Abstract
The polar flagellar motor of Vibrio alginolyticus rotates using Na+ influx through the stator, which is composed of 2 subunits, PomA and PomB. About a dozen stators dynamically assemble around the rotor, depending on the Na+ concentration in the surrounding environment. The motor torque is generated by the interaction between the cytoplasmic domain of PomA and the C-terminal region of FliG, a component of the rotor. We had shown previously that mutations of FliG affected the stator assembly around the rotor, which suggested that the PomA-FliG interaction is required for the assembly. In this study, we examined the effects of various mutations mainly in the cytoplasmic domain of PomA on that assembly. All mutant stators examined, which resulted in the loss of motor function, assembled at a lower level than did the wild-type PomA. A His tag pulldown assay showed that some mutations in PomA reduced the PomA-PomB interaction, but other mutations did not. Next, we examined the ion conductivity of the mutants using a mutant stator that lacks the plug domain, PomA/PomBΔL(Δ41–120), which impairs cell growth by overproduction, presumably because a large amount of Na+ is conducted into the cells. Some PomA mutations suppressed this growth inhibition, suggesting that such mutations reduce Na+ conductivity, so that the stators could not assemble around the rotor. Only the mutation H136Y did not impair the stator formation and ion conductivity through the stator. We speculate that this particular mutation may affect the PomA-FliG interaction and prevent activation of the stator assembly around the rotor.
INTRODUCTION
Bacterial flagella confer a motile ability for cells and have a rotary motor in their base. It is known that the motor is driven by the influx of coupling ions down the gradient of electrochemical potential between the outside and the inside of cells (2, 23). The flagella of Escherichia coli and Salmonella are H+ driven, and the polar flagellum of Vibrio alginolyticus is Na+ driven. A flagellar motor consists of a rotor and stator parts. The rotor has a rotational ring structure, and the stators, which are embedded in the membrane and have ion conductivity, assemble around the rotor (15). The stator of V. alginolyticus consists of the subunits PomA and PomB for its Na+-driven motor, whereas the stators of E. coli and Salmonella consist of MotA and MotB for their H+-driven motor (3, 10, 20). PomA (MotA) is a protein with four transmembrane (TM) domains and a large cytoplasmic loop between the second and third TM regions (1, 32). PomB (MotB) has a single TM domain which is located near its N-terminal region and has a large C-terminal periplasmic domain which interacts with peptidoglycan to anchor the stator (1, 4). PomA (MotA) and PomB (MotB) are thought to form a hexamer complex in a 4:2 ratio to give ion conductivity (28).
The torque is generated by the interaction between the large cytoplasmic region of PomA and FliG, one of the rotor components. In the H+-driven motor of E. coli, it has been shown that charged residues of MotA and FliG are important for torque generation (26, 33). On the other hand, the Na+-driven motor of V. alginolyticus may not require the corresponding charged residues for torque generation (29, 31). PomB (MotB) has an essential charged residue, aspartic acid, in its TM domain (24, 34). It was recently shown by attenuated total reflection Fourier-transform infrared (ATR-FTIR) spectroscopy that Na+ binds to that aspartic acid (Asp24) of PomB (21).
We recently showed that overproduction of a mutant stator complex, which is composed of PomA and an in-frame deletion variant of PomB, PomBΔL(Δ41–120), inhibited cell growth. The deleted region contains the periplasmic linker region that includes the “plug” segment, which is thought to work to prevent excessive Na+ influx. That growth defect caused by the plug-deleted stator is probably due to a high Na+ influx through the mutant stator (14). This mutant stator complex is quite useful to detect the Na+-conducting activity of the stator in vivo, as well as the in vitro reconstitution system using the purified stator complex.
It was shown previously that in the H+-driven motor of E. coli, the stator complex is not fixed, but at least 11 stator complexes dissociate and associate dynamically around the rotor (13, 19). In the Na+-driven flagellar motor of marine Vibrio and Shewanella oneidensis, the stator complex is also not fixed and the association of the stator complexes around the rotor is dependent on Na+, the coupling ion for the force generation (5, 18). This dynamic property might be an adaptation to environmental conditions in which the coupling ion is abundant. In V. alginolyticus, it was suggested that the interaction between the T ring, which was recently found as a novel periplasmic basal structure of the motor composed of MotX and MotY, and the periplasmic region of PomB as well as the interaction between FliG and the cytoplasmic region of PomA is important for the stator assembly around the rotor (12, 22). From those lines of evidence, we can infer that Na+ affects those interactions.
In this study, we focused on mutations of the PomA cytoplasmic domain and investigated the ability of stator assembly around the rotor and formation of the PomA/PomB complex. Furthermore, the ion-conducting activity was estimated using PomBΔL(Δ41-120) with the mutant PomA by the suppression of the growth inhibition. We discuss the defects in motor function caused by each of the mutations.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Mutations were introduced into pomA in pHFGAB, pHFGBA2, and pTSK37 using the QuikChange site-directed mutagenesis procedure, according to the protocol described by Stratagene. V. alginolyticus was cultured at 30°C in VC medium (0.5% [wt/vol] polypeptone, 0.5% [wt/vol] yeast extract, 0.4% [wt/vol] K2HPO4, 3% [wt/vol] NaCl, 0.2% [wt/vol] glucose) or in VPG medium (1% [wt/vol] polypeptone, 0.4% [wt/vol] K2HPO4, 3% [wt/vol] NaCl, 0.5% [wt/vol] glycerol). Chloramphenicol was added to a final concentration of 2.5 μg · ml−1.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| V. alginolyticus strains | ||
| VIO5 | VIK4 laf (Rif+ Pof+ Laf−) | 17 |
| NMB191 | VIO5 ΔpomAB (Rifr Laf− Pom−) | 30 |
| Plasmids | ||
| pBAD33 | Cmr; PBAD | 8 |
| pHFGBA2 | His6-gfp-pomB and pomA in pBAD33 | 7 |
| pHFGAB | His6-gfp-pomA and pomB in pBAD33 | 7 |
| pTSK37 | pomA and pomBΔ41-120 in pBAD33 | 14 |
| pHFAB | pomA and pomB in pBAD33 | 7 |
Rifr, rifampin resistant; Cmr, chloramphenicol resistant; Pof+, normal polar flagellar formation; Laf−, defective in lateral flagellar formation; PBAD, arabinose-inducible promoter.
Detection of proteins.
Overnight cultures grown in VC medium were inoculated at a 100-fold dilution into VPG medium containing 0.006% (wt/vol) arabinose and were cultured for 4 h to produce green fluorescent protein (GFP)-PomA and PomB or GFP-PomB and PomA or were inoculated into VPG medium containing 0.2% (wt/vol) arabinose and were cultured at 30°C for 7 h to produce PomA and PomBΔL(Δ41–120). Cells were harvested by centrifugation and were then suspended in sodium dodecyl sulfate (SDS) loading buffer containing 5% (vol/vol) β-mercaptoethanol to an optical density at 660 nm of 10 and boiled for 5 min at 95°C. Samples were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes, and immunoblotting was performed using anti-PomA (PomA1312) (30) and anti-PomB (PomBC2 B0455) antibodies (25).
Observation of stator localization with fluorescence microscopy.
Overnight cultures of NMB191 cells harboring the plasmid pHFGAB or pHFGBA2 were inoculated at a 100-fold dilution into VPG medium containing 0.006% (wt/vol) arabinose, and cells were grown for 4 h at 30°C. Cells were then harvested by centrifugation and resuspended in TMN500 (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 5 mM glucose, 500 mM NaCl) and incubated for about 10 min. Cells were fixed to the glass surface of the slide using 0.1% (wt/vol) poly-l-lysine and were observed by epifluorescence microscopy as described previously (5). Images were recorded using a digital charge-coupled device (CCD) camera (Hamamatsu Photonics; Orca-AG) and software (Scanalytics; IP lab ver. 3.9.5r2). All images were obtained with a 1-min integration time using the software. The percentage of polar localization was evaluated by dividing the number of cells that have a fluorescent dot at the cell pole by the total number of the cells in a field.
Detection of PomA-PomB interaction using the coelution assay.
The coelution assay was performed as described previously with slight modifications (16). Briefly, NMB191 cells harboring the plasmid pHFGBA2 were cultured at 30°C for 4 h in VC medium containing 0.02% arabinose, harvested by centrifugation, and sonicated, and membrane fractions were obtained by ultracentrifugation. Solubilization of total cell membranes (5 mg · ml−1 proteins) was achieved by addition of the detergent Cymal-5 (Anatrace, Maumee, OH) to a final concentration of 1% (wt/vol) and incubation at room temperature for 1 h. Insoluble materials were removed by ultracentrifugation, and the supernatant was mixed with nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) and then incubated for 1 h at 4°C with gentle mixing. Ni-NTA beads were washed three times with a buffer containing 5 mM imidazole. The His6-GFP-PomB/PomA complex was then eluted with a buffer containing 200 mM imidazole and was analyzed by immunoblotting.
Measurement of growth inhibition of PomA/PomBΔL(Δ41–120)-overproducing cells.
Overnight cultures of NMB191 cells harboring the plasmid pTSK37 were inoculated at a 100-fold dilution into VPG medium containing 0.2% (wt/vol) arabinose, and cells were grown with shaking (180 rpm) for 8 h at 30°C. The optical density at 660 nm was measured every hour.
RESULTS
Phenotypes resulting from mutations in PomA.
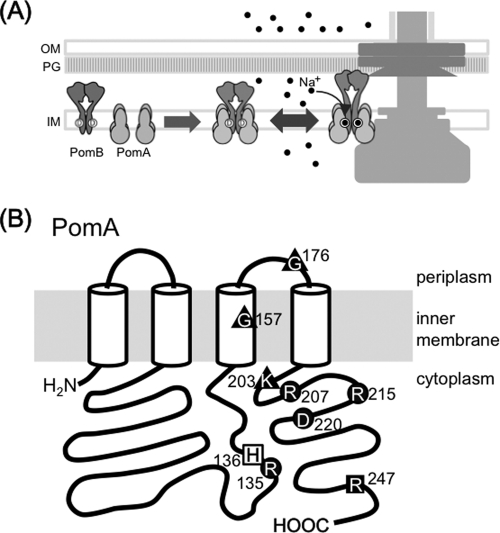
The flagellar stator complexes need to be properly assembled around the rotor to generate a force (Fig. 1A). The abnormal or lost interaction between stators and a rotor should affect motor function and swimming behavior. We assumed that mutations in the PomA cytoplasmic domain may affect such interactions; hence, we selected such mutations from the mutant collections of our laboratory. We expected that those mutations might also give a dominant effect against the wild-type stator. The PomA cytoplasmic mutants, R135Q, H136Y, K203E, R207E, R215E, D220K, and R247C; the TM3 mutant, G157D; and the periplasmic mutant, G176E, which were isolated in previous studies, were confirmed for their swimming ability in VPG medium and their motility on soft agar plates, as well as dominant effects over wild-type PomA (6, 11, 16) (Fig. 1B; Table 2). These mutant phenotypes were the same as or similar to those reported previously (11, 16) (data not shown). Expression of the stator complex was examined by immunoblotting, which confirmed that the expression level of each mutant protein is almost the same as that of the wild type (data not shown).
Fig 1.
Diagram of the flagellar motor and the stator component PomA. (A) A schematic model of the stator assembly around the rotor in the Na+-driven flagellar motor of the polar flagellum in V. alginolyticus. The circles with the letter D inside indicate the Na+ binding sites of PomB. The black dots represent sodium ions. OM, outer membrane; PG, peptidoglycan layer; IM, inner membrane. (B) PomA point mutations studied in this report were mapped on a schematic diagram and are indicated as black circles with residue numbers. ■, mutation disrupts PomA protein expression; ▲, mutations disrupt PomA/B complex formation; ●, mutations disrupt ion conduction; □, mutation keeps both PomA/B complex-forming ability and ion conductivity. All of these mutations show a nonmotile phenotype except for R207E, which is slow-motile, and reduced stator polar localization rates, around 30% or less, whereas the wild type shows a rate reduction of more than 80%.
Table 2.
Characterization of Pom mutants
| Mutation in PomA | PomAB |
+GFPe |
||||
|---|---|---|---|---|---|---|
| Motilitya | Growthb | Dominancec | Complexd | Dominancec | Complexd | |
| None (wild type) | +++ | ++ | NT | ++ | NT | ++ |
| R135Q | − | − | ++ | NT | + | ++ |
| H136Y | − | + | ++ | NT | + | ++ |
| G157D | − | NT | ++ | NT | + | − |
| G176E | − | NT | ++ | NT | + | +/− |
| K203E | − | NT | − | − | − | − |
| R207E | + | − | − | ++ | − | ++ |
| R215E | − | − | + | ++ | + | ++ |
| D220K | − | − | ++ | ++ | + | ++ |
| R247C | − | +/− | + | NT | + | (++) |
| PomB-D24N | − | − | − | NT | − | ++ |
The size of the swarm ring produced by each strain was measured on VPG soft agar plates. +++, cells carrying the mutant PomA swarmed as much as did the wild-type PomA and PomB; +, cells swarmed less than half as much as did the wild type; −, cells did not swarm.
The growth inhibitions were evaluated from additional data in Fig. 5A. −, growth inhibition was not observed (strains grew like the vector control); +/−, slightly inhibited; +, significantly inhibited; ++, strongly inhibited by the wild-type PomA and PomBΔL(Δ41-120); NT, not tested.
The dominant effect of the mutant PomA or PomB against the VIO5 wild-type cells was evaluated by measuring the swarm ring size. ++, strong dominance (no swarming); +, weak dominance (smaller swarming ring); −, no dominance (swarming like the vector control); NT, not tested.
From the previous report (16) and from Fig. 4, the complex formation of PomA and PomB or PomA and GFP-PomB was evaluated. ++, normal stator complex formation was observed as in the wild-type PomA and PomB; +/−, weaker stator complex formation was observed than that with the wild type; −, no complex formation was observed; (++), the stator complex formation was detected, but the PomA protein was decreased in whole cells; NT, not tested.
GFP-PomB was expressed with PomA.
Localization of the stator complex with the PomA mutations.
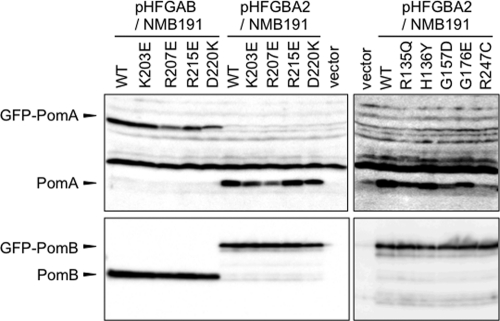
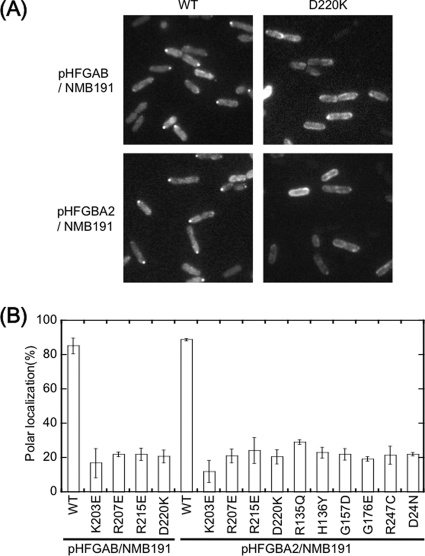
The ability of each mutant stator for polar localization was examined using GFP-fused stator components. The nine mutations were separately introduced into the pomA gene in the plasmid pHFGBA2, which produces PomA and GFP-fused PomB upon arabinose induction. In addition, four mutations (K203E, R207E, R215E, and D220K) were introduced into the pomA gene in the plasmid pHFGAB, which produces GFP-PomA/PomB upon arabinose induction. When NMB191 cells containing pHFGAB or pHFGBA2 encoding the wild-type stator with GFP were induced by 0.006% arabinose, fluorescent dots were observed at the cell poles as previously reported (5). Under this condition, the stator proteins were produced from the plasmids, and the protein amounts were estimated by immunoblotting (Fig. 2). GFP-PomB was expressed at the wild-type level even in the presence of mutant PomA; however, the amount of PomA was reduced a little in the K203E, R207E, G157D, G176E, and R135Q mutants and was reduced significantly in the R247C mutant. PomA expression was not affected in the H136Y, R215E, and D220K mutants. In pHFGAB, which encodes GFP-PomA/PomB, the amount of PomB was constant with all mutant GFP-PomA proteins as well as with the wild-type GFP-PomA. The amount of GFP-PomA was reduced a little in the R207E and D220K mutants but was similar to the wild-type level in the K203E and R215E mutants. When we observed the cells producing GFP-PomA/PomB or PomA/GFP-PomB, the localization of PomA mutations to the cell poles, indicated by GFP, was reduced to 30% or less compared to around 80% with the wild-type PomA (Fig. 3A and B).
Fig 2.
Protein expression of GFP-fused stator protein. GFP-PomA/PomB and GFP-PomB/PomA were expressed in NMB191 cells from plasmids pHFGAB and pHFGBA2, respectively. Protein expression was induced by 0.006% (wt/vol) arabinose. PomA and PomB were detected by immunoblotting using anti-PomA and anti-PomB antibodies.
Fig 3.
Observation of GFP-fused stator assembly at the cell poles. GFP-PomA/PomB from pHFGAB or GFP-PomB/PomA from pHFGBA2 was expressed in NMB191 cells and was observed by fluorescence microscopy. (A) Images for wild-type (WT) PomA and PomA-D220K. (B) Percentages of polar localization of fluorescence dots.
Detection of the interaction between PomA and PomB.
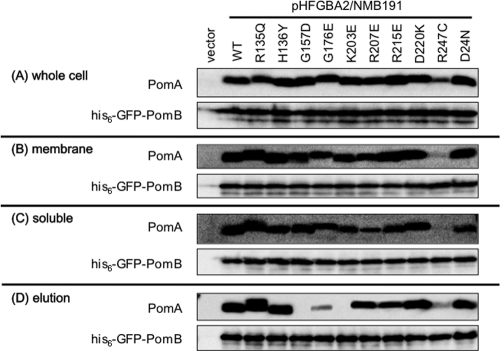
To examine the PomA-PomB interaction of PomA mutants, we performed a coelution assay. NMB191 cells containing pHFGAB2 carrying the pomA mutation and its derivatives were cultured in the presence of 0.02% arabinose and sonicated, and the membrane fraction was solubilized with the detergent Cymal-5. In this study, Cymal-5 gave better results in purifying the PomAB complex than did CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, which we had used before (27). His6-GFP-PomB was then purified using Ni-NTA agarose, and samples were subjected to SDS-PAGE followed by Western blot analyses (Fig. 4). As seen in the elution fractions, the amount of copurified PomA protein was drastically reduced in the K203E mutant as reported previously (16) and was almost not detectable in the G157D mutant. A small amount of PomA was copurified in the G176E mutant. Similar amounts of PomA were copurified in the wild type and the R135Q, H136Y, R207E, R215E, and D220K mutants and in the PomB-D24N mutant whose mutation site is the Na+ binding site located in the TM region of PomB. We were not able to evaluate the interaction of PomA-R247C and PomB because the expression level of the mutant PomA was low even in the whole-cell fraction.
Fig 4.
Detection of PomA-PomB interaction by the coelution assay. His6-GFP-PomB/PomA from pHFGBA2 was expressed with 0.02% (wt/vol) arabinose induction in NMB191 cells. PomA-PomB interaction was detected by the coelution assay followed by immunoblot analysis using anti-PomA and anti-PomB antibodies.
Estimation of ion permeability of the stator complex.
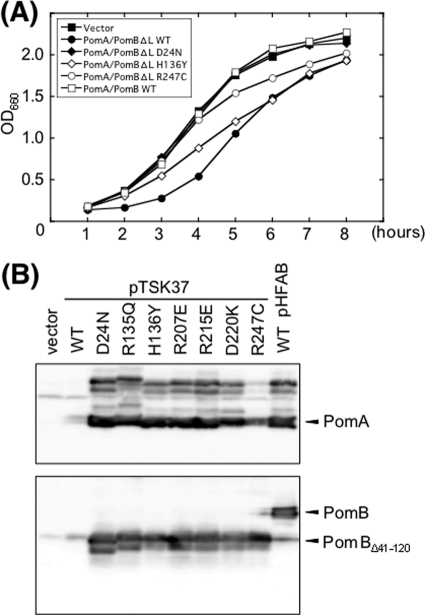
We have previously shown that Na+ ions are necessary to assemble the stator complexes around the rotor and that an Asn mutation of the Na+ binding residue, PomB-D24, prevents the polar localization of the stator complexes (5). We speculated that the mutations of PomA used in this study might also prevent the ion conductivity and the polar localization of the stator complexes. To estimate the ion conductivity of the stator, we used a plug-deleted stator consisting of PomA and PomBΔL(Δ41–120). A previous study showed that the growth of NMB191 cells was inhibited by producing the plug-deleted stator with arabinose induction but that the growth was rescued by the addition of the PomB-D24N mutation (14) (Fig. 5A). This suggested that the Na+ conductivity and the growth inhibition were directly correlated in the plug-deleted stator. Thus, we introduced a series of point mutations (R135Q, H136Y, R207E, R215E, D220K, and R247C) into the pomA gene that is on the plasmid that produces the plug-deleted stator, after which cell growth was measured. Those mutations suppressed the growth inhibition in a similar manner as with the PomB-D24N mutation, except for the R247C and H136Y mutations (data not shown; Table 2). The growth inhibition was observed after 4 to 5 h of incubation for the H136Y mutation and after 6 to 7 h of incubation for the R247C mutation (Fig. 5A). The amounts of stator proteins, PomA and PomBΔL(Δ41–120), produced were examined by immunoblotting (Fig. 5B). Protein levels from the plasmid carrying PomA/PomBΔL(Δ41–120) carrying no pomA mutation were much lower than those from the plasmid carrying a mutant PomA. Among these mutations, only the R247C mutation resulted in a smaller amount of PomA than did the other mutations.
Fig 5.
Measurement of growth inhibition due to overproduction of PomA/PomBΔL(Δ41–120). (A) The growth rate of each strain was measured, and representative examples are shown. NMB191 cells carrying the empty vector pBAD33 (■), pHFAB producing PomA/PomB (□), and pTSK37 producing the plug-deleted stator, PomA/PomBΔL(Δ41–120) (●), and with PomB-D24N (♦), with PomA-H136Y (♢), or with PomA-R247C (○) were grown in the presence of 0.2% (wt/vol) arabinose, and optical densities at 660 nm (OD660) were measured every hour. (B) Protein expression of the stator protein. PomA and PomB or PomBΔL(Δ41–120) expressed from pHFAB or pTSK37, respectively, were expressed in NMB191 cells. Each plasmid was induced with 0.2% (wt/vol) arabinose. PomA and PomB were detected by immunoblotting using anti-PomA and anti-PomB antibodies.
DISCUSSION
In order for the flagellar motor to work properly, the stator complexes and force-generating units have to be assembled around the rotor. The electrochemical potential energy of the ions is converted into mechanical force to drive the flagellar motor. This conversion is believed to occur by the interaction between the cytoplasmic domain of PomA, a stator component, and the C-terminal region of FliG, a rotor component (15). We have reported that construction of the polar flagellar basal body or the rotor of V. alginolyticus is likely to be directed by FlhF, which is a putative GTPase and is similar to the bacterial signal recognition proteins FtsY and Ffh (9). Without the basal body, the PomA/B stator complexes cannot localize at the cell poles, and the proper stator complex formation is necessary for the polar localization of the stators (7). Even in the presence of the basal body, the polar localization of the PomA/B complex is lost when the T ring structure composed of the Na+ motor-specific components, MotX and MotY, is missing (22). In V. alginolyticus, the stator complexes of the Na+-driven flagellar motor dynamically associate and dissociate around the rotor depending on the Na+ concentration in the environment (5). The Na+ binding and/or flow may be required for the assembly of the stators.
We have shown in a previous study that some fliG mutants, which have mutations in their C-terminal region, do not have defects in flagellation but do have impaired motor function (12). These fliG mutants prevent the polar localization of the PomA/B complex, which suggests that the interaction between FliG and PomA is necessary to fix the PomA/B stator complex around the rotor. We speculated that the PomA-FliG interaction induced a large conformational change at the periplasmic region of PomB and bound to the T ring and the peptidoglycan layer (12). In this study, we demonstrate that the cytoplasmic domain of PomA is also important for the stator to assemble around the rotor. PomA mutants that carry a single amino acid substitution in their cytoplasmic domain are not able to localize the stator complex around the rotor (Fig. 3A and B). Those single amino acid substitutions may affect various processes, such as the production of PomA, the interaction with PomB, and/or the ion conductivity of the stator complex. Western blot analysis showed that the protein expression levels of some PomA mutants are a little reduced, and the PomA-R247C mutant was not detectable upon 0.006% (wt/vol) arabinose induction (Fig. 2). In this mutant, the defect of the stator assembly around the rotor may be explained by this reduced protein level.
We also examined the interaction between PomA and PomB in the pomA mutations (Fig. 4). The coelution assay indicated that the interaction between PomA and PomB is lost or strongly reduced with the PomA-G157D, G176E, and K203E mutants. We hypothesize that these mutant PomA proteins are not able to form functional stator complexes by associating with PomB, and therefore, the stators do not assemble around the rotor. Because PomB is less stable in the absence of PomA (30), the defect of the interaction may also affect the stability of the mutant protein.
The cytoplasmic domain of PomA (MotA) and the C-terminal region of FliG are thought to interact, and this interaction is required for torque generation. In the H+-driven motors of E. coli, it is inferred that the conserved charged residues of FliG and MotA interact, which is important for torque generation (26, 33). Mutation of the cytoplasmic domain at L131 and T132 in PomA affects the temperature sensitivity of the mutants at the conserved charged residues of PomA (6). From these lines of evidence, we speculate that residues R88, E89, E96, E97, and E99, which are conserved charged residues, and L131 and T132 are located close to each other.
All PomA mutations suppressed the growth inhibition that occurs upon overproduction of PomA/PomBΔL(Δ41–120), which lacks a plug domain, although the suppression was weak in the R247C and H136Y mutants (Fig. 5). The growth recovery of the R135Q, R207E, R215E, and D220K mutants was to the same extent as that in the PomB-D24N mutant, which lacks the ion binding site and is expected to lose its ion conductivity (Table 2). We think that the ion conductivity was lost in those four mutant PomA proteins. It has been suggested that R215 and G139 are located close together in the PomA structure because the R215E mutation of the C-terminal cytoplasmic domain was suppressed by the G139D mutation in the loop region between the second and third TM domains (16). This region might be important to transmit the conformational change caused by the ion binding or flow through the channel composed of the TM regions of PomA and PomB.
In contrast, the growth inhibition was still observed with the PomA-H136Y and R247C mutations. This may be because the stator complexes with those mutations are still capable of ion transport (Fig. 5A). Remarkably, in the PomA-H136Y mutation, the protein production was normal (Fig. 2B and 5B), the stator complex formation was normal (Fig. 4), and the ion conductivity remained (Fig. 5A). Therefore, the H136Y mutation in the PomA cytoplasmic domain is most likely to directly disturb the interaction with FliG. We speculate that the region around H136 is a site of interaction of PomA with the C-terminal region of FliG to generate torque. It is also possible that this mutation inhibits a structural change of the stator complex to interact with a rotor when Na+ ions bind or flow in it. Incidentally, decreased amounts of the wild-type PomA and PomBΔL(Δ41–120) proteins were detected compared to all mutant proteins (Fig. 5B). This implies that the transcription and/or translation of the pomA and pomB genes is inhibited by the high Na+ concentration in the cytoplasm or, alternatively, that PomA and PomB proteins are easily degraded by the high-Na+ condition.
Finally, we want to emphasize that the ion conductivity of the stator complex is affected by some of the PomA cytoplasmic domain mutations. This implies that the ion binding and ion flow strongly correlate with the structure of the cytoplasmic domain of PomA. The charged residues R88, E89, E96, E97, and E99 and the residues L131 and T132, in addition to H136, may be located on or near an interaction surface against the C-terminal region of FliG. Our future studies will be directed at testing this speculation.
ACKNOWLEDGMENTS
We thank Takashi Terauchi for helpful discussion. We also thank Maya Kitaoka for critically reviewing and editing the manuscript.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan and from the Japan Science and Technology Corporation (to M.H. and S.K.).
Footnotes
Published ahead of print 17 February 2012
REFERENCES
- 1. Asai Y, et al. 1997. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J. Bacteriol. 179: 5104– 5110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blair DF. 2003. Flagellar movement driven by proton translocation. FEBS Lett. 545: 86– 95 [DOI] [PubMed] [Google Scholar]
- 3. Braun TF, Al-Mawsawi LQ, Kojima S, Blair DF. 2004. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 43: 35– 45 [DOI] [PubMed] [Google Scholar]
- 4. Chun SY, Parkinson JS. 1988. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science 239: 276– 278 [DOI] [PubMed] [Google Scholar]
- 5. Fukuoka H, Wada T, Kojima S, Ishijima A, Homma M. 2009. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol. Microbiol. 71: 825– 835 [DOI] [PubMed] [Google Scholar]
- 6. Fukuoka H, Yakushi T, Homma M. 2004. Concerted effects of amino acid substitutions in conserved charged residues and other residues in the cytoplasmic domain of PomA, a stator component of Na+-driven flagella. J. Bacteriol. 186: 6749– 6758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fukuoka H, Yakushi T, Kusumoto A, Homma M. 2005. Assembly of motor proteins, PomA and PomB, in the Na+-driven stator of the flagellar motor. J. Mol. Biol. 351: 707– 717 [DOI] [PubMed] [Google Scholar]
- 8. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177: 4121– 4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kojima M, et al. 2011. Conversion of mono-polar to peritrichous flagellation in Vibrio alginolyticus. Microbiol. Immunol. 55: 76– 83 [DOI] [PubMed] [Google Scholar]
- 10. Kojima S, Blair DF. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43: 26– 34 [DOI] [PubMed] [Google Scholar]
- 11. Kojima S, Kuroda M, Kawagishi I, Homma M. 1999. Random mutagenesis of the pomA gene encoding a putative channel component of the Na+-driven polar flagellar motor of Vibrio alginolyticus. Microbiology 145: 1759– 1767 [DOI] [PubMed] [Google Scholar]
- 12. Kojima S, Nonoyama N, Takekawa N, Fukuoka H, Homma M. 2011. Mutations targeting the C-terminal domain of FliG can disrupt motor assembly in the Na+-driven flagella of Vibrio alginolyticus. J. Mol. Biol. 414: 62– 74 [DOI] [PubMed] [Google Scholar]
- 13. Leake MC, et al. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443: 355– 358 [DOI] [PubMed] [Google Scholar]
- 14. Li N, Kojima S, Homma M. 2011. Characterization of the periplasmic region of PomB, a Na+-driven flagellar stator protein in Vibrio alginolyticus. J. Bacteriol. 193: 3773– 3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li N, Kojima S, Homma M. 2011. Sodium-driven motor of the polar flagellum in marine bacteria Vibrio. Genes Cells 16: 985– 999 [DOI] [PubMed] [Google Scholar]
- 16. Obara M, Yakushi T, Kojima S, Homma M. 2008. Roles of charged residues in the C-terminal region of PomA, a stator component of the Na+-driven flagellar motor. J. Bacteriol. 190: 3565– 3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Okunishi I, Kawagishi I, Homma M. 1996. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol. 178: 2409– 2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paulick A, et al. 2009. Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol. Microbiol. 71: 836– 850 [DOI] [PubMed] [Google Scholar]
- 19. Reid SW, et al. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. U. S. A. 103: 8066– 8071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sato K, Homma M. 2000. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275: 5718– 5722 [DOI] [PubMed] [Google Scholar]
- 21. Sudo Y, et al. 2009. Interaction between Na+ ion and carboxylates of the PomA-PomB stator unit studied by ATR-FTIR spectroscopy. Biochemistry 48: 11699– 11705 [DOI] [PubMed] [Google Scholar]
- 22. Terashima H, Fukuoka H, Yakushi T, Kojima S, Homma M. 2006. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na-driven flagella and required for stator formation. Mol. Microbiol. 62: 1170– 1180 [DOI] [PubMed] [Google Scholar]
- 23. Terashima H, Kojima S, Homma M. 2008. Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270: 39– 85 [DOI] [PubMed] [Google Scholar]
- 24. Terashima H, Kojima S, Homma M. 2010. Functional transfer of an essential aspartate for the ion-binding site in the stator proteins of the bacterial flagellar motor. J. Mol. Biol. 397: 689– 696 [DOI] [PubMed] [Google Scholar]
- 25. Terauchi T, Terashima H, Kojima S, Homma M. 2011. A conserved residue, PomB-F22, in the transmembrane segment of the flagellar stator complex, has a critical role in conducting ions and generating torque. Microbiology. 157: 2422– 2432 [DOI] [PubMed] [Google Scholar]
- 26. Yakushi T, Yang J, Fukuoka H, Homma M, Blair DF. 2006. Roles of charged residues of rotor and stator in flagellar rotation: comparative study using H+-driven and Na+-driven motors in Escherichia coli. J. Bacteriol. 188: 1466– 1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yonekura K, Maki-Yonekura S, Homma M. 2011. The structure of the flagellar motor protein complex PomAB: implications for the torque-generating conformation. J. Bacteriol. 193: 3863– 3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yorimitsu T, Kojima M, Yakushi T, Homma M. 2004. Multimeric structure of the PomA/PomB channel complex in the Na+-driven flagellar motor of Vibrio alginolyticus. J. Biochem. 135: 43– 51 [DOI] [PubMed] [Google Scholar]
- 29. Yorimitsu T, Mimaki A, Yakushi T, Homma M. 2003. The conserved charged residues of the C-terminal region of FliG, a rotor component of the Na+-driven flagellar motor. J. Mol. Biol. 334: 567– 583 [DOI] [PubMed] [Google Scholar]
- 30. Yorimitsu T, Sato K, Asai Y, Kawagishi I, Homma M. 1999. Functional interaction between PomA and PomB, the Na+-driven flagellar motor components of Vibrio alginolyticus. J. Bacteriol. 181: 5103– 5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yorimitsu T, Sowa Y, Ishijima A, Yakushi T, Homma M. 2002. The systematic substitutions around the conserved charged residues of the cytoplasmic loop of Na+-driven flagellar motor component PomA. J. Mol. Biol. 320: 403– 413 [DOI] [PubMed] [Google Scholar]
- 32. Zhou J, Fazzio RT, Blair DF. 1995. Membrane topology of the MotA protein of Escherichia coli. J. Mol. Biol. 251: 237– 242 [DOI] [PubMed] [Google Scholar]
- 33. Zhou J, Lloyd SA, Blair DF. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 95: 6436– 6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou J, et al. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 180: 2729– 2735 [DOI] [PMC free article] [PubMed] [Google Scholar]