbidity of a McFarland standard of 3 (approximately 108 CFU/ml). One milliliter of each bacterial suspension was mixed, and 50 μl of this suspension was added into each hydroxyapatite disk-containing well. To follow the development and detect the presence of the different bacteria in the biofilm, DNA was extracted following the protocol proposed for the ToTALLY RNA kit (Applied Biosystems).
DNA and RNA extraction.
Samples for the biofilms were immediately transferred to screw-cap tubes containing the lysis buffer and bead beaten for 1 min at maximum speed. Planktonic phase was recovered by centrifugation at 14,000 × g and analyzed in parallel. DNA and RNA were extracted simultaneously following the protocol of the mirVana miRNA isolation kit for RNA and the ToTALLY RNA kit (Applied Biosystems, Ambion) for DNA. For RNA analysis, genomic DNA was removed using a Turbo DNA-free kit (Applied Biosystems, Ambion).
qPCR.
qPCR was performed to quantify the different bacteria in the biofilm as described by Frias-Lopez et al. (19). Additionally, concentrations of different bacteria in the planktonic phase (control) were also assessed. Specific primers for qPCR were designed using PerlPrimer against genes with a single copy in each of the genomes (Table 1). To quantify the number of genome copies, standards of the different bacteria were labeled with acridine orange (75) and counted under a fluorescence microscope to assess bacterial concentration. DNA was extracted as described above.
Table 1.
Bacterial strains and primers used in the study
| Bacterial strain or RT-qPCR primer | Forward primer 5′–3′ | Reverse primer 5′–3′ | Target gene (source or reference) |
|---|---|---|---|
| Bacterial strains | |||
| Actinomyces naeslundii (MG1) | GAGAAGAACTCCCTATCCATACC | CTTCATGGGTTGGGATTTCCT | Target is fimA gene, a fimbrial protein (this work) |
| Fusobacterium nucleatum (ATCC10953) | CTTATACATAGAGGACACAGAC | TTTCCAACAACATCTCCCTG | HprK gene (30) homologue of HPr kinase/phosphorylase (this work) |
| Lactobacillus casei (ATCC 334) | TGAATATTTACACGGTTCGGCA | GGGCCTTAGTTTCATCCGAC | Target is the phosphoglucomutase gene (10) (this work) |
| Streptococcus mitis (NCTC 12261, ATCC 49456) | CGTATCGGTCGTCTTGCT | TTGCATAACTGGATCTGTAAGGTC | Target is the glyceraldehyde-3-phosphate gene (this work) |
| Veillonella parvula (ATCC17745) | ACAGTTACTTTATAGTCTGTACC | TGGTCAATTGGCTAAACGTCAA | Target is the chaperone dnaK gene (29) (this work) |
| Aggregatibacter actinomycetemcomitans (HK1651) | ACGCAGACGATTGACTGAATTTAA | GATCTTCACAGCTATATGGCAGCTA | Target is lktC gene part of the leukotoxin operon lktBAC (54) |
| Porphyromonas gingivalis (ATCC 33277) | CCTACGTGTACGGACAGAGCTATA | AGGATCGCTCAGCGTAGCATT | Target is the Arg-gingipain gene (54) |
| RT-qPCR primers | |||
| Universal | CGCTAGTAATCGTGGATCAGAATG | TGTGACGGGCGGTGTGTA | 16S rRNA (79) |
| Actinomyces naeslundii (MG1) | ATCTCCAAGGTTCTGCACGACGAGTA | ATGTTGATGGTGATACCGCGCTGA | ANA_0022 (this work) |
| Aggregatibacter actinomycetemcomitans (HK1651) | AAGAACTTAACGCTTGGGCGATGC | TAACGCTTCTTGTGCAACACTGGC | AA00470 (this work) |
| Veillonella parvula (ATCC17745) | AAAGCCTTGGGCCATTCTCTGTTG | CCAAGGCCTCTTGTTCTTGCATCA | HMPREF1035_1004 (this work) |
| Porphyromonas gingivalis (ATCC 33277) | AGAAAGCCAGCCATTGTTGCATGG | TGTTCGGGACACCTGACTGTTCAT | PGN_0074 (this work) |
Doubling time was calculated using the following formula: td = (t2−t1) × log (2)/log(q2/q1), where q2 represents the final number of cells, q1 represents the initial number of cells added, and (t2−t1) represents the interval of incubation. Statistical differences in doubling time were assessed using the exact Wilcoxon rank sum test in R (http://www.r-project.org/).
RNA amplification and Illumina sequencing.
Prior to amplification, rRNA was remove using the MICROBExpress kit from Ambion (Applied Biosystems) according to the manufacturer's instructions. RNA amplification was performed on 100 ng of total RNA using the MessageAmp II-bacteria kit (Applied Biosystems) by following the manufacturer's instructions. Sequencing was performed at the Biopolymers Facility at Harvard Medical School. The samples were prepped using the NuGen mRNA-Seq kit into next-generation libraries with Illumina adapters. The libraries were then checked for quality control via the bioanalyzer to ensure correct sizing and then run on a qPCR assay, which will only amplify library fragments with the Illumina adapters correctly ligated. The libraries are then clustered on a single-read flow-cell using the Illumina cBot, and then the sequencing was run for 72 cycles using an Illumina GAII sequencer.
Bioinformatic selection of intergenic regions (IGRs) from genomes.
Genomes were downloaded from the HOMD database server (http://www.homd.org/). IGRs were extracted using the open reading frame (ORF) coordinates from the corresponding .ppt files of the different genomes. A Perl script writing for this purpose extracted IGRs with a 5-bp overlap over the adjacent ORFs on both the 3′ and 5′ ends. For further analysis, we selected for only IGRs larger than 50 bp.
Short read sequence alignment and bioinformatic analysis of the results.
Trimmed non-rRNA cDNA reads from the species in the biofilm and planktonic phases were searched against the databases of their own genomes using three different algorithms, i.e., SSAHA2 (55), BLAT (31), and MegaBLAST (82), and assigned to the best-hit gene. The process was sequential: first reads were searched using SSAHA2, the nonassigned reads were then searched using BLAT, and, finally, the nonassigned reads left were searched using MegaBLAST. The same procedure was initially used to remove rRNA sequences.
SSAHA2 options were ssaha2-solexa, -skip 2, -diff 0, -kmer 13, -disk 1, -output psl; BLAT options were the default for the program; and MegaBLAST options were megablast-d DB, -i <input>, -n T, -m8, -o <output>, -v1, -b1, -a2 for rRNA analysis and megablast-d DB, -i <input>, -n T, -m8, -o <output>, -e 1e-5, -W 35, -v1, -b1, -a8 for ORF and IGR analysis.
For testing the accuracy of the different algorithms in our database, we split the genomes of the organisms in the community into fragments of 80 bp using the “splitter” function from EMBOSS with an overlap of 10 bp and add overlapping to size.
Normalization of raw results was performed by scaling to library size, and only genes with matches in the biofilm and planktonic phase were used for analysis. We used a cutoff value of at least an 8-fold difference in gene expression to be considered significant.
RT-qPCR confirmation of gene expression for specific genes.
All RT-qPCR measurements were performed in triplicate. Fifty nanograms of RNA, in all cases, were subjected to RT-qPCR using an iCycler 584BR (Bio-Rad Laboratories) with iScript one-step RT-PCR kit with SYBR green (Bio-Rad Laboratories). cDNA synthesis and PCR amplification were carried out in the same tube. PCR conditions included 10 min at 50°C for cDNA synthesis, followed by denaturation at 95°C for 5 min, and then 55 cycles of 95°C for 10 s, 60°C for 30 s minute, and 72°C for 30 s, followed by melting curve analysis. Fluorescence data were captured during annealing reactions, and specificity of the amplification was confirmed using melting curve analysis. Data were collected and recorded by iCycler iQ software (Bio-Rad Laboratories) and initially determined as a function of threshold cycle (CT). CT was defined as the cycle at which the fluorescence intensity in a given reaction tube rose above background, which was calculated as 10 times the mean standard deviation (SD) of fluorescence in all wells over the baseline cycles. Levels of increased 16S RNA copies were expressed relative to control levels, calculated as 2Δ−(CTexp − CTcontrol). Primers are listed in Table 1.
COGs classification.
Orthologous genes were classified by Clusters of Orthologous Groups (COGs) functional category (http://www.ncbi.nlm.nih.gov/COG/grace/fiew.cgi) according to their assignment in the COGs database. If a protein had more than one COGs category, all were used. Later we calculated the frequencies of the different functional categories in the healthy and pathogenic biofilms. To assess enrichment of certain functional categories, we calculated the ratio of frequencies for healthy versus pathogenic biofilm and vice versa.
Gene set enrichment analysis.
A parametric analysis of gene set enrichment (PAGE) (33) was performed to detect groups of genes and pathways that were differentially expressed during biofilm formation. PAGE analysis was performed based on sets of genes selected for the present work based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology classification of proteins (http://www.genome.jp/kegg/). Before performing PAGE analysis, we assessed the normal distribution of the results using Q-Q normal plots in R. KEGG numbers were obtained either from genome information at NCBI or by using the KEGG automatic annotation server (KAAS) (http://www.genome.jp/tools/kaas/).
Prediction of possible interactions between IGRs and genomic DNA.
To predict potential targets for the differentially expressed IGRs, we used the stand-alone version of IntaRNA (9), which predicts interactions between two bacterial RNA molecules. Overexpressed IGRs were used as input, and the respective genomes of the bacteria used in the experiments were the target sequences. The running conditions were the default except that window size was adjusted to the minimum sequence size in the respective genomes and only output results with an energy below −15 kcal/mol were considered.
RESULTS
Biofilm species composition.
We first verified the presence of all of the bacteria in the biofilm and examined their relative amounts on the hydroxyapatite (HA) disks used as a biofilm support in our experiments. qPCR analysis of each species in the biofilm was used to calculate their abundance (see Table S1 in the supplemental material). In the case of the pathogenic biofilm, after 24 h of incubation of a biofilm consisting of the species associated with health, 2 pathogens were added (P. gingivalis and A. actinomycetemcomitans) and the biofilm was incubated for an additional period of 6 h under anaerobic conditions. Table S1 summarizes the doubling times of the different strains growing in the biofilm and the supernatant. In general, estimates of generation times for cells on the biofilm were shorter than for the planktonic phase (see Table S1). The doubling times in the supernatant ranged from 1.56 to 3.03, while in the biofilm they were less variable, ranging from 0.92 to 1.38. Except for the planktonic phase of Veillonella parvula and Streptococcus mitis, there were no major differences in growth rate after addition of the pathogens. Therefore, changes in the expression patterns in the biofilm should not be attributed to changes in growth rate when the pathogens were present.
Effect of periodontopathogens in the metatranscriptome of a healthy biofilm.
Oral biofilms (dental plaque) play a major role in the development of periodontal disease, hence the interest in characterizing specific patterns of gene expression associated with the establishment of dental plaque. In this section we compare the expression profiles of the biofilms (healthy and pathogenic) versus their corresponding planktonic phases.
Given the novelty of using next-generation sequencing techniques for this kind of analysis, the first step was to confirm that 80 bp (average size of the sequences obtained by Illumina sequencing in the experiments) was enough to distinguish between the transcripts from the different species used in our biofilms. BLAST analysis of 80-bp fragments created from the genomes of the organisms used in the biofilms showed that the BLAST results against our database matched the expected origin of the organisms in 100% of cases.
Most sequences identified in the actual results from our experiment were rRNA, and a large fraction of genes was being expressed at any given time (Table 2). Independently of the total number of sequences, we observed that between 38% and 66% of all ORFs in the metagenome were being expressed at any given time. The number of IGRs expressed at any given time was lower, ranging from 9% to 25% of the total number of IGRs in the metagenome. However, the lower value (9%) could be due to the fact that the number of total sequences in this fraction was also low (203,215 hits) and we may not have reached saturation (Table 2).
Table 2.
Number of different ORFs and IGRs identified in the experimentsa
| Characteristic | No. of rRNA hits, biofilms | Total no. of hits, biofilm ORFs | Total no. of hits, supernatant ORFs | No. of unique ORFs identified, biofilm | No. of unique ORFs identified | Total no. of unique ORFs in the community | No. of rRNA hits, supernatant | Total no. of hits, biofilm IGRs | Total no. of hits, supernatant IGRs | No. of unique IGRs identified, biofilm | No. of unique IGRs identified, supernatant | Total no. of unique IGRs in the community |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Healthy | 5,157,293 | 1,331,097 | 593,118 | 7,982 (66%) | 7,520 (62%) | 12,054 | 11,649,872 | 765,970 | 452,726 | 2,146 (24%) | 2,236 (25%) | 8,919 |
| Pathogenic | 14,025,715 | 578,919 | 744,494 | 6,923 (38%) | 9,714 (53%) | 18,146 | 21,036,949 | 203,215 | 826,255 | 1,122 (9%) | 2,247 (18%) | 12,318 |
rRNA sequences were discarded from the final analysis. We present the results for protein-coding ORFs and small noncoding RNA IGRs. In parentheses, the fraction of ORFs or IGRs that were identified when compared with the total number in the metagenome.
In our analysis, we took a conservative approach and we only considered ORFs or IGRs whose matches appeared in both fractions (biofilm and planktonic phase) and were up- or downregulated more than an 8-fold difference. We confirmed the differences in expression in the libraries by RT-qPCR. We selected 4 genes that were highly expressed in our libraries and performed RT-qPCR from 3 individual disks that were treated as in the original experiment, but this time, we did not mix them to extract the RNA. Figure S1 in the supplemental material shows the results of the RT-qPCR, and the 4 genes showed upregulation in the biofilm fraction. Moreover, the standard deviation of the 3 biological replicates was very low.
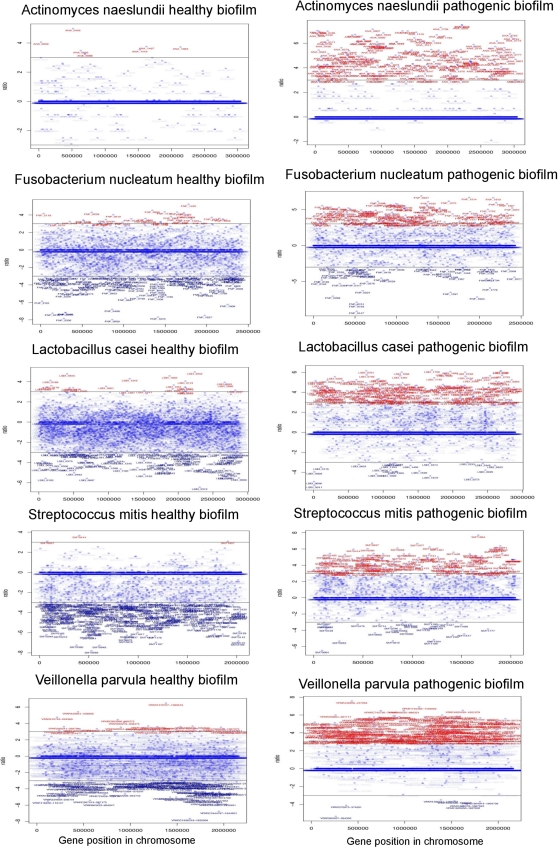
We added the two periodontal pathogens P. gingivalis and A. actinomycetemcomitans to create a pathogenic biofilm and then compared biofilm levels of expression against the corresponding planktonic phase levels with and without pathogens. Our first observation was a change in the metabolism of the healthy components of the biofilm compared to the planktonic phase (Fig. 1). The list of upregulated genes was much larger than in the case of the healthy biofilm by itself (see Tables S2 and S3 in the supplemental material). Likewise, a similar change was observed in the profiles of expression of the IGRs (see Fig. S2 in the supplemental material). The whole community seems to undergo a change in both regulation and metabolism when new organisms are incorporated to the biofilm.
Fig 1.
Expression profile of the ORFs comparing biofilm and free-living bacteria gene expression in healthy and pathogenic biofilms. A line at −3 and +3 shows the cutoff selected for the experiments (8-fold difference). In red are the genes whose ratios of gene expression were higher than 3 in the biofilm versus planktonic phase. In dark blue (bottom of the graph) are the genes whose ratios of gene expression were lower than −3 in the biofilm versus planktonic phase (were upregulated in the planktonic phase).
There were genes up- and downregulated in the biofilm fraction independent of whether it was a healthy or a pathogenic biofilm. We observed certain commonalities in the levels of expression of these genes independent of whether the pathogens were present or not (Table 3), which in the case of upregulated genes may represent a subset of key proteins to the maintenance of the biofilm. Among those there were genes related to multidrug resistance efflux pump (FNP_0510 and FNP_0890) and lipid biosynthesis (LSEI_2119 and LSEI_2111). We also identified transcription factors that are involved in controlling diverse physiological processes (ANA_0022 and HMPREF1035_1004).
Table 3.
Common changes in ORFs up- and downregulated in both biofilms: healthy and pathogenica
| Organism and ORF tag | Healthy Δlog 2 expression | Pathogenic Δlog 2 expression | Function |
|---|---|---|---|
| Upregulated ORFs | |||
| Actinomyces naeslundii | |||
| ANA_0022 | 4.06 | 4.17 | Translation elongation factor Tu (tuf) |
| ANA_1497 | 3.75 | 6.19 | DNA-directed RNA polymerase, beta subunit (rpoB) |
| ANA_1963 | 3.72 | 4.27 | 65-K antigen mbaA (GROEL2) |
| ANA_1419 | 3.54 | 6.89 | 2-Oxoglutarate dehydrogenase E1 component (sucA) |
| Fusobacterium nucleatum | |||
| FNP_0239 | 4.30 | 3.19 | Possible transposase |
| FNP_0510 | 4.03 | 3.68 | Possible AcrR family transcriptional regulator |
| FNP_0890 | 3.60 | 3.38 | MOP/MATE family multidrug resistance efflux pump NorM |
| Fusobacterium nucleatumplasmid | |||
| FNP_PFN3G07 | 3.66 | 5.64 | Relaxase |
| Lactobacillus casei | |||
| LSEI_0981 | 3.54 | 5.08 | Pseudogene |
| LSEI_2119 | 3.40 | 3.54 | 3-Oxoacyl-(acyl-carrier-protein) synthase III |
| LSEI_2111 | 3.06 | 3.40 | Biotin carboxylase |
| Streptococcus mitis | |||
| SM12261_0414 | 3.66 | 5.95 | Oxidoreductase, short chain dehydrogenase/reductase family |
| Veillonella parvula | |||
| HMPREF1035_1004 | 6.49 | 5.54 | LysR substrate binding domain protein |
| HMPREF1035_0421 | 4.47 | 3.85 | Putative cobaltochelatase, CobN subunit |
| HMPREF1035_0817 | 3.47 | 3.56 | Conserved hypothetical protein |
| HMPREF1035_1551 | 3.30 | 4.85 | VanW-like protein |
| HMPREF1035_0824 | 3.08 | 3.01 | Conserved domain protein |
| HMPREF1035_1988 | 3.08 | 3.27 | Monogalactosyldiacylglycerol synthase, C-terminal domain protein |
| Downregulated ORFs | |||
| Fusobacterium nucleatum | |||
| FNP_1258 | −3.45 | −3.36 | Heat shock protein DnaK (Hsp70) |
| FNP_1297 | −3.41 | −6.50 | 2-Nitropropane dioxygenase |
| FNP_1442 | −3.04 | −3.07 | Rod shape-determining protein |
| FNP_2009 | −3.28 | −3.32 | GTP-binding protein |
| FNP_2257 | −3.81 | −3.96 | O-antigen GlcNac transferase |
| FNP_2288 | −3.84 | −7.03 | Peroxiredoxin |
| FNP_2427 | −4.92 | −3.90 | Phenylalanine-tRNA ligase |
| Lactobacillus casei | |||
| LSEI_1093 | −3.20 | −4.17 | UDP-glucose pyrophosphorylase; K00963 UTP-glucose-1-phosphate uridylyltransferase (EC 2.7.7.9) |
| LSEI_1114 | −3.70 | −3.12 | Cold shock protein; K03704 cold shock protein (beta-ribbon, CspA family) |
| LSEI_1193 | −4.64 | −3.06 | Hypothetical protein |
| LSEI_1520 | −3.28 | −3.87 | GatB/YqeY domain-containing protein; K09117 hypothetical protein |
| LSEI_1674 | −4.33 | −3.08 | HD superfamily hydrolase; K06950 |
| Streptococcus mitis | |||
| SM12261_0064 | −3.65 | −6.50 | LysM domain protein |
| SM12261_0263 | −3.18 | −5.34 | DAK2 domain protein |
| SM12261_0668 | −3.58 | −4.33 | Ribosomal protein L31 |
| SM12261_0714 | −3.54 | −3.27 | Membrane protein, putative |
| SM12261_0796 | −5.42 | −3.56 | ABC transporter, ATP-binding protein |
| SM12261_0815 | −3.20 | −5.31 | Triosephosphate isomerase |
| SM12261_0883 | −3.38 | −3.84 | DNA-directed RNA polymerase, omega subunit, putative |
| SM12261_1004 | −4.41 | −3.40 | Chaperonin, 60 kDa |
| SM12261_1191 | −5.29 | −3.58 | Pneumococcal surface protein A |
| SM12261_1192 | −5.05 | −3.04 | Pneumococcal surface protein A |
| SM12261_1195 | −4.34 | −5.42 | Ribosomal protein S9 |
| SM12261_1249 | −3.06 | −3.55 | Ribosomal protein S2 |
| SM12261_1274 | −3.62 | −3.32 | Putative TIM-barrel protein, nifR3 family subfamily |
| SM12261_1397 | −4.77 | −4.60 | PspC domain family |
| SM12261_1517 | −3.51 | −4.45 | Phosphomevalonate kinase |
These are genes that were up- or downregulated more than 3× log 2 in both experiments. The numeric values show the log 2 level of up- or downregulation in both cases.
In the case of downregulated genes, we identified several chaperones (FNP_1258, LSEI_1114, and SMT1004) as well as proteins involved in cell wall metabolism (FNP_1142, FNP_2257, and SMT0064) or membrane-associated proteins (SMT0714, SMT1191, and SMT1192).
As mentioned above, the addition of periodontal pathogens to the biofilm increased the number of upregulated genes. When we compared biofilm versus planktonic phase in the healthy community, the total of upregulated ORFs was 97, but when we did the same comparison after adding the pathogens the number of upregulated ORFs was 962. While in the case of downregulated ORFs, the opposite is true: 675 ORFs where downregulated in the healthy biofilm and only 89 were downregulated in the presence of the pathogens.
In the case of genes that were specifically upregulated in healthy biofilm but not in the pathogenic biofilm (see Table S2 in the supplemental material), most ORFs were hypothetical proteins but also ORFs predicted to encode transporters (FNP_0336, FNP_0337, FNP_0622, FNP_1222, FNP_1291, and LSEI_0065) and transcriptional regulators (FNP_0988, FNP_1409, LSEI_2120, and LSEI_2242). More telling was the pattern of downregulated genes (see Table S2). There was a large number of downregulated chaperones in all organisms present in the community (FNP_1256, FNP_1259, LSEI_0687, LSEI_2800, LSEI_0636, SMT0648, SMT1009, SMT0281, SMT0282, SMT1007, SMT1005, HMPREF1035_1494, HMPREF1035_1496, and HMPREF1035_0031) as well as putative cell division proteins such as FtsZ or FtsA (FNP_1635, FNP_0784, SMT2115, SMT2113, SMT2129, SMT2124, SMT2104, SMT2112, SMT2114, SMT1522, SMT2135, SMT1523, and HMPREF1035_1084). Additionally, phosphotransferase system (PTS) proteins in both Lactobacillus casei and Streptococcus mitis were highly downregulated (LSEI_2700, LSEI_0370, LSEI_2073, SMT1148, SMT0181, SMT1149, SMT1765, and SMT0182).
In the pathogenic biofilm, we were able to identify large groups of proteins with related functions given the larger number of genes upregulated (see Table S3 in the supplemental material). Looking only at genes that were exclusively upregulated in the pathogenic biofilm, the most upregulated ORFs for A. naeslundii (128-fold overexpression or more) were all putative molecular chaperones such as DnaK, ClpB, ClpC, or LAC ORF (ANA_2043, ANA_2044, ANA_1739, and ANA_2020). In the case of F. nucleatum, most upregulated ORFs were hypothetical proteins, 79 out of 150 proteins had no known function.
Lactobacillus casei presented a large number of ABC-type transport systems upregulated, most of them related to peptide transport (LSEI_1344, LSEI_2063, LSEI_1892, LSEI_0175, LSEI_1207, LSEI_2446, LSEI_2447, LSEI_1261, LSEI_1345, LSEI_2628, LSEI_0308, LSEI_0991, LSEI_0999, LSEI_0704, LSEI_0022, LSEI_1814, and LSEI_2466), and also a large number of putative transposases (LSEI_0237, LSEI_0361, LSEI_1068, LSEI_1333, LSEI_1509, LSEI_1842, LSEI_2461, and LSEI_0347), even one from L. casei plasmid (LSEI_A09).
We found a similar pattern in the specific upregulated genes of S. mitis in the pathogenic biofilm: ABC transporters (SMT1856, SMT1393, SMT1052, SMT0394, SMT0604, SMT1308, SMT1460, SMT1392, SMT0080, and SMT1570) and phosphate (SMT1432 and SMT1431) and sugar (SMT1424, SMT1871, SMT1746, and SMT1844) ABC transporters were highly upregulated, as well as transposases (SMT0439, SMT0520, SMT0689, SMT1941, SMT0994, SMT0820, SMT0821, and SMT1123). Mur ligase family proteins (SMT2108, SMT2116, SMT2125, and SMT2130), which are involved in peptidoglycan biosynthesis, were also upregulated.
Finally, in V. parvula, the presence of pathogens upregulated a large number of hypothetical proteins but also proteins that have been associated with virulence in other organisms such as hemagglutinins (HMPREF1035_0018 and HMPREF1035_0017), Hep/Hag repeat protein (HMPREF1035_0731, HMPREF1035_0445, HMPREF1035_0440, HMPREF1035_0447, HMPREF1035_0441, HMPREF1035_0442, and HMPREF1035_0034) and transcriptional regulators of the MarR family (HMPREF1035_0560).
In the case of downregulated proteins, after adding the pathogens, we did not see any clear pattern in the function of the ORFs. Only in the case of F. nucleatum, most of the downregulated ORFs were annotated as hypothetical proteins (FNP_0147, FNP_0149, FNP_0151, FNP_1776, FNP_2364, FNP_0529, and FNP_1367).
Differences in gene expression of the periodontal pathogens under planktonic and biofilm conditions.
Differences of gene expression in the periodontopathogens themselves comparing the planktonic fraction against the fraction attached to the biofilm were also analyzed (see Fig. S3 and Table S4 in the supplemental material). One striking observation was that A. actinomycetemcomitans had no downregulated genes that we could observe and a large number of upregulated genes in the biofilm phase: 355 ORFs were upregulated more than 8-fold in the biofilm phase compared to the planktonic phase (see Table S3). A large number were ribosomal proteins (AA01067, AA00362, AA00565, AA01228, AA01236, AA00361, AA01368, AA02039, AA02880, AA01229, AA02099, and AA00369). Two of the more upregulated ORFs have putative functions related to pathogenesis or cytotoxicity in other organisms. AA00777, a soluble lytic murein transglycosylase that was 256-fold upregulated, has homology with a soluble lytic murein transglycosylase in Neisseria that is involved in cytotoxin production (14), while AA01976, a colicin V production protein that was 168-fold upregulated, has homology with proteins involved in the production of virulence factors in Escherichia coli (7). As in the case of A. naeslundii, in the presence of the pathogens, we found an overexpression of molecular chaperones such as Hsp60, Hsp70, and DnaJ in the biofilm (AA01284, AA00657, and AA01057).
Contrary to the large number of overexpressed genes in A. actinomycetemcomitans, in the case of P. gingivalis, we observed overexpression of only 26 ORFs in the biofilm fraction, most of them being hypothetical proteins (see Table S3 in the supplemental material). A large number of proteins were downregulated (193 ORFs), but as in the case of the upregulated ORFs, most of them were annotated as hypothetical proteins.
COGs functional categories and gene set enrichment analysis.
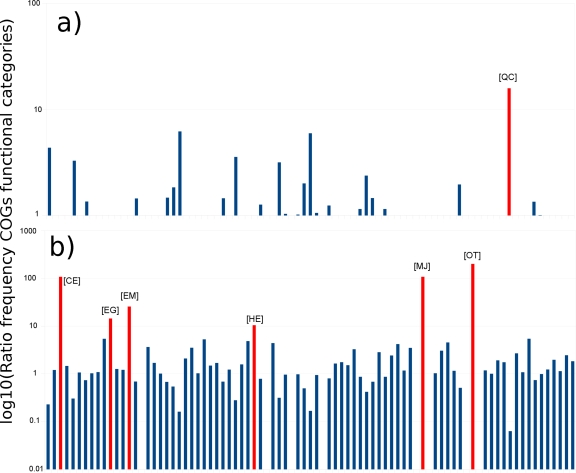
In another set of analysis, we proceed to determine sets of genes or metabolic pathways that were upregulated in the biofilm compared to the planktonic phase. In this case, we do not focus on individual genes from individual organisms but on whole metabolic pathways or activities from the whole microbial communities to obtain an overall picture of the metabolic changes in the biofilms. Similarly to what we observed with ORFs and IGRs, COGs categories were overexpressed in the presence of the periodontopathogens (Fig. 2). Only one group of functional categories was downregulated in the presence of the pathogens: [QC] for secondary-metabolite biosynthesis, transport and catabolism, and energy production and conversion; as though in the presence of pathogens all the members of the community stopped the production of secondary metabolites and focus their effort in other tasks. In contrast, upregulated functional categories in the presence of pathogens seem to be involved primarily in growth and division (Fig. 2).
Fig 2.
Summary of COGs functional categories in the biofilms. The plot shows the log 10 values for the ratio of frequencies of the different functional categories based on the COGs classification. Only the categories with a higher ratio than 10 are labeled. (a) Ratio of the frequencies of biofilm/biofilm + pathogens, shows the functional categories overrepresented in the biofilm without the pathogens. (b) Ratio of the frequencies of biofilm + pathogens/biofilm, shows the functional categories overrepresented in the biofilm with the pathogens. In red, we show the categories with a ratio higher than 10. [QC], secondary metabolites biosynthesis, transport and catabolism, and energy production and conversion; [CE], energy production and conversion and amino acid transport and metabolism; [EG], amino acid transport and metabolism and carbohydrate transport and metabolism; [EM], amino acid transport and metabolism and cell wall/membrane/envelope biogenesis; [HE], coenzyme transport and metabolism and amino acid transport and metabolism; [MJ], cell wall/membrane/envelope biogenesis and translation, ribosomal structure and biogenesis; [OT], posttranslational modification, protein turnover, chaperones and signal transduction mechanisms.
On a second set of analysis, we selected sets of genes based on the KEGG orthology and used PAGE (33) to identify sets of genes that were enriched or depleted, with statistical significance, in the biofilms in relation to free-living bacteria and between healthy and pathogenic biofilms.
Table 4 presents a summary of the results of PAGE analysis on healthy and pathogenic biofilm gene expression profiles. As in the case of individual genes, the expression profiles of healthy and pathogenic biofilms were very different. The number of significant gene sets was larger for downregulated than for upregulated genes. Some of the gene sets had only a few genes (e.g., linoleic acid metabolism, bisphenol A degradation, etc.), while certain gene sets were represented by a large number of genes (e.g., ABC transporters, nitrogen metabolism, fructose and mannose metabolism, etc.). The gene sets defined as photosynthesis or photosynthesis proteins also comprised genes involved in the respiratory chain and vitamin metabolism. In fact, the upregulated genes were the different subunits (a, b, c) of the ATP synthase complex and the cobaltochelatase gene (cobN), which is involved in the biosynthesis of coenzyme B12.
Table 4.
PAGE comparing biofilm versus planktonic phasea
| Biofilm | Gene set | Z score | Fold change | No. of genes | P value |
|---|---|---|---|---|---|
| Upregulated | |||||
| Healthy | ko00195 photosynthesis | 1.918 | 0.994 | 8 | 0.055 |
| ko00194 photosynthesis proteins | 1.918 | 0.994 | 8 | 0.055 | |
| ko02010 ABC transporters | 1.761 | 0.269 | 92 | 0.078 | |
| ko00630 glyoxylate and dicarboxylate metabolism | 1.684 | 0.582 | 18 | 0.092 | |
| ko01001 protein kinases | 1.662 | 0.559 | 19 | 0.096 | |
| Pathogenic | ko02020 two-component system | 3.328 | 0.619 | 77 | 0.001 |
| ko00940 phenyl propanoid biosynthesis | 2.251 | 2.121 | 3 | 0.024 | |
| ko02022 two-component system | 2.124 | 0.517 | 45 | 0.034 | |
| ko03000 transcription factors | 2.040 | 0.375 | 79 | 0.041 | |
| ko00910 nitrogen metabolism | 2.038 | 0.513 | 42 | 0.042 | |
| ko00591 linoleic acid metabolism | 1.991 | 2.297 | 2 | 0.047 | |
| ko00363 bisphenol A degradation | 1.991 | 2.297 | 2 | 0.047 | |
| ko00633 trinitrotoluene degradation | 1.920 | 1.567 | 4 | 0.055 | |
| ko02060 PTS | 1.890 | 0.465 | 44 | 0.059 | |
| Downregulated | |||||
| Healthy | ko00520 amino sugar and nucleotide sugar metabolism | −3.663 | −0.849 | 40 | 0 |
| ko00010 glycolysis/gluconeogenesis | −4.072 | −1.039 | 33 | 0 | |
| ko03440 homologous recombination | −3.178 | −1.042 | 20 | 0.001 | |
| ko00980 metabolism of xenobiotics by cytochrome P450 | −3.287 | −3.407 | 2 | 0.001 | |
| ko02060 PTS | −3.479 | −0.787 | 42 | 0.001 | |
| ko00982 drug metabolism-cytochrome P450 | −3.139 | −4.601 | 1 | 0.002 | |
| ko00604 glycosphingolipid biosynthesis-ganglio series | −2.933 | −4.299 | 1 | 0.003 | |
| ko00531 glycosaminoglycan degradation | −2.933 | −4.299 | 1 | 0.003 | |
| ko00641 3-chloroacrylic acid degradation | −2.994 | −2.194 | 4 | 0.003 | |
| ko00500 starch and sucrose metabolism | −2.812 | −0.765 | 29 | 0.005 | |
| ko00511 other glycan degradation | −2.84 | −1.574 | 7 | 0.005 | |
| ko00071 fatty acid metabolism | −2.759 | −1.43 | 8 | 0.006 | |
| ko03030 DNA replication | −2.704 | −0.991 | 16 | 0.007 | |
| ko00624 1- and 2-methyl naphthalene degradation | −2.594 | −1.437 | 7 | 0.009 | |
| ko00061 fatty acid biosynthesis | −2.531 | −1.071 | 12 | 0.011 | |
| ko00550 peptidoglycan biosynthesis | −2.439 | −0.688 | 27 | 0.015 | |
| ko00740 riboflavin metabolism | −2.347 | −1.216 | 8 | 0.019 | |
| Biofilm | ko03110 chaperones and folding catalysts | −2.311 | −0.619 | 30 | 0.021 |
| ko00281 geraniol degradation | −2.315 | −1.959 | 3 | 0.021 | |
| ko00650 butanoate metabolism | −2.222 | −0.627 | 27 | 0.026 | |
| ko03018 RNA degradation | −2.187 | −0.857 | 14 | 0.029 | |
| ko00903 limonene and pinene degradation | −2.155 | −1.58 | 4 | 0.031 | |
| ko00930 caprolactam degradation | −2.022 | −2.964 | 1 | 0.043 | |
| ko00830 retinol metabolism | −1.964 | −2.036 | 2 | 0.049 | |
| ko00240 pyrimidine metabolism | −1.931 | −0.396 | 51 | 0.053 | |
| ko00072 synthesis and degradation of ketone bodies | −1.814 | −1.329 | 4 | 0.07 | |
| ko03032 DNA replication proteins | −1.797 | −0.466 | 32 | 0.072 | |
| ko00051 fructose and mannose metabolism | −1.786 | −0.449 | 34 | 0.074 | |
| ko02040 flagellar assembly | −1.769 | −2.593 | 1 | 0.077 | |
| ko00970 aminoacyl-tRNA biosynthesis | −1.7 | −0.455 | 30 | 0.089 | |
| ko00900 terpenoid backbone biosynthesis | −1.689 | −0.554 | 20 | 0.091 | |
| Pathogenic | ko00240 pyrimidine metabolism | −4.135 | −0.91 | 55 | 0 |
| ko03012 translation factors | −4.708 | −2.131 | 13 | 0 | |
| ko03011 ribosome | −6.072 | −1.349 | 54 | 0 | |
| ko03010 ribosome | −6.072 | −1.349 | 54 | 0 | |
| ko00230 purine metabolism | −3.23 | −0.613 | 74 | 0.001 | |
| ko03020 RNA-polymerase | −3.014 | −1.555 | 10 | 0.003 | |
| ko00710 carbon fixation in photosynthetic organisms | −2.85 | −0.992 | 22 | 0.004 | |
| ko00523 polyketide sugar unit biosynthesis | −2.657 | −2.168 | 4 | 0.008 | |
| ko00970 aminoacyl-tRNA biosynthesis | −2.548 | −0.747 | 31 | 0.011 | |
| ko03430 mismatch repair | −2.558 | −0.89 | 22 | 0.011 | |
| ko03400 DNA repair and recombination proteins | −2.297 | −0.402 | 87 | 0.022 | |
| ko00520 amino sugar and nucleotide sugar metabolism | −2.299 | −0.559 | 45 | 0.022 | |
| ko01002 peptidases | −1.926 | −0.39 | 65 | 0.054 | |
| ko03018 RNA degradation | −1.879 | −0.767 | 16 | 0.06 | |
| ko00983 drug metabolism, other enzymes | −1.829 | −0.944 | 10 | 0.067 | |
| ko00521 streptomycin biosynthesis | −1.765 | −1.018 | 8 | 0.078 | |
| ko00740 riboflavin metabolism | −1.679 | −0.913 | 9 | 0.093 | |
| ko00790 folate biosynthesis | −1.661 | −0.752 | 13 | 0.097 |
Gene sets are based on the KEGG orthology. Results show gene sets that were significantly upregulated in the biofilm fraction in relation to the planktonic fraction. “ko” numbers shown in the table correspond to KEGG metabolic pathways.
There was an agreement between patterns of expression based on PAGE results and just looking at groups of genes being up- or downregulated. For instance, as mentioned above, we found a large number of downregulated genes in the healthy biofilm that were identified as chaperones and we showed by PAGE analysis that the gene set ko03110 chaperones and folding catalysts, with 30 genes, was significantly downregulated (P value = 0.02) (Table 4). A large number of genes identified as members of PTS were downregulated in L. casei and S. mitis, and the gene set ko02060 PTS was also identified as significantly downregulated in the healthy biofilm (Table 4). In contrast with these results, the same ko02060 PTS gene set was significantly upregulated in the presence of the pathogens.
Prediction of potential interactions between IGRs and the ORFs in the bacterial genomes.
After identifying upregulated IGRs in the biofilms, we predicted the possible interactions between those and the ORFs in the genomes of the bacteria used in the experiments. To this end, we used IntaRNA, a program that predicts accurately interactions between two RNA molecules. The interactions are predicted by an energy-based approach that is based on the unfolding and hybridization energies and that a seed region is required to initiate the RNA-RNA interaction (9).
First, IGRs upregulated more than 8-fold in the biofilm versus planktonic phase were selected to be used as potential small noncoding RNAs against their corresponding genomes as predicted targets. Only interactions with energy values below −15 kcal/mol were considered in the analysis. In the case of upregulated IGRs that were present in both biofilms (healthy and pathogenic), 3 mRNA-predicted targets were identified as upregulated in both biofilms: ANA_1497, a DNA-directed RNA polymerase beta subunit (rpoB); ANA_1963, a 65-K antigen mbaA (GROEL2); and ANA_1419, alpha-2-oxoglutarate dehydrogenase (sucA). Likewise, we identified 3 IGRs whose predicted targets were downregulated in both biofilms: SMT0815, a triosephosphate isomerase; SMT1004, a chaperonin of 60 kDa; and SMT1249, ribosomal protein S2 (Table 3).
Table S5 in the supplemental material summarizes the predicted target ORFs that were also identified as differentially expressed in the ORF libraries. There were more potential predicted target genes identified, but we focused only on the ones that assuredly presented differences in expression in the biofilm community. A large number of predicted targets agreed with the ORFs identified as downregulated in S. mitis, with 35 predicted targets that are most likely regulated by the small noncoding RNAs upregulated in our libraries. The pathogenic biofilm had less predicted targets that appeared also in the differentially expressed ORFs, only 4 for the upregulated genes and 3 for the downregulated (see Table S5). In the case of V. parvula, we found predicted IGRs that targeted HMPREF1035_0440 and HMPREF1035_0441, two Hep/Hag repeat proteins that in other organisms have been linked to virulence (72). These proteins were upregulated; therefore, the small noncoding RNAs were stabilizing the transcripts. The Hep/Hag domain is a seven-residue repeat that makes up the majority of the sequence of a family of bacterial hemagglutinins and invasins (42).
DISCUSSION
Multispecies biofilm models of dental plaque have been widely used for studying the dynamics of attachment and colonization of bacteria on different surfaces (23, 70, 74). Expression profiles of oral monospecies biofilms have also been widely studied to assess profiles of expression of oral pathogens under different environmental conditions (66, 67, 77, 78). However, studies on the profiles of expression of oral organisms in multispecies biofilm models are still limited (40). The use of metatranscriptomic analysis of biofilm models opens a complete new set of possibilities for studying gene expression profiles of whole microbial communities, facilitating the discovery of genes or groups of genes of importance under different environmental conditions.
In this study, we identified transcripts of ORFs and IGRs that are associated with the biofilm community in a healthy biofilm model as well as on the effect that two important periodontal pathogens (A. actinomycetemcomitans and P. gingivalis) (24) have on a healthy biofilm. Metatranscriptome results can be evaluated at different levels of complexity, starting by comparing gene expression at the level of individual genes and going up to comparisons of gene sets (groups of genes with similar functions or cell localizations) that belong to defined gene orthologues (e.g., COGs or KEGG orthology).
We found a higher growth rate of the organisms in the biofilm than in the planktonic phase. The planktonic phase reached an extremely high number of cells, whose only explanation may be the high initial number of cells in the inoculum or the fact that multispecies communities of oral bacteria grow better than isolated species (60). Nonetheless, doubling times in the planktonic phase were in agreement with previous estimation of doubling times for oral bacteria (5, 15). Calculations for doubling time on the biofilm considered that, at time zero, there were 0 bacteria attached to the hydroxyapatite surface, which could overestimate the rate of growth but will give a good assessment of whether the addition of pathogens had any effect on the overall growth of the biofilm. Moreover, initial attachment cannot be distinguished from growth in the initial stages of biofilm formation, hence the short doubling time of the pathogens in the biofilm. Furthermore, we observed the presence of a large planktonic population that may influence biofilm gene expression profiles by depleting nutrients, limiting exposure of the biofilm to certain environmental stresses and altering the concentration of quorum-sensing molecules to which the biofilms are exposed. With these limitations in mind, we proceed to analyze expression profiles in biofilm and planktonic phases from our experiments.
The existence of differences in gene expression between the free-living and biofilm components of a monospecies biofilm of oral bacteria is well established (47, 68, 69). When looking at the list of genes overexpressed in the healthy biofilm in relation to the planktonic phase, certain ORFs that have been previously associated with biofilm formation were identified. In a proteomic study of A. naeslundii surface proteins, Paddick et al. showed that groEL2 was significantly upregulated in biofilm-grown cells (57). F. nucleatum showed a large number of upregulated genes in the biofilm. Two of them were putative transposases, which have been shown as being expressed in vivo by F. nucleatum (43). Similarly, homologues of the most highly expressed gene in V. parvula, a LysR substrate binding domain protein, has been associated in biofilm formation in other bacteria. Mutants of this gene in Burkholderia cenocepacia exhibit absence of extracellular matrix and reduced biofilm formation (56). L. casei showed a high number of hypothetical proteins as being overexpressed. Two of the highly expressed proteins in L. casei are associated with biotin metabolism, probably linked to lipid production, and interestingly, we observed two ORFs that have been annotated as pseudogenes as being highly upregulated in the biofilm. These genes were also upregulated in the presence of the pathogens added to the biofilm, which indicates their importance in biofilm formation under most environmental conditions.
Regardless of the few similarities observed in the expression of specific genes in the individual bacteria, compared with previous studies, the general rule is that most of the upregulated genes in the multispecies biofilm seem not to be upregulated in monospecies biofilms of the same organisms (64, 65, 74, 75). This could be due to the different growth conditions but also could be the result of the interactions between the organisms in the community. More interesting were the results of the downregulated ORFs in the healthy biofilm. Looking at the specific genes downregulated in the healthy biofilm but not in the pathogenic biofilm, we identified a large number of chaperones from all organisms in the biofilm, which can indicate a low level of stress in the system.
The most surprising finding was observing that the presence of the periodontal pathogens P. gingivitis and A. actinomycetemcomitans in a multispecies biofilm model alters dramatically the patterns of gene expression of the healthy community biofilm, and this is not due to changes in growth rates. We have shown that generation times on the biofilms were similar for the different bacteria independent of the presence of periodontopathogens in the community. Wen et al. have obtained similar results growing S. mutans in a dual-species biofilm model and studying expression of selected genes by RT-qPCR (76). The presence of another organism in the biofilm altered biofilm formation and expression of virulence factors in S. mutans, and those changes were specific depending on the cocultivated organism used. For instance, luxS in S. mutans was downregulated in the presence of either Streptococcus oralis or L. casei, but nothing happened in the presence of Streptococcus sanguinis (76). Recently, Hajishengallis et al. have shown that P. gingivalis even at low colonization levels changes the amount and composition of the commensal community (25).
In the healthy biofilm, most upregulated ORFs were hypothetical proteins, transporters and transcriptional regulators, which have also been observed in microarray analyses of monospecies biofilms of Streptococcus mutans (68) and P. gingivalis (47). When the pathogens were added, a more complex picture arose. We identified again a large number of hypothetical proteins being upregulated, but there were specific features not seen in the healthy biofilm alone. Putative molecular chaperones were highly upregulated in A. naeslundii. In one recent study, groEL/groES chaperones were induced in an S. mutans biofilm in the presence of starch (35). Similarly, Acinetobacter baumannii needs the chaperone CsuA to form biofilms on plastic (51). Moreover, contrary to what was observed in the healthy biofilm, there was no downregulation of any group of chaperones. This switch from a metatranscriptome where chaperones are downregulated to one where they are upregulated may be the result of stress due to the presence of the pathogenic newcomers to the system.
ABC transporters were upregulated in L. casei and S. mitis, which is a common observation in studies on oral monospecies biofilms (35, 68, 77, 78). More interesting was the fact that in Lactobacillus casei and Streptococcus mitis there were a large number of putative transposases upregulated in the pathogenic biofilm. This phenomenon has also been observed in a biofilm of Treponema denticola, another important periodontal pathogen (53). In that study, the authors observed an upregulation of a family of putative transposases, concluding that there is a higher potential for genetic mobility in T. denticola when growing as a biofilm. Whether this genetic mobility in the multispecies biofilms is intraspecies or interspecies has yet to be proven. Additionally, there was an upregulation of genes encoding several putative virulence factors in V. parvula, which was also observed by Mitchell et al. in their work on T. denticola biofilm expression profiles (53).
When looking at the expression profiles of the periodontal pathogens, in P. gingivalis, most up- and downregulated ORFs encoded hypothetical proteins, which agrees with a similar microarray study on P. gingivalis W50, where the largest fraction of up- and downregulated genes during biofilm growth encoded hypothetical proteins (47). In the case of A. actinomycetemcomitans, we did not identify any ORFs as downregulated based on the cutoff values used in this work. The most upregulated ORFs were similar to genes encoding virulence factors of proteins with cytotoxic activity in other organisms: a soluble lytic murein transglycosylase (14) and a colicin V production protein (7). A. actinomycetemcomitans also showed upregulation of putative molecular chaperones in the biofilm.
The whole community can be viewed as a “metaorganism,” and in this case, we should consider its metatranscriptome as a unity, looking at various functional categories that are differentially expressed under different environmental conditions. The analysis of COGs showed an increase in the number of different COGs upregulated in the biofilm when pathogens were added. This finding is consistent with the observation of higher expression of different genes when pathogens are present.
Expression of genes involved in secondary-metabolite production, such as polyketide antibiotics or bacteriocins (see Table S3 in the supplemental material), is suppressed when the pathogens are added, and it has been shown that interference of secondary metabolites suppress biofilm formation in other systems (17). Although we do not know the mechanism by which this happens, the fact that this functional category is downregulated in the presence of pathogens could partially explain why there is a significant increase in biomass that could not be attributed to the addition of new nutrients.
The COGs functional categories upregulated in the presence of pathogens fundamentally agreed with the conclusions presented above. COGs associated with transport ([EM], [HE], [EG], [CE]) were upregulated, as well as functional categories associated with cell growth and division ([MJ]). Interestingly, one of the functional categories upregulated corresponds to chaperones ([OT]).
Gene set enrichment analysis has been widely used to analyze microarray data sets by focusing on gene sets, that is, groups of genes that share common biological functions, chromosomal locations, or regulations, rather than focusing on individual genes (32, 71, 72). We defined our gene sets on the KEGG pathways definitions and applied a modified gene set enrichment analysis method based on a parametric statistical analysis model (33). Using this approach, we identified sets of genes that were enriched or depleted in the healthy and pathogenic biofilms. The “two-component system” sets of the pathogenic biofilm (ko02020 and ko02022) presented the largest number of genes and shared features of the “protein kinases” in the healthy biofilm. Two-component signal transduction systems play an important role in biofilm formation in monospecies biofilms (44, 47).
One of the clearly defining gene sets of the pathogenic biofilm was the “nitrogen metabolism” set, with genes from A. naeslundii, F. nucleatum, L. casei, V. parvula, and A. actinomycetemcomitans. P. gingivalis is a well-known proteolytic organism that is capable of liberating amino acids from the proteins in the medium and makes them available for uptake by other species present in the biofilm. Gharbia et al. demonstrated that the capacity of hydrolyzing by Fusobacterium spp. increases by 30% in the presence of P. gingivalis (20).
Glutamine synthetase, an enzyme catalyzing formation of glutamine from glutamate and ammonium ion, is one of the most important enzymes in nitrogen metabolism. This enzyme has been shown to play a key role in biofilm formation in Mycobacterium bovis (13). Moreover, the loss of this enzyme in Bacillus subtilis did not decrease growth or cellular motility but had dramatic effects on biofilm formation (12). Finally, the gene set PTS, which is related to the use of different sugars, was also upregulated during biofilm formation in the pathogenic biofilm. In Vibrio cholerae, the phosphoenolpyruvate phosphotransferase system is essential for biofilm formation (27, 28). As the PTS is highly conserved among bacteria, it may be relevant to a number of biofilm-forming organisms. Accordingly, PTS was one of the significantly downregulated gene sets in the healthy biofilm according to the PAGE analysis. The glycolysis/gluconeogenesis gene set was also significantly downregulated in the healthy biofilm. Downregulation of glycolytic genes in biofilms has also been observed in S. mutans (62).
Finally, we attempted to link the expression profiles of IGRs with the changes in metabolic activity observed based on the ORF patterns of expression. Small noncoding RNAs transcribed in the IGRs of the prokaryotic genomes play an important role in metabolism regulation (11), and they can act as regulators of gene expression in all prokaryotes in which they have been studied. They are often defined as noncoding transcripts that act in trans to control the translation or stability of their target mRNA (11, 46). However, recent reports show that they could act in a significantly different way: in cis-acting chromosomally encoded antisense small noncoding RNAs or even as regulatory and translated RNAs (4, 46). Our working hypothesis is that we could shed light on the role of small noncoding RNA in the regulation ORFs identified as being differentially expressed in our experiments. Moreover, we could select for potential roles of small noncoding RNAs in a targeted manner based on the metatranscriptomic results. We identified IGRs whose targets were genes that were differentially expressed according to the ORF libraries. These results indicate the potential importance of those IGRs on the transcriptional regulation of the mRNA for those genes. Furthermore, in several cases, more than one single IGR had those mRNAs as targets, which suggests the importance of small noncoding RNAs in the regulation of such targets. When IGRs had targets that were downregulated they probably had to do with facilitating the degradation of those mRNAs, while in the case of upregulated proteins, the small noncoding RNAs are probably stabilizing the mRNAs. Even when we could not find a direct link between the IGRs and ORFs in our libraries, the targets agreed with the general results from the functional orthologues analyzed. For instance, we found that COGs related to secondary metabolism were overexpressed in healthy versus pathogenic biofilm. Targets found for the IGRs in the pathogenic biofilm contained a large number of genes related to the production of bacteriocins in L. casei (LSEI_2375, LSEI_2374, and LSEI_2381).
In the present work, we have shown that the addition of periodontal pathogens to a healthy biofilm multispecies model has a drastic effect in changing the gene expression profiles of the organisms present in the healthy community. Moreover, we have shown that using metatranscriptomic analysis, changes in gene expression profiles can be accurately evaluated, focusing either on changes at the gene level or treating the whole community as a “metacommunity” and analyzing the transcriptome as a whole and identifying potential important small noncoding RNAs involved in the regulation of important genes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the Colgate Palmolive Company.
We thank Tsute Chen at the Forsyth Institute for providing us with the Perl script to identify genomic IGRs and extracting the PTT files used. We also thank Mary-Ellen Davey for critical discussions and reading of the manuscript, which greatly improved its quality.
Footnotes
Published ahead of print 10 February 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Al-Ahmad A, et al. 2009. Bacterial colonization of enamel in situ investigated using fluorescence in situ hybridization. J. Med. Microbiol. 58: 1359– 1366 [DOI] [PubMed] [Google Scholar]
- 2. Albandar JM, Brunelle JA, Kingman A. 1999. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988–1994. J. Periodontol. 70: 13– 29 [DOI] [PubMed] [Google Scholar]
- 3. Andersson AF, et al. 2006. Global analysis of mRNA stability in the archaeon Sulfolobus. Genome Biol. 7: R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Argaman L, et al. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11: 941– 950 [DOI] [PubMed] [Google Scholar]
- 5. Beckers HJ, van der Hoeven JS. 1982. Growth rates of Actinomyces viscosus and Streptococcus mutans during early colonization of tooth surfaces in gnotobiotic rats. Infect. Immun. 35: 583– 587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bik EM, et al. 2010. Bacterial diversity in the oral cavity of ten healthy individuals. ISME J. 4: 962– 974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blanco JE, Blanco M, Mora A, Blanco J. 1997. Production of toxins (enterotoxins, verotoxins, and necrotoxins) and colicins by Escherichia coli strains isolated from septicemic and healthy chickens: relationship with in vivo pathogenicity. J. Clin. Microbiol. 35: 2953– 2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown LJ, Brunelle JA, Kingman A. 1996. Periodontal status in the United States, 1988–1991: prevalence, extent, and demographic variation. J. Dent. Res. 75: 672– 683 [DOI] [PubMed] [Google Scholar]
- 9. Busch A, Richter AS, Backofen R. 2008. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 24: 2849– 2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cai H, Rodríguez BT, Zhang W, Broadbent JR, Steele JL. 2007. Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Microbiology 153: 2655– 2665 [DOI] [PubMed] [Google Scholar]
- 11. Caron M-P, Lafontaine DA, MassÉ E. 2010. Small RNA-mediated regulation at the level of transcript stability. RNA Biol. 7: 140– 144 [DOI] [PubMed] [Google Scholar]
- 12. Chagneau C, Saier MHJ. 2004. Biofilm-defective mutants of Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 8: 177– 188 [DOI] [PubMed] [Google Scholar]
- 13. Chandra H, Basir SF, Gupta M, Banerjee N. 2010. Glutamine synthetase encoded by glnA-1 is necessary for cell wall resistance and pathogenicity of Mycobacterium bovis. Microbiology 156: 3669– 3677 [DOI] [PubMed] [Google Scholar]
- 14. Cloud KA, Dillard JP. 2002. A lytic transglycosylase of Neisseria gonorrhoeae is involved in peptidoglycan-derived cytotoxin production. Infect. Immun. 70: 2752– 2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Jong MH, van der Hoeven JS, JHvan OS, Olijve JH. 1984. Growth of oral Streptococcus species and Actinomyces viscosus in human saliva. Appl. Environ. Microbiol. 47: 901– 904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Diaz PI, et al. 2006. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol. 72: 2837– 2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dickschat JS. 2010. Quorum sensing and bacterial biofilms. Nat. Prod. Rep. 27: 343– 369 [DOI] [PubMed] [Google Scholar]
- 18. Foster JS, Kolenbrander PE. 2004. Development of a multispecies oral bacterial community in a saliva-conditioned flow cell. Appl. Environ. Microbiol. 70: 4340– 4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frias-Lopez J, et al. 2008. Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. U. S. A. 105: 3805– 3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gharbia SE, Shah HN, Welch SG. 1989. The influence of peptides on the uptake of amino acids in Fusobacterium; predicted interactions with Porphyromonas gingivalis. Curr. Microbiol. 19: 231– 235 [Google Scholar]
- 21. Grenier D, Mayrand D. 1986. Nutritional relationships between oral bacteria. Infect. Immun. 53: 616– 620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guggenheim B, Guggenheim M, Gmür R, Giertsen E, Thurnheer T. 2004. Application of the Zürich biofilm model to problems of cariology. Caries Res. 38: 212– 222 [DOI] [PubMed] [Google Scholar]
- 23. Guggenheim M, Shapiro S, Gmür R, Guggenheim B. 2001. Spatial arrangements and associative behavior of species in an in vitro oral biofilm model. Appl. Environ. Microbiol. 67: 1343– 1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haffajee AD, Socransky SS. 2005. Microbiology of periodontal diseases: introduction. Periodontol. 2000 38: 9– 12 [DOI] [PubMed] [Google Scholar]
- 25. Hajishengallis G, et al. 2011. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10: 497– 506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hojo K, Nagaoka S, Ohshima T, Maeda N. 2009. Bacterial interactions in dental biofilm development. J. Dent. Res. 88: 982– 990 [DOI] [PubMed] [Google Scholar]
- 27. Houot L, Chang S, Absalon C, Watnick PI. 2010. Vibrio cholerae phosphoenolpyruvate phosphotransferase system control of carbohydrate transport, biofilm formation, and colonization of the germfree mouse intestine. Infect. Immun. 78: 1482– 1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Houot L, Watnick PI. 2008. A novel role for enzyme I of the Vibrio cholerae phosphoenolpyruvate phosphotransferase system in regulation of growth in a biofilm. J. Bacteriol. 190: 311– 320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jumas-Bilak E, et al. 2004. Veillonella montpellierensis sp. nov., a novel, anaerobic, Gram-negative coccus isolated from human clinical samples. Int. J. Syst. Evol. Microbiol. 54: 1311– 1316 [DOI] [PubMed] [Google Scholar]
- 30. Karpathy SE, et al. 2007. Genome sequence of Fusobacterium nucleatum subspecies polymorphum—a genetically tractable fusobacterium. PLoS One 2: e659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12: 656– 664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim S-W, Rai D, Aguiar R. 2011. Gene-set enrichment analysis unveils the mechanism for the phosphodiesterase 4B control of glucocorticoid response in B-cell lymphoma. Clin. Cancer Res. 17: 6723– 6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim S-Y, Volsky DJ. 2005. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics 6: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kinniment SL, Wimpenny JW, Adams D, Marsh PD. 1996. Development of a steady-state oral microbial biofilm community using the constant-depth film fermenter. Microbiology 142 ( Pt 3): 631– 638 [DOI] [PubMed] [Google Scholar]
- 35. Klein MI, et al. 2010. Dynamics of Streptococcus mutans transcriptome in response to starch and sucrose during biofilm development. PLoS One 5: e13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kolenbrander PE. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54: 413– 437 [DOI] [PubMed] [Google Scholar]
- 37. Kolenbrander PE, et al. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66: 486– 505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kolenbrander PE, Egland PG, Diaz PI, Palmer RJ., Jr 2005. Genome-genome interactions: bacterial communities in initial dental plaque. Trends Microbiol. 13: 11– 15 [DOI] [PubMed] [Google Scholar]
- 39. Koo H, Xiao J, Klein MI, Jeon JG. 2010. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J. Bacteriol. 192: 3024– 3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kuboniwa M, et al. 2009. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 9: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. 2007. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71: 653– 670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lawrence PK, Kittichotirat W, McDermott JE, Bumgarner RE. 2010. A three-way comparative genomic analysis of Mannheimia haemolytica isolates. BMC Genomics 11: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee H-R, et al. 2011. In-vivo-induced antigenic determinants of Fusobacterium nucleatum subsp. nucleatum. Mol. Oral Microbiol. 26: 164– 172 [DOI] [PubMed] [Google Scholar]
- 44. Li Y-H, et al. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 184: 6333– 6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin X, Lamont RJ, Wu J, Xie H. 2008. Role of differential expression of streptococcal arginine deiminase in inhibition of fimA expression in Porphyromonas gingivalis. J. Bacteriol. 190: 4367– 4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu JM, Camilli A. 2010. A broadening world of bacterial small RNAs. Curr. Opin. Microbiol. 13: 18– 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lo AW, et al. 2009. Comparative transcriptomic analysis of Porphyromonas gingivalis biofilm and planktonic cells. BMC Microbiol. 9: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marsh PD. 2006. Dental plaque as a biofilm and a microbial community—implications for health and disease. BMC Oral Health 6 ( Suppl 1): S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McKee AS, McDermid AS, Ellwood DC, Marsh PD. 1985. The establishment of reproducible, complex communities of oral bacteria in the chemostat using defined inocula. J. Appl. Bacteriol. 59: 263– 275 [DOI] [PubMed] [Google Scholar]
- 50. McNab R, et al. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185: 274– 284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McQueary CN, Actis LA. 2011. Acinetobacter baumannii biofilms: variations among strains and correlations with other cell properties. J. Microbiol. 49: 243– 250 [DOI] [PubMed] [Google Scholar]
- 52. Meuric V, Gracieux P, Tamanai-Shacoori Z, Perez-Chaparro J, Bonnaure-Mallet M. 2008. Expression patterns of genes induced by oxidative stress in Porphyromonas gingivalis. Oral Microbiol. Immunol. 23: 308– 314 [DOI] [PubMed] [Google Scholar]
- 53. Mitchell HL, et al. 2010. Treponema denticola biofilm-induced expression of a bacteriophage, toxin-antitoxin systems and transposases. Microbiology 156: 774– 788 [DOI] [PubMed] [Google Scholar]
- 54. Morillo JM, et al. 2004. Quantitative real-time polymerase chain reaction based on single copy gene sequence for detection of periodontal pathogens. J. Clin. Periodontol. 31: 1054– 1060 [DOI] [PubMed] [Google Scholar]
- 55. Ning Z, Cox AJ, Mullikin JC. 2001. SSAHA: a fast search method for large DNA databases. Genome Res. 11: 1725– 1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. O'Grady EP, Nguyen DT, Weisskopf L, Eberl L, Sokol PA. 2011. The Burkholderia cenocepacia LysR-type transcriptional regulator ShvR influences expression of quorum-sensing, protease, type II secretion, and afc genes. J. Bacteriol. 193: 163– 176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Paddick JS, et al. 2006. Effect of biofilm growth on expression of surface proteins of Actinomyces naeslundii genospecies 2. Appl. Environ. Microbiol. 72: 3774– 3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Paster BJ, et al. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183: 3770– 3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Paster BJ, Olsen I, Aas JA, Dewhirst FE. 2006. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2000 42: 80– 87 [DOI] [PubMed] [Google Scholar]
- 60. Periasamy S, Chalmers NI, Du-Thumm L, Kolenbrander PE. 2009. Fusobacterium nucleatum ATCC 10953 requires Actinomyces naeslundii ATCC 43146 for growth on saliva in a three-species community that includes Streptococcus oralis 34. Appl. Environ. Microbiol. 75: 3250– 3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Poretsky RS, et al. 2005. Analysis of microbial gene transcripts in environmental samples. Appl. Environ. Microbiol. 71: 4121– 4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rathsam C, et al. 2005. Up-regulation of competence—but not stress-responsive proteins accompanies an altered metabolic phenotype in Streptococcus mutans biofilms. Microbiology 151: 1823– 1837 [DOI] [PubMed] [Google Scholar]
- 63. Rickard AH, et al. 2006. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 60: 1446– 1456 [DOI] [PubMed] [Google Scholar]
- 64. Saito Y, et al. 2008. Stimulation of Fusobacterium nucleatum biofilm formation by Porphyromonas gingivalis. Oral Microbiol. Immunol. 23: 1– 6 [DOI] [PubMed] [Google Scholar]
- 65. Selinger DW, Saxena RM, Cheung KJ, Church GM, Rosenow C. 2003. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 13: 216– 223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shemesh M, Tam A, Aharoni R, Steinberg D. 2010. Genetic adaptation of Streptococcus mutans during biofilm formation on different types of surfaces. BMC Microbiol. 10: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shemesh M, Tam A, Kott-Gutkowski M, Feldman M, Steinberg D. 2008. DNA-microarrays identification of Streptococcus mutans genes associated with biofilm thickness. BMC Microbiol. 8: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shemesh M, Tam A, Steinberg D. 2007. Differential gene expression profiling of Streptococcus mutans cultured under biofilm and planktonic conditions. Microbiology 153: 1307– 1317 [DOI] [PubMed] [Google Scholar]
- 69. Simionato MR, et al. 2006. Porphyromonas gingivalis genes involved in community development with Streptococcus gordonii. Infect. Immun. 74: 6419– 6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Standar K, et al. 2010. Setup of an in vitro test system for basic studies on biofilm behavior of mixed-species cultures with dental and periodontal pathogens. PLoS One 5: e13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Subramanian A, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102: 15545– 15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Thomas MA, Yang L, Carter BJ, Klaper RD. 2011. Gene set enrichment analysis of microarray data from Pimephales promelas (Rafinesque), a non-mammalian model organism. BMC Genomics 12: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tomlin KL, et al. 2005. Quorum-sensing mutations affect attachment and stability of Burkholderia cenocepacia biofilms. Appl. Environ. Microbiol. 71: 5208– 5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Walker C, Sedlacek MJ. 2007. An in vitro biofilm model of subgingival plaque. Oral Microbiol. Immunol. 22: 152– 161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Watson SW, Novitsky TJ, Quinby HL, Valois FW. 1977. Determination of bacterial number and biomass in the marine environment. Appl. Environ. Microbiol. 33: 940– 946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wen ZT, Yates D, Ahn S-J, Burne RA. 2010. Biofilm formation and virulence expression by Streptococcus mutans are altered when grown in dual-species model. BMC Microbiol. 10: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wen ZT, et al. 2011. Transcriptome analysis of LuxS-deficient Streptococcus mutans grown in biofilms. Mol. Oral Microbiol. 26: 2– 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yamanaka T, et al. 2009. Gene expression profile and pathogenicity of biofilm-forming Prevotella intermedia strain 17. BMC Microbiol. 9: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yoshida A, et al. 2003. Development of a 5′ fluorogenic nuclease-based real-time PCR assay for quantitative detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. J. Clin. Microbiol. 41: 863– 866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yoshimura M, et al. 2008. Proteome analysis of Porphyromonas gingivalis cells placed in a subcutaneous chamber of mice. Oral Microbiol. Immunol. 23: 413– 418 [DOI] [PubMed] [Google Scholar]
- 81. Zaura E, Keijser BJF, Huse SM, Crielaard W. 2009. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 9: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7: 203– 214 [DOI] [PubMed] [Google Scholar]
- 83. Zijnge V, et al. 2010. Oral biofilm architecture on natural teeth. PLoS One 5: e9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.