Abstract
TonB systems transduce the proton motive force of the cytoplasmic membrane to energize substrate transport through a specific TonB-dependent transporter across the outer membrane. Vibrio vulnificus, an opportunistic marine pathogen that can cause a fatal septicemic disease in humans and eels, possesses three TonB systems. While the TonB1 and TonB2 systems are iron regulated, the TonB3 system is induced when the bacterium grows in human serum. In this work we have determined the essential roles of the leucine-responsive protein (Lrp) and cyclic AMP (cAMP) receptor protein (CRP) in the transcriptional activation of this system. Whereas Lrp shows at least four very distinctive DNA binding regions spread out from position −59 to −509, cAMP-CRP binds exclusively in a region centered at position −122.5 from the start point of the transcription. Our results suggest that both proteins bind simultaneously to the region closer to the RNA polymerase binding site. Importantly, we report that the TonB3 system is induced not only by serum but also during growth in minimal medium with glycerol as the sole carbon source and low concentrations of Casamino Acids. In addition to catabolite repression by glucose, l-leucine acts by inhibiting the binding of Lrp to the promoter region, hence preventing transcription of the TonB3 operon. Thus, this TonB system is under the direct control of two global regulators that can integrate different environmental signals (i.e., glucose starvation and the transition between “feast” and “famine”). These results shed light on new mechanisms of regulation for a TonB system that could be widespread in other organisms.
INTRODUCTION
In Gram-negative bacteria, small molecules cross the outer membrane (OM) by passive diffusion through transmembrane porins; however, substrates that either are poorly permeative through porins (those greater than 600 Da) or are present at very low concentrations require energized transport for their translocation across the OM (54). The TonB complex transduces the proton motive force (PMF) of the cytoplasmic membrane to energize substrate transport through a specific TonB-dependent transporter (TBDT) across the OM (50). In Escherichia coli, this TonB system is a cytoplasmic transmembrane complex composed of the proteins TonB, ExbB, and ExbD that spans the periplasm (45, 50). Our group has described that in Vibrio species, the complex with the TonB2 protein includes a fourth protein, TtpC (59). Until recently, TonB-dependent transport was shown exclusively for the physiological uptake of iron complexes and vitamin B12 (cobalamin). In recent years there have been descriptions of TBDTs involved in the transport of maltodextrins, zinc, nickel, and sucrose in different species, which suggests the possibility that a broader range of nutrients could be transported through this complex (2, 34, 42, 53, 58). In Neisseria gonorrhoeae, an iron-regulated TBDT protein, TdfF, is important in the intracellular growth of this pathogen, and its role remains to be elucidated (24).
Vibrio vulnificus is an opportunistic marine pathogen that can cause a fatal septicemic disease in humans and eels (23, 27). This estuarine bacterium preferentially affects individuals with underlying hepatic diseases and other compromised conditions, such as hemochromatosis and beta thalassemia. In humans, this pathogen frequently causes fatal sepsis with a very rapid progress, resulting in a mortality rate of more than 50% within a few days (27). We have described the role of the TonB1 and TonB2 complexes in V. vulnificus virulence in iron-overloaded mice (1). These two systems are involved in the transport of both the hydroxamate-type siderophore and vulnibactin, the two endogenously produced iron chelators siderophores, as well as heme and hemoglobin (1; A. F. Alice and J. H. Crosa, unpublished results). As expected, the operons coding for those complexes were expressed preferentially under iron-limiting conditions under the control of the Fur protein (1). We have also identified a TonB3 complex in this pathogen that is specifically induced when V. vulnificus grows in human serum with the addition of ferric ammonium citrate (1). This system consists of seven genes arranged in one operon, including a gene that codes for a putative TBDT as well as those encoding the essential proteins of the complex, TtpC3, ExbB3, ExbD3, and TonB3. Moreover, this cluster of genes has homologues in Vibrio parahaemolyticus as well as in Vibrio alginolyticus.
Since it was clear that both the expression of this operon and its genetic arrangement show significant differences from the conventional iron-regulated TonB complexes, we attempted to unveil the mechanisms of transcription activation of the TonB3 system. This knowledge can be used to determine the nature of the substrate(s) that is transported by this system. In this work we describe the essential roles of two global regulators, Lrp and cyclic AMP (cAMP) receptor protein (CRP), in the activation of this operon under conditions that bacteria find under glucose starvation and in the transition between “feast” and “famine.” In addition, we also demonstrate that the TonB3 system is involved in V. vulnificus invasion in iron-overloaded mice.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this work are listed in Table 1. Bacterial strains were grown routinely in Trypticase soy broth with the addition of 1.5% NaCl (TSBS) (V. vulnificus) or in LB broth (Escherichia coli) supplemented with antibiotics as appropriate: ampicillin at 400 μg/ml and chloramphenicol at 2 μg/ml (V. vulnificus) or ampicillin at 100 μg/ml, chloramphenicol at 30 μg/ml, and kanamycin at 50 μg/ml (E. coli). Thiosulfate-citrate-bile salts-sucrose (TCBS) agar was used as a selective medium for V. vulnificus in the conjugations. M9 was used as a minimal medium (16), and when needed, different carbon sources other than glucose were used at 0.5% (wt/vol). Casamino Acids (CAA) or l-leucine was added at the concentrations indicated in the figures, the figure legends, and elsewhere in the text. Human serum samples from at least two healthy donors were pooled, heat inactivated, centrifuged, divided into aliquots, and stored at −80°C. In some experiments, commercial pooled normal human serum was used (catalog number IPLA-SER; Innovative Research, Novi, MI), with results similar to those with sera obtained from other sources. Ferric ammonium citrate (FAC) (200 μg/ml) was added to allow V. vulnificus growth (3).
Table 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant genotype or characteristics | Reference or source |
|---|---|---|
| V. vulnificus strains | ||
| CMCP6 | Wild type | J. Rhee |
| ALE-LAC | CMCP6 ΔlacZ | 33 |
| AA-100 | ΔlacZ Δlrp | This study |
| AA-101 | ΔlacZ Δcrp | This study |
| Δ498 | ΔlacZ Δ528-396 | This study |
| AA-9 | ΔtonB1 ΔtonB2 | 1 |
| AA-12 | ΔtonB1 ΔtonB2 ΔtonB3 ΔlacZ | 1 |
| E. coli strains | ||
| Top 10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| S17-1λpir | λ-pir lysogen; thi pro hsdR hsdM+recA RP4-2 Tc::Mu-Km::Tn7(Tpr Smr) | 57 |
| BL21(DE3) Star | F−ompT hsdSB (rB− mB−) gal dcm rne131 (DE3) | Invitrogen |
| MM294 | F−endA1 hsdR17 supE44 thi-1 spoT1 λ− harboring plasmid pRK2013 | 37 |
| Plasmids | ||
| pCR-Blunt II-TOPO | Blunt-end cloning vector, Kmr | Invitrogen |
| pMMB208 | Broad-host-range expression vector, Cmr Ptac | 40 |
| pTL61T | Contains a promoterless lacZ gene used for transcriptional gene fusions studies, Ampr | 31 |
| pDM4 | Suicide vector with oriR6K, CmrsacB | 39 |
| pET200 | Expression vector, Kmr | Invitrogen |
| pRK2013 | Helper plasmid, Kmr | 22 |
| p732 (p42TL) | pTL61T containing the VV1_0842 promoter region from 42Pst-42Bam fused to lacZ | 1 |
| p632 | pTL61T containing the fragment 42-632-42Bam fused to lacZ | This study |
| p498 | pTL61T containing the fragment 42-498-42Bam fused to lacZ | This study |
| p400 | pTL61T containing the fragment 42-400-42Bam fused to lacZ | This study |
| p300 | pTL61T containing the fragment 42-300-42Bam fused to lacZ | This study |
| p548 | pTL61T containing the fragment 42-548-42Bam fused to lacZ | This study |
| pdint | pTL61T containing the fragment 42-632-42Bam with a deletion between positions −233 and −217 fused to lacZ | This study |
| pMMLrp | pMMB208 derivative containing the lrp gene | This study |
| pMMCrp | pMMB208 derivative containing the crp gene | This study |
| pETLrp | pET200 derivative containing the lrp gene | This study |
| pETCrp | pET200 derivative containing the crp gene | This study |
| p632-477 | PCR fragment amplified with primers 42-632 and 42-477Bam cloned in pCR-Blunt II-TOPO | This study |
| p632-300 | PCR fragment amplified with primers 42-632 and 42-300Rev cloned in pCR-Blunt II-TOPO | This study |
| p300-42B | PCR fragment amplified with primers 42-300 and 42-Bam cloned in pCR-Blunt II-TOPO | This study |
DNA manipulations and sequence analysis.
Plasmids were prepared using the QIAprep miniprep kit (Qiagen, Valencia, CA). Touchdown PCRs were performed with either Taq polymerase (Invitrogen, Carlsbad, CA) or Vent polymerase (New England BioLabs Inc., Ipswich, MA) using the following conditions: 95°C for 4 min; 30 cycles of 95°C for 30 s, 63°C for 1 min (the temperature of this step was lowered 0.3°C for each cycle), and 72°C for 1 min; and a final extension step at 72°C for 10 min. Digestions and ligations were performed according to the manufacturer's instructions (New England BioLabs Inc., Ipswich, MA). DNA sequencing reactions were carried out by the Vollum Sequencing Core Facility (Oregon Health and Science University [OHSU]).
Construction and complementation of the V. vulnificus mutants.
In-frame deletions of the entire coding sequences of genes were generated using splicing by overlap extension (SOE) PCR (55). Upstream and downstream regions (approximately 700 to 800 bp) flanking each gene were amplified with specific primers, and both fragments were mixed and ligated in a new PCR with primers 1 and 2 for each gene (see Table S1 in the supplemental material). The amplified fragment was cloned in the pCR-Blunt II vector (Invitrogen, Carlsbad, CA), digested with appropriate restriction enzymes, and subcloned in the suicide vector pDM4 previously digested with the same restriction enzymes. E. coli S17-1λpir transformed with the pDM4 derivatives was conjugated with V. vulnificus, and exconjugants were selected on TCBS agar with chloramphenicol. Clones were subsequently screened for chloramphenicol sensitivity and sucrose resistance, and deletions within the genes of interest were confirmed by PCR.
For the complementation experiments, each gene was amplified by PCR with primers containing restriction sites as indicated in Table S1 in the supplemental material. The fragments were cloned in the pCR-Blunt II vector (Invitrogen, Carlsbad, CA), sequenced, and then subcloned in pMMB208 under the control of the Ptac promoter. The constructs were transferred to V. vulnificus strains by triparental conjugation using the helper plasmid pRK2013. In order to induce transcription of the cloned genes, 0.1 mM or 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to the medium.
RNA isolation and cDNA synthesis.
Strains were grown to the determined optical density at 600 nm (OD600) in minimal medium or for various times in human serum with the addition of ferric ammonium citrate (HS-FAC). Total RNA was extracted from each strain using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions, treated with RNase-free DNase, quantified, and stored at −80C. Usually, 1 μg of RNA thus treated was used for cDNA synthesis with the Quantitect reverse transcription kit (Qiagen, Valencia, CA).
qRT-PCR.
cDNA synthesized as described above was used in quantitative real-time PCRs (qRT-PCRs) with Power SYBR green PCR master mix (Applied Biosystems, Carlsbad, CA) and 500 nM (each) forward and reverse primers (see Table S1 in the supplemental material). Relative expression was determined by calculating 2−ΔΔCT using the 23S rRNA gene as an internal control. Reactions were conducted in triplicate in a StepOne real-time PCR system (Applied Biosystems, Carlsbad, CA). Statistical analysis was performed by one-way analysis of variance (ANOVA) with Bonferroni's posttest.
Overexpression and purification of the V. vulnificus recombinant proteins.
The DNA fragments coding for Lrp and CRP were amplified using primers described in Table S1 in the supplemental material and individually cloned into the expression vector pET200 (Invitrogen, Carlsbad, CA). A recombinant plasmid with the correct sequence was used for expression of each protein in E. coli BL21(DE3) Star. In general, 200 ml of LB supplemented with kanamycin was inoculated with an overnight culture of the strain harboring the plasmid of interest, and when the OD600 reached 0.6, 1 mM IPTG was added to induce the expression of the protein. The culture was incubated for another 4 h at 37°C, centrifuged, washed, and resuspended in lysis buffer (50 mM Na2HPO4, 300 mM NaCl, 10 mM imidazole, pH 8.0). Cells were sonicated and centrifuged, and the supernatant was passed through an Ni-nitrilotriacetic acid resin (Qiagen, Valencia, CA) according to the manufacturer's instructions. After the elution from the column, each protein was dialyzed overnight at 4°C against buffer D (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM MgCl2, 2 mM dithiothreitol [DTT], 10% glycerol) and the concentration determined with a bicinchoninic acid assay kit (Pierce, Rockford, IL). The proteins were aliquoted and stored at −80°C.
Electrophoretic mobility shift assay (EMSA).
The gel mobility shift assay was performed using the Roche second-generation digoxigenin (DIG) gel shift kit (Roche, Indianapolis, IN). DNA fragments were amplified by PCR and 3′ end labeled with DIG-11-ddUTP using terminal transferase as described by the manufacturer. After the labeling efficiency was determined, each of the labeled probes (4 nM) was incubated with increasing amounts of the protein of interest in binding buffer A [20 mM HEPES (pH 7.6), 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM DTT, 0.2% (wt/vol) Tween 20, 30 mM KCl, and 1 μg/μl poly(dI-dC)] and incubated for 20 min at 37°C. When CRP was analyzed, 0.2 mM cAMP was added to binding buffer A. For competition experiments, a 4 nM concentration of the labeled probe was incubated with increasing amounts (usually 400 nM) of the unlabeled specific probe. Samples were separated on 6% polyacrylamide gels (Invitrogen, Carlsbad, CA) and run in 0.5× Tris-borate-EDTA (TBE) for 90 min at 100 V. The DNA-protein complexes were transferred to positively charged Hybond N+ nylon membranes (GE Healthcare, Piscataway, NJ) and the chemiluminescent signal detected according to the manufacturer's instructions (Roche, Indianapolis, IN) by using an ImageQuant LAS4000 camera (GE Healthcare, Piscataway, NJ).
DNase I footprinting experiments.
DNase I footprinting experiments were performed on 32P-end-labeled fragments using either [γ-32P]ATP (7,000 Ci/mmol; MP Biomedicals, Solon, OH) or [α-32P]dATP (3,000 Ci/mmol; Perkin-Elmer, Waltham, MA). For 5′ labeling of probe C, a PCR fragment amplified with primers M13F and M13R using plasmid p300-42B as a template was labeled with T4 polynucleotide kinase, digested with SacI, and purified from 5% native polyacrylamide gels. For 3′ labeling of probe D, a PCR fragment amplified using plasmid p632-300 with primers 632Pst-M13F was digested with SpeI-EcoRV, end labeled with DNA polymerase I large (Klenow) fragment (Invitrogen, Carlsbad, CA) as indicated by the manufacturer, and gel purified as described above. Binding reactions (with 20-μl mixtures) were carried out under the same conditions as described for EMSAs, and DNase I (Promega) treatment was performed for 1 min at room temperature. All samples were analyzed in either 5% or 6% polyacrylamide–8 M urea gels, run at 60 W, dried, and scanned in a Storm 825 PhosphorImager (GE Healthcare, Piscataway, NJ). Gels were calibrated with Maxam-Gilbert G+A reactions prepared as described previously (10).
β-Galactosidase assays.
The upstream region of the VV1_0842 (TonB3 cluster) gene was amplified with primers indicated in Table S1 in the supplemental material, cloned in the pCR-Blunt II vector (Invitrogen, Carlsbad, CA), sequenced, and subcloned in the pTL61T vector previously digested with the corresponding restriction enzymes, generating the plasmids described in Table 1. The empty vector and plasmids thus constructed were transferred to a V. vulnificus ΔlacZ wild-type strain or the various mutant strains by triparental conjugation. Bacterial strains harboring the plasmids with the lacZ fusions or the empty vector were grown under the conditions described for each figure. Two hundred microliters of each culture was centrifuged, resuspended in buffer Z, and used in the assay. β-Galactosidase activities from cultures growing in minimal medium were determined as described previously (38). Due to the turbidity of the human serum used in this work, when samples obtained from this source were analyzed, the counts of CFU ml−1 at each time point were used to normalize the β-galactosidase activities instead of the OD600 (1). Values of β-galactosidase activities obtained from cells growing in human serum are comparable; however, they were not comparable to those obtained from cells growing in minimal medium due to the different units used to normalize them (CFU and OD600, respectively). The levels of β-galactosidase from lacZ fusions were compared using one-way ANOVA with Tukey's posttest. All experiments were repeated at least three times (with some of them performed at least 10 times) with two replicates each, and the results were expressed as means ± standard deviations (SD) for each group examined. All tests were performed with GraphPad Prism 4.0, and statistical significance was defined as a P value of <0.05.
Fishing ligand experiment.
The protocol used for the identification of the proteins bound to probe A was adapted from a method previously described by Nordhoff et al. (46). The crude extracts used in these experiments were prepared as follows: 10 ml of a culture of V. vulnificus CMCP6 growing in HS-FAC for 6 h was centrifuged, resuspended in binding buffer A (with the omission of Tween 20), sonicated, and centrifuged. The protein concentration was measured as described above, and the sample was kept at −20 C. Four micrograms of a biotinylated PCR fragment amplified with primers 42-632 (biotinylated) and 42-477Bam was incubated with 400 μg Dynabeads M-280–Streptavidin (Invitrogen, Carlsbad, CA) for 15 min at room temperature in binding and washing buffer B-W (5 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, 1 M NaCl). The DNA-beads were washed 3 times with buffer B-W and incubated for 20 min at 37°C using gentle rotation with 150 μg of total proteins from the V. vulnificus crude extract obtained as described above in the presence of 1 μg poly(dI-dC) and 0.2% (wt/vol) Tween 20. After the incubation, the mixture was washed four times with buffer W (20 mM Tris-HCl [pH 7.5], 0.1 mM DTT, 100 μM NaCl, 1 mM EDTA, 1× protease inhibitor). The first wash also included 1 μg of poly(dI-dC). The magnetically separated beads were resuspended in 40 μl of 1% SDS and boiled for 10 min. The supernatant was analyzed in the Proteomic Shared Resource at OHSU. A negative-control sample was also prepared in the same manner as described above but with the omission of the biotinylated DNA, which was replaced with the same volume of buffer. At least two positive and two negative samples were analyzed independently, and proteins were identified with false-discovery rate of 3%.
Virulence experiments.
Competitive index (CI) experiments were performed as described previously (1). Briefly, bacterial strains were cultured in TSBS, and then equal volumes were mixed and serial dilutions performed. A dilution (usually containing ca. 5,000 to 8,000 CFU) was injected subcutaneously (s.c.) in 4- to 6-week-old CD-1 mice (Charles River Laboratories, Wilmington, MA). In the iron-overloaded mouse model, 900 μg of FAC is injected intraperitoneally (i.p.) 30 min prior to the bacterial challenge (1). It has been previously demonstrated that mice are more susceptible to V. vulnificus infections when iron is injected i.p. before the inoculation of the bacterial cells (66). Animals were checked after approximately 10 h of inoculation, and when they showed signs of the disease (e.g., lethargy, slow movements, or lack of appetite), they were euthanized with CO2 according to IACUC regulations. A skin sample (1 cm by 1 cm around the inoculation site), spleen, and liver were aseptically extracted and homogenized by using a Seward Stomacher lab blender in the presence of phosphate-buffered saline (PBS) (1 ml for skin and spleen and 2 ml for liver). Serial dilutions were performed in PBS, and the dilutions were plated on TSBS plus 1.5% agar (TSAS) plates supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) to determine the CFU of the strains under analysis. CI values were obtained as described in reference 61. The statistical analysis was performed by using one-way ANOVA with Bonferroni's posttest with GraphPad Prism 4.0 software.
RESULTS
The TonB3 system is involved in V. vulnificus invasion in iron-overloaded mice.
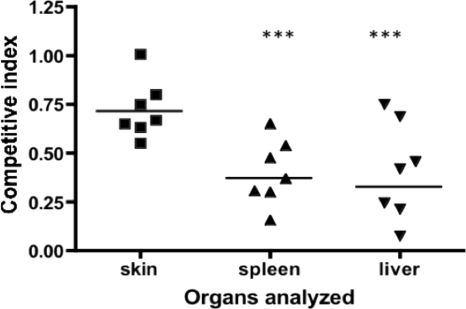
The TonB3 system in V. vulnificus is unique in both its genetic organization and the conditions under which the expression is induced. As shown in Fig. 1, this operon includes a gene that codes for a putative TonB-dependent receptor (TBDR) (VV1_0842) as well as those encoding the TonB3 protein and the accessory proteins TtpC3, ExbB3, and ExbD3 (VV1_0847, VV1_0844, VV1_0845, and VV1_0846, respectively). We have previously shown that this operon was specifically induced when V. vulnificus was grown in human serum with the addition of FAC (HS-FAC) (1). In this pathogen the TonB1 and TonB2 systems are important virulence factors; however, the ΔtonB3 mutant strain did not show any change in the 50% lethal dose (LD50) compared to that of the wild-type strain (1). To determine if this system has any role during the invasion of V. vulnificus and progress of infection, we performed competitive index (CI) assays, which we have described as a very sensitive test to examine the contribution of a gene to the virulence of this pathogen when LD50 values are similar to that of the wild-type strain (1). A CI test between the ΔtonB1 ΔtonB2 double mutant (TonB3+) and the tonB triple mutant (TonB3−) was carried out in the iron-overloaded mouse model as described in Materials and Methods (s.c. inoculations). We used a ΔtonB1 ΔtonB2 genetic background for these experiments because we do not know if these two systems could complement a tonB3 mutation in vivo. The results obtained with livers and spleens from infected animals showed that the strain in which the three tonB systems were mutated is affected in the invasion of V. vulnificus, highlighting a possible role for the TonB3 cluster in this process (Fig. 2).
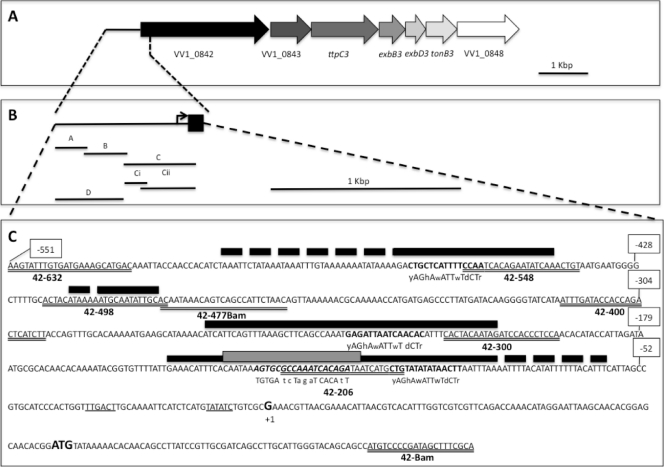
Fig 1.
Organization of the tonB3 operon and the regulatory region. (A) Genetic arrangement of the tonB3 operon. The gene names are based on homology to the counterpart of the TonB2 systems of V. vulnificus and other vibrios. (B) Close-up of the regulatory region analyzed in this work. The different lines indicate the approximate locations of the probes used throughout this work. The length of the promoter region and the sizes of the probes in this panel are not drawn to scale. (C) Sequence of the promoter region from position −551 (relative to the transcription start site [+1]) to position +81 of the partial coding sequence of the VV1_0842 gene. The likely −10 and −35 elements are underlined, and the identified start point of the transcription and the predicted start codon are shown in bold. The extents of the protection identified by DNase I footprinting for each protein are indicated with black (Lrp) and gray (CRP) boxes, and the locations of the predicted binding sites as determined by similarity to consensus motifs (13, 17, 28) are indicated in bold. Binding site positions that could not be delimited by DNase I footprinting are indicated with dotted lines. The positions of the primers used for amplification of the probes used in EMSA and DNase I footprintings as well as those used in the construction of the lacZ fusions are indicated with double underlines. Relevant nucleotide positions relative to the transcription start site are also indicated.
Fig 2.
Competitive indices for tonB mutant strains. Mixtures of the strains analyzed (0.1 ml) were injected s.c. in iron-overloaded animals as described in Materials and Methods. When animals showed signs of the disease, organs were removed and homogenized and bacterial cells plated on TSBS plus 1.5% agar (TSAS)–X-Gal plates. Bars represent the geometric mean CI value for each organ. Experiments were performed twice. ***, P < 0.001 based on one-way ANOVA with Bonferroni's posttest.
A region located far upstream of the promoter is essential for full activation of the tonB3 operon.
Since the TonB3 system may have a role in V. vulnificus infection and is specifically induced when the bacteria are growing in human serum, we attempted to determine the function of this system under those conditions. Taking into account that we could not detect a full induction of the expression of this operon in either rich medium or CM9 minimal medium (1), we assumed that some component(s) of the human serum was specifically important for this system induction. Considering the complexity of the components of human serum, we hypothesized that by determining the identity of the activator(s) involved in that particular induction, we could understand the role of this system in V. vulnificus physiology.
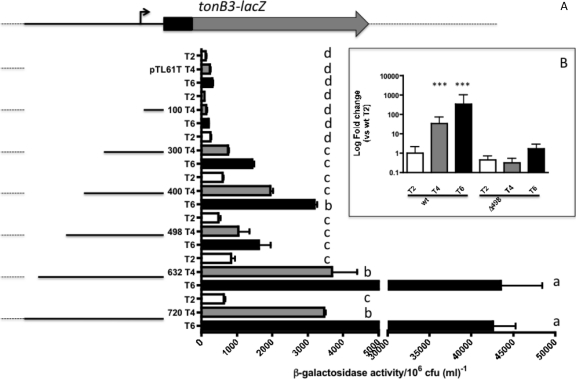
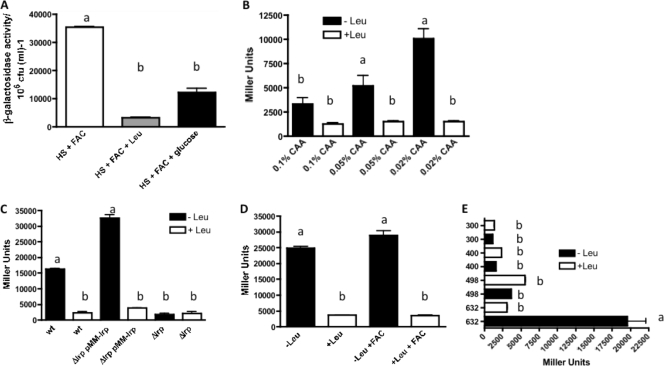
We first determined the minimal region of the promoter needed for its expression by constructing various transcriptional fusions of the promoter region of this operon to the lacZ gene, which were conjugated into a V. vulnificus lacZ mutant strain. The expression was analyzed at various times during growth, with the maximum expression observed after 6 h, when the cells reached the late log phase (1). Figure 3A shows that three different levels of activity were measured after 6 h of growth. In one group there was very low activity, as in the case of the empty vector pTL61T and the p100 construct; the absence of expression in the latter construct corresponds with the deletion of the −35 region and confirms our previous results, where we determined the start point of the transcription being at bp −81 from the ATG ((1) (Fig. 1). A second group of constructs, from bp −218 (p300) to −420 (p498) relative to the transcription start site, show an intermediate level of activity. Interestingly, constructs including the region from nucleotide −551 to −420 (constructs p632 and p720) were shown to be essential to achieve the full activation of this promoter observed in HS-FAC. Further confirmation of these results was obtained by using a strain with a deletion of the region from bp −528 to −396 in the chromosome (here named Δ498). RNA extracted from this strain growing in HS-FAC was used to analyze the expression of the tonB3 gene by qRT-PCR, which was compared to that of the wild type. As expected, the transcription of this operon was induced in the wild-type strain growing under these conditions; however, the Δ498 derivative showed a lower level of expression, confirming the results obtained with the lacZ fusions (Fig. 3B).
Fig 3.
Expression of various tonB3 promoter deletions in human serum. (A) The deletions of the promoter region of the TonB3 operon were cloned in front of the promoterless lacZ gene in the pTL61T vector and plasmids conjugated into the ΔlacZ strain. Cells were grown in human serum plus FAC (200 μg/ml) and samples taken at different times to determine β-galactosidase activities (white bars [T2], 2 h; gray bars [T4], 4 h; and black bars [T6], 6 h after inoculation). Due to the turbidity of the human serum, the OD600 was not recorded and activity was normalized to 106 CFU ml−1. The results shown are the means from three to seven independent experiments with standard deviations. Those bars showing no significant difference are labeled with the same letters, and those showing significant difference (P < 0.05) are labeled with different letters, based on one-way ANOVA with Tukey's posttest. (B) Expression of the tonB3 gene in the wild-type (wt) and Δ498 strains growing in HS-FAC as determined by qRT-PCR. RNA extracted from the wild-type and Δ498 strains growing in HS-FAC was reverse transcribed and cDNAs used in qRT-PCR as described in Materials and Methods. Relative expression of the tonB3 gene was determined by calculating 2−ΔΔCT using the 23S rRNA gene as an internal control. ***, P < 0.001 based on one-way ANOVA with Bonferroni's posttest for values for the wild type compared to the mutant strain.
The Lrp protein is needed for the activation of the tonB3 operon.
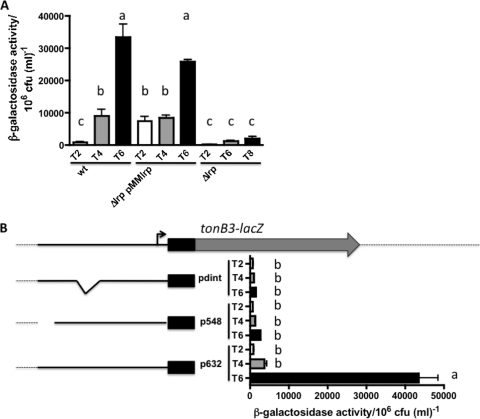
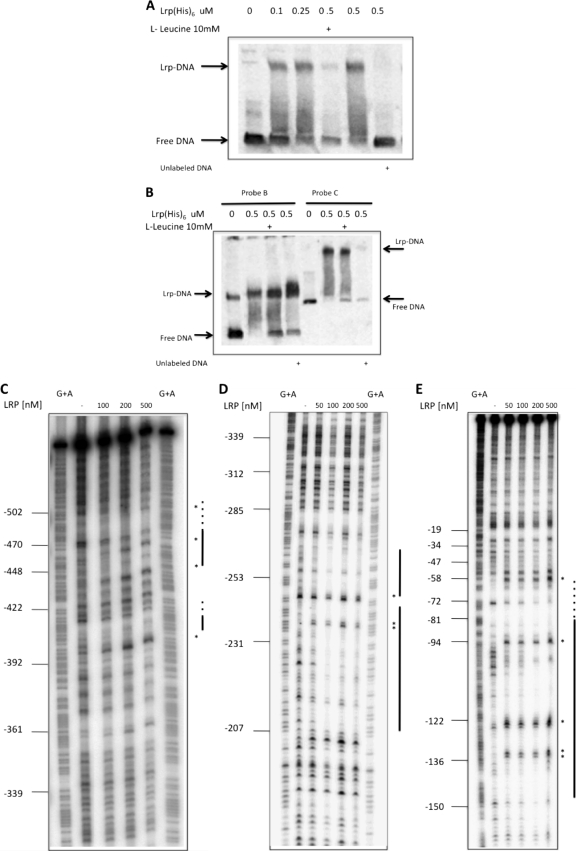
The results described above suggested that the region from bp −551 to −420 was acting in cis in the activation of the tonB3 promoter. We hypothesized that a transcriptional regulator(s) can bind this region and activate the expression of the tonB3 operon. To determine the identity of this protein(s), we performed fishing ligand experiments as described in Materials and Methods by using cell extracts obtained from V. vulnificus growing in HS-FAC (inducing conditions). A total of 158 proteins were identified between the positive and negative samples. Peptides corresponding to the open reading frame (ORF) VV1_2951 (encoding the leucine-responsive regulator Lrp) were highly represented in the positive sample, while they were absent in the negative controls. This protein belongs to a group of seven transcription factors that in E. coli control approximately 50% of the regulated genes (5). To confirm the role of this protein in the activation of this operon, we studied the tonB3-lacZ transcriptional fusion (construct p632) under inducing conditions in both the Δlrp mutant and the complemented strains. Compared with that in the wild-type strain, we could observe a significant reduction in the activity of this promoter in the mutant strain, which is restored to wild-type values in the complemented strain (Fig. 4A). It is worth noting that when we overexpressed the Lrp protein with 1 mM IPTG, the protein level reached was toxic for the bacterial cells; however, lower levels of IPTG (0.1 mM) allowed sufficient expression to observe full activation of the operon without affecting the viability of the cells (data not shown). To address whether the role of Lrp in the activation was a result of a direct binding of this protein to the 632-to-498 region, we purified the Lrp(His)6 protein and used it in EMSAs with the corresponding labeled probe. A shift in the mobility of the probe was detected only when the protein was added to the reaction mixture in the presence of poly(dI-dC) as a nonspecific competitor. This shift could be reversed when unlabeled competitive DNA was used, confirming the specificity of the binding observed (Fig. 5A). Usually the amino acid l-leucine might act as a coeffector of Lrp, and it potentiates, overcomes, or has no effect on the function of Lrp upon its target genes (9, 44, 60). We then tested the possibility that the presence of this amino acid in the binding buffer of the EMSAs could modify the binding of Lrp to this DNA fragment. As can be observed in Fig. 5A, Lrp does not bind to the target DNA in the presence of l-leucine, suggesting a role for this amino acid in the regulation of this operon. Additionally, we also tested whether this protein could bind other regions of the promoter. In Fig. 5B we show that Lrp could bind to a probe that includes the transcription start point as well as the putative binding site of the RNA polymerase (probe C, from nucleotide −218 to +81 with respect to the ATG). Similarly, when the area comprising nucleotides −420 to −218 (probe B) was used, we observed a specific shift in the mobility of the probe upon the binding of Lrp. Furthermore, the presence of l-leucine affected the observed binding of Lrp to these two DNA regions (Fig. 4B).
Fig 4.
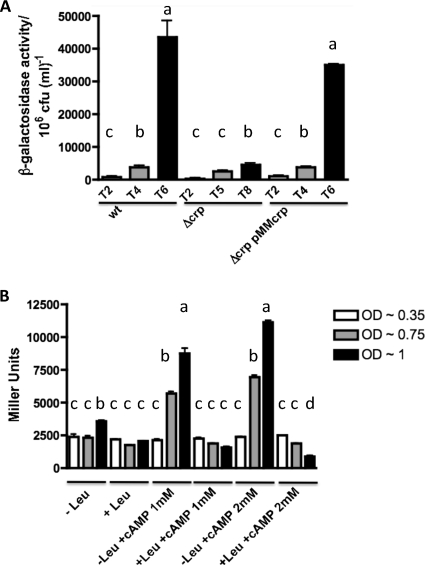
The lrp gene is essential for tonB3 induction in HS-FAC. (A) Wild-type, Δlrp, and Δlrp pMMlrp strains harboring plasmid p632 containing the tonB3-lacZ fusion were grown in HS-FAC. Samples were removed at various time points and analyzed as described for Fig. 3A. (B) Constructs p548 and pdint were conjugated into the V. vulnificus ΔlacZ strain, samples were removed at various times, and β-galactosidase activity was measured as described above. The bars represent means ± standard deviations (n = 3). Bars showing no significant difference are labeled with the same letters, and those showing a significant difference (P < 0.01 for panel A and P < 0.001 for panel B) are labeled with different letters, based on one-way ANOVA with Tukey's posttest.
Fig 5.
Analysis of the binding of the Lrp(His)6 protein to the tonB3 promoter region. (A and B) EMSA with different promoter regions of this operon that were amplified, labeled as described in Materials and Methods (probe concentration, 4 nM), and incubated with increasing concentrations of Lrp(His)6 as indicated in each panel. (A) Probe A amplified with primers 42-632 and 42-477Bam; (B) probe B amplified with primers 42-498 and 42-300Rev and probe C amplified with primers 42-300 and 42Bam. Probes are illustrated in Fig. 1B. In each panel it is indicated when 10 mM l-leucine was added to the binding buffer as well as when unlabeled probe DNA (400 nM) was used for competition. Free labeled DNA and Lrp(His)6-DNA complexes are indicated. (C, D, and E) DNase I footprinting analysis of the tonB3 promoter with purified Lrp(His)6. End-labeled probes D (C and D) and C (E) were incubated with various concentrations of Lrp(His)6 as indicated in each panel and treated with DNase I. Gels were calibrated using G+A sequencing reactions (G+A), and relevant positions are indicated. The locations of DNA binding sites for Lrp(His)6 are shown by black lines, and putative binding sites with intrinsic resistance to DNase I are shown with dotted lines. Hypersensitive sites due to Lrp(His)6 binding are labeled with asterisks.
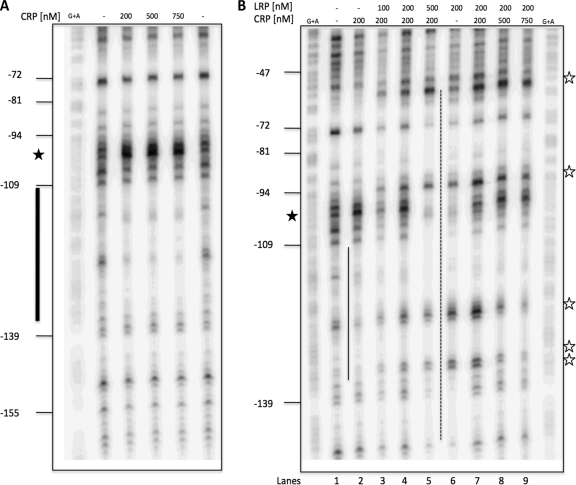
We next determined the binding sites of Lrp in the tonB3 promoter region by performing DNase I footprinting assays. When a fragment that includes the far-upstream region as well as the middle region (probe D) was used together with purified Lrp protein, we observed high levels of protection at positions −509 to −443 and −415 to −398 (Fig. 5C), which overlaps a sequence with similarity to the Lrp consensus (Fig. 1) (13, 17). The protection observed also lead to phased hypersensitivity (26) that is commonly observed with this protein (64). However, it should be noted that due to the intrinsic resistance to DNase I of the region comprising approximately nucleotides −509 to −477 and −415 to −410, it was difficult to delimit the binding site for Lrp at those locations. Interestingly, another large protected region was observed in the middle region, which spans positions −265 to −208 with hypersensitive sites that are consistent with the DNA bending or wrapping described for this protein (Fig. 5D) (64). A sequence with similarity to the Lrp consensus was also identified at this position (Fig. 1) (13, 17). The analysis of the proximal region showed an additional Lrp binding site at position −147 to approximately −59, which lies right upstream of the −35 region, suggesting that this protein could interact with the RNA polymerase (Fig. 5E). Again it is worth noting the presence of a region with intrinsic resistance to DNase I between positions −81 and −59. As expected, the protected region overlapped a sequence with high similarity to the Lrp consensus sequence (Fig. 1) (13, 17). The binding of the Lrp protein to all those regions is consistent with our EMSA results.
Considering the previous results, two new lacZ transcriptional fusions were constructed and analyzed in cells growing in HS-FAC. In one of them we deleted a DNA fragment between positions −551 and −461 (construct p548), while in the second one we deleted nucleotides −233 to −217 (construct pdint). According to our footprinting experiments, Lrp binds to these regions of the tonB3 promoter, and when they are deleted, the induction of the expression of the lacZ fusions is abolished (Fig. 4B). Altogether, these results confirm the role of Lrp in the control of the transcription of the tonB3 operon.
Activation of the tonB3 operon in minimal medium under starvation conditions.
It was clear from our EMSA experiments that l-leucine affected the Lrp binding to the DNA target regions. To determine if this amino acid has any role in the expression of the tonB3 promoter in vivo, we studied its expression in cells growing in HS-FAC supplemented with 10 mM l-leucine. It is worth mentioning that the concentration of l-leucine in sera from healthy individuals is 123 μmol liter−1 (range, 98 to 148 μmol liter−1) (51). The activity of the tonB3 promoter obtained under these conditions was reduced at least 10-fold compared to that of the wild type growing in serum without the amino acid supplementation (Fig. 6A).
Fig 6.
Effects of l-leucine and glucose on tonB3 promoter expression. (A) The V. vulnificus ΔlacZ strain harboring the p632 construct was grown in HS-FAC or in the same medium with the addition of 10 mM l-leucine or 0.5% glucose. Samples were removed at various times, and β-galactosidase activity was measured as described for Fig. 3A. Values obtained at 6 h after the inoculation are shown. (B) The V. vulnificus ΔlacZ strain harboring the p632 construct was grown in M9 with 0.5% glycerol and various concentrations of CAA in the presence or absence of 10 mM l-leucine up to an OD600 of ∼0.7, samples were removed, and β-galactosidase activity was measured as described in Materials and Methods. (C) Wild-type, Δlrp, and Δlrp pMMlrp strains harboring plasmid p632 containing the tonB3-lacZ fusion were grown in M9–0.5% glycerol–0.02% CAA in the presence or absence of 10 mM l-leucine, and β-galactosidase activity was measured as described above. (D) The V. vulnificus ΔlacZ strain harboring the p632 construct was grown in M9–0.5% glycerol–0.02% CAA or in the same medium with the addition of 10 mM l-leucine. When indicated, 200 μg/ml FAC was added. Samples were removed at an OD600 of ∼0.7 and β-galactosidase activity measured as described above. (E) V. vulnificus ΔlacZ strains harboring the p300, p400, p498, or p632 construct were grown in M9–0.5% glycerol–0.02% CAA or in the same medium with the addition of 10 mM l-leucine. Samples were removed at an OD600 of ∼0.7 and β-galactosidase activity measured as described above. The bars represent means ± standard deviations (n = 3). Those bars showing no significant difference are labeled with the same letters, and those showing a significant difference (P < 0.001 for panels A, C, D, and E and P < 0.05 for panel B) are labeled with different letters, based on one-way ANOVA with Tukey's posttest.
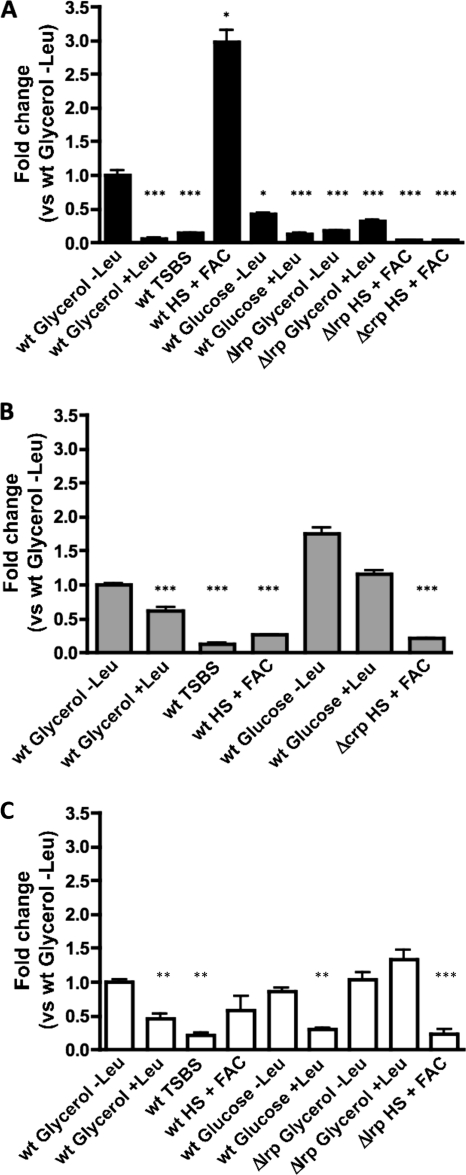
Although we observed induction of this operon when V. vulnificus was grown in HS-FAC, our attempts to obtain similar levels of expression in both rich and minimal media in the past were unsuccessful (1). With the discovery of the roles of Lrp and l-leucine in the control of the transcription of this operon, we explored its transcription under conditions where Lrp is usually expressed at high levels and/or the presence of this amino acid could play any regulatory role. In E. coli the expression of Lrp is repressed in rich medium as well as in minimal medium with 1% CAA (11, 29, 32, 44). We determined the expression level of the lrp gene in cells growing in TSBS, determining that it was approximately 3-fold lower than that in cells growing in HS-FAC (P > 0.05; no significant difference was observed) (see Fig. 8B); however, in agreement with our previous observations, the expression of the tonB3 gene in TSBS was ∼33-fold lower than that observed in HS-FAC (P < 0.001) (see Fig. 8A). Since the Lrp levels appeared not to be the main reason for these lower levels of expression of the tonB3 operon in rich medium, we assumed that the presence of l-leucine (and other amino acids) in TSBS possibly could be the reason for the low level of expression observed in that medium.
Fig 8.
Expression of the tonB3, lrp, and crp genes under various conditions. Total RNA was extracted from the different strains growing under the indicated conditions. The mRNA levels (represented by 2−ΔΔCT values) of various genes in the different strains then were measured by qRT-PCR and expressed as the fold change from that measured in the wild-type strain growing in M9–glycerol–0.02% CAA without the addition of l-leucine. (A) tonB3; (B) lrp; (C) crp. The bars represent means ± standard deviations (n = 3). The significance of differences was analyzed by one-way ANOVA with Bonferroni's posttest. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We tested this hypothesis by analyzing the expression of the tonB3-lacZ fusion in cells growing in M9 minimal medium with various carbon sources and CAA concentrations. Lowering the CAA concentration in minimal medium with glycerol as a carbon source was sufficient to obtain high levels of expression of the tonB3 mRNA, which were comparable to, although still ∼3-fold lower than, those observed in HS-FAC (P < 0.05) (see Fig. 8A). Likewise, high levels of β-galactosidase were detected in the tonB3-lacZ fusion strain analyzed in this medium, with the highest levels measured when the cells were growing in the presence of glycerol and 0.02% CAA (Fig. 6B). In complete agreement with our previous finding in HS-FAC, when l-leucine was added to the minimal medium, the expression was repressed (Fig. 6B). In addition, when the Δlrp mutant strain was analyzed in this medium, the expression levels of the tonB3 promoter were reduced at least 10-fold compared to those of the wild type, and adding l-leucine only had a marginal effect (Fig. 6C). When the complemented strain was analyzed under similar conditions, β-galactosidase activity was restored to even higher levels than in the wild type (Fig. 6C), indicating that overexpression of this gene activates the expression of the tonB3 promoter at higher levels than in the wild type. It is also worth noting that the Δlrp mutant strain has longer doubling times in this medium than the wild type (∼3.5 h versus ∼2 h), suggesting that the Lrp protein has an important role in the physiology of V. vulnificus under these conditions or that a cofactor needed in the mutant is missing in the minimal medium used (32). As expected, the complemented strain showed a doubling time similar to that of the wild type. The results detailed above showed for the first time a complete agreement between the results obtained from V. vulnificus cells growing in HS-FAC and those obtained from cells growing in M9 with glycerol as the sole carbon source and low CAA concentrations. Furthermore, analysis of the expression of the tonB3-lacZ fusion in M9 glycerol with low CAA and with the addition of 200 μg/ml FAC confirmed that high iron concentrations do not play any role in the expression of this operon (Fig. 6D). Analysis of the p498, p400, and p300 lacZ fusion constructs in minimal medium with glycerol and low CAA gave results similar to those obtained with cells growing in HS-FAC (i.e., low levels of activation) (Fig. 6E).
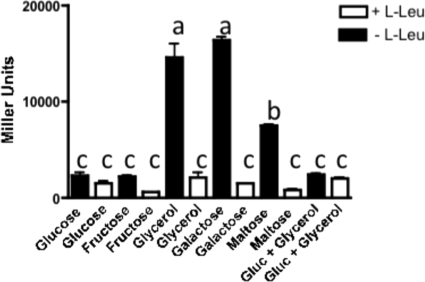
As mentioned above, we tested the expression of this operon in cells growing with various carbon sources; importantly, in the presence of glucose the expression was reduced at least 5- to 7-fold (P < 0.001) compared to that in the cells growing with glycerol as the sole carbon source (Fig. 7 and 8A). In addition, the expression of the tonB3-lacZ fusion was reduced only 2-fold when l-leucine was added to the medium (Fig. 7 and 8A). Similar results were obtained with fructose, another phosphoenolpyruvate phosphotransferase transport system (PTS) sugar, as the sole carbon source (Fig. 7). However, when galactose or maltose (non-PTS sugars) were used as carbon sources in minimal medium, there was an increase in the activity of the tonB3-lacZ fusion, which in the case of galactose was identical to that observed when glycerol was used as the carbon source (Fig. 7). Furthermore, when the cells were growing in the presence of glucose and glycerol, the activity was 5- to 7-fold (P < 0.001) lower than that observed in glycerol alone, and no effect was observed when l-leucine was present in the medium (Fig. 7). In agreement with these results, the addition of glucose to the HS-FAC reduced the expression of the tonB3 promoter almost 3-fold (P < 0.001) at 6 h, when the maximum expression is observed in the absence of this sugar (Fig. 6A). Altogether, these results suggested the involvement of a mechanism of catabolite repression in the expression of the tonB3 operon and/or in the expression of the lrp gene. To address whether the expression of the lrp gene is affected by the presence of glucose in the medium, we carried out qRT-PCR assays from RNA obtained from cells growing in M9 with low CAA and with either glucose or glycerol as a carbon source. There were no significant changes in the lrp transcript level when measured in the presence of glucose and compared to that with glycerol, indicating that this gene is not under catabolite repression control (Fig. 8B).
Fig 7.
Expression of the tonB3 promoter with various carbon sources. The V. vulnificus ΔlacZ strain harboring the p632 construct was grown in M9 with various carbon sources (0.5% each) and 0.02% CAA in the presence or absence of 10 mM l-leucine and β-galactosidase activity measured as described in Materials and Methods. The bars represent means ± standard deviations (n = 3). Those bars showing no significant difference are labeled with the same letters, and those showing a significant difference (P < 0.01) are labeled with different letters, based on one-way ANOVA with Tukey's posttest.
CRP is also involved in the activation of the tonB3 operon.
The results described in the previous section strongly suggested the possibility that the tonB3 operon is under catabolite repression control. It is known that the cAMP receptor protein (CRP) is involved in the activation and/or repression of various genes and operons in response to the presence of glucose in the medium (6, 28, 56). To test the hypothesis that this protein also acts directly on the promoter region of this operon and/or indirectly through the regulation of lrp, we evaluated the expression of the tonB3 operon and the lrp gene in a Δcrp mutant strain. Unfortunately, the CMCP6 V. vulnificus Δcrp mutant strain cannot grow with glycerol as the sole carbon source in minimal medium, so we could not directly address the expression of those genes in cells growing under those conditions. However, this mutant strain was able to grow in HS-FAC, although the doubling time was longer than that of the wild type (∼2.5 h versus ∼1.5 h), so we were able to evaluate the expression of the tonB3 system under this condition. The expression of the tonB3 promoter was significantly lower in the Δcrp mutant than in the wild type as measured by β-galactosidase activity (Fig. 9A). As expected, the levels of the tonB3 mRNA determined by qRT-PCR were lower in this mutant strain (Fig. 8A). Furthermore, the complemented strain showed levels of expression similar to those of the wild type (Fig. 9A), confirming the role of this protein in the regulation of this promoter at the transcription initiation level. The expression of the lrp gene was not significantly different in this Δcrp mutant strain growing in HS-FAC than in the wild type growing in the same medium (P > 0.05) (Fig. 8B). This is important, as in E. coli lrp is responsive to both growth rate and CRP (29, 41). Additionally the expression of the crp gene in the wild-type strain growing in minimal medium with glycerol and low CAA is similar to that in the same strain growing in HS-FAC (Fig. 7C). However, we observed lower levels of crp mRNA when l-leucine was added to the minimal medium (Fig. 8C). This effect, which was relieved in a Δlrp mutant strain growing under the same conditions, suggests a possible role for Lrp in the control of crp transcription.
Fig 9.
Role of CRP in tonB3 promoter expression. (A) Wild-type, Δcrp, and Δcrp pMMcrp strains harboring plasmid p632 containing the tonB3-lacZ fusion were grown in HS-FAC. Samples were removed at various times as indicated and analyzed as described for Fig. 3A. (B) The V. vulnificus ΔlacZ strain harboring the p632 construct was grown in M9 with 0.5% glucose and 0.02% CAA in the presence or absence of 10 mM l-leucine. When indicated, 1 mM or 2 mM cAMP was also added to the medium. Samples were removed at the indicated OD600 and β-galactosidase activity measured as described in Materials and Methods. The bars represent means ± standard deviations (n = 3). Those columns showing no significant difference are labeled with the same letters, and those showing a significant difference (P < 0.05) are labeled with different letters, based on one-way ANOVA with Tukey's posttest.
We further confirmed the role of CRP in the activation of the tonB3 operon by analyzing its expression in minimal medium with glucose as a carbon source in the presence of exogenously added cAMP. In the presence of glucose, the intracellular levels of cAMP are low (20); hence, it is expected that CRP does not bind to the binding sites located in the regulatory regions of the genes under its control (28). Here, when we added cAMP to the minimal medium containing glucose, we could recover expression from the tonB3-lacZ fusion as well as the l-leucine regulation at the levels observed in the presence of glycerol (Fig. 9B).
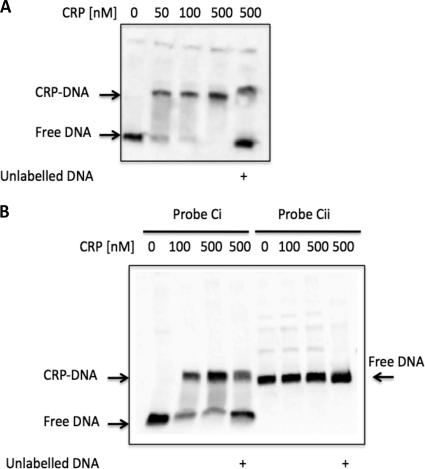
To address whether the observed effect on the expression of the tonB3 gene is a consequence of the direct action of the CRP on the promoter region of this operon, we performed EMSAs with purified CRP(His)6. Figure 10A shows the shift detected when probe C (primers 42-300/42Bam) was incubated with increasing concentrations of this protein in the presence of cAMP in the binding buffer. This shift was reversed in the presence of unlabeled competing DNA. We further demonstrated that CRP does not bind to probe A (far-upstream region) or probe B (middle region), indicating that this protein binds only in the region closer to the start point of the transcription (data not shown). When we split probe C in two, Ci (42-300/42-206Rev) and Cii (42-206-For/42Bam), we established a shift only in probe Ci (Fig. 10B). An in silico analysis of this region showed the presence of a putative binding site with high identity to the sequence consensus for this protein centered at bp −122.5 from the start point of the transcription (Fig. 1C). The identification of the binding site of this protein was done by performing DNase I footprinting with probe C. A footprint was identified between the nucleotides −136 and −107, which overlaps with the identified consensus sequence (28) (Fig. 11A).
Fig 10.
EMSA analysis of the promoter region of the TonB3 operon with CRP(His)6. The different promoter regions of this operon were amplified and labeled as described in Materials and Methods and incubated with increasing concentrations of CRP(His)6 in the presence of 0.2 mM cAMP as indicated in each panel. (A) Probe C (42-300/42Bam); (B) probes Ci (42-300/42-206Rev) and Cii (42-206-For/42Bam). Probes are illustrated in Fig. 1B. Indicated are free labeled DNA and CRP(His)6-DNA complexes. Probes were used at 4 nM, and in each panel it is indicated when unlabeled probe DNA (400 nM) was used for competition.
Fig 11.
DNase I footprint analysis of the tonB3 promoter region with purified CRP(His)6 and Lrp(His)6 proteins. End labeled probe C was incubated with various concentrations of CRP(His)6 (A) or CRP(His)6 and Lrp(His)6 (B) as indicated and treated with DNase I. Gels were calibrated using G+A sequencing reactions (G+A), and relevant positions are indicated. The locations of DNA binding sites for CRP(His)6 are shown by full black lines, while those that correspond to Lrp(His)6 are shown by dotted lines. Hypersensitive sites produced by CRP(His)6 binding are labeled with black stars, and those due to Lrp(His)6 binding are labeled with white stars.
Since in our footprinting studies with purified Lrp(His)6 we observed a protection in the same position where cAMP-CRP binds to the DNA, we wished to determine whether these proteins can bind to this DNA fragment simultaneously. Competitive EMSA experiments were not sensitive enough to distinguish between Lrp-DNA complexes and possible Lrp-CRP-DNA complexes (data not shown). However, by using the binding signatures of each protein, we evaluated the titration of Lrp against a constant concentration of CRP. Similar analyses were previously performed by other authors with a different system (4). In general, as the Lrp concentration increased, the signatures characteristic of CRP binding disappeared, being replaced by signatures characteristic of complete Lrp binding (Fig. 11B, compare lane 2 with lanes 3, 4, and 5). This replacement, however, occurred gradually. At lower Lrp concentrations (Fig. 11B, lanes 3 and 4), signatures associated with both CRP and Lrp binding coexisted. Although Lrp-associated protection and hypersensitivity at positions −59 and −94 were present, the CRP-associated hypersensitivity site (−99) remained apparent even at intermediate Lrp concentrations (Fig. 11B, lanes 3 and 4). At high Lrp concentrations, all signatures and protection corresponded to this protein alone (Fig. 11B, compare lanes 5 and 6). We then performed the same type of footprinting analysis with increasing concentrations of CRP and constant Lrp concentrations. At lower concentrations of CRP, signatures associated with this protein were already present and coexisted with those of Lrp (Fig. 11B, lanes 7 and 8). Interestingly, high concentrations of CRP were not enough to completely displace Lrp, as hypersensitive signatures of the latter protein were still visible at 0.75 μM CRP (sites −59 and −94; Fig. 11B, lane 9). These results argue that when present at higher concentrations (0.5 μM), Lrp may displace CRP from its site in this promoter; however, CRP could displace Lrp from one of the binding sites (approximately bp −147 to −109) but could not displace it from the binding site residing downstream of the CRP binding site even at high concentrations (Fig. 11B, lanes 8 and 9). Importantly, these results also support the possibility that Lrp and CRP bind simultaneously to form a CRP-Lrp-DNA complex that could drive transcription under the conditions studied in this work.
DISCUSSION
TonB systems are responsible for transducing the energy from the proton motive force from the inner membranes to the outer membranes of Gram-negative bacteria for the transport of iron-siderophore complexes, cobalamin, or colicins by TBDT. Recently other substrates for the TonB systems, including zinc, nickel, maltodextrins, and sucrose, have also been identified in different species (2, 34, 42, 53, 58). In addition to the control of the expression of the TBDTs involved in iron-siderophore transport by the Fur protein, other regulatory mechanisms have been described, including AraC-like transcriptional regulators (FetR and MpeR), extracytoplasmic sigma factors (e.g., FecI, PvdS, and MbaS), a riboswitch (e.g., btuB), small RNAs (e.g., omrA and omrB), and the nickel-sensing transcriptional regulator NikR (see reference 45 for a review).
In this work we show that two global regulators, Lrp and CRP, regulate expression of the TonB3 operon, which includes the genes encoding the TBDT protein as well as the structural proteins TtpC3, ExbB3, ExbD3, and TonB3, in response to l-leucine and glucose starvation. We also demonstrated that theTonB3 system is involved in V. vulnificus invasion in the iron-overloaded mouse model. Recently, it was demonstrated that the gene encoding the heme receptor HupA in V. vulnificus has a binding site for CRP in the promoter region and that this protein controls the transcription of hupA when the cells are growing at 40°C; however, Fur (and iron) is the main regulator under physiological conditions (47). Likewise, in the same pathogen the iron-regulated vulnibactin receptor is also under the control of CRP (14). Zhang et al. (69) also showed the possible involvement of CRP in the control of the transcription of the fecA, fepA, cirA, and fiu genes in a fur mutant strain of E. coli. It is still not clear whether a direct or an indirect mechanism controls the changes in the expression observed in the crp mutant in the last two cases (14, 69).
Lrp is a protein that has an important role in the cell physiology, and it has been designated a physiological barometer (21). Its main function is to control the expression of target genes and operons according to the nutritional status of the cell, and in E. coli it controls approximately 400 genes, at least 130 of which involve direct interactions (13, 60). This protein ties bacterial metabolism to environmental signals, mediating transitions between “feast” and “famine.” Therefore, Lrp upregulates specific genes during famine and downregulates other genes during feast (9, 13, 60). Most of the genes regulated by Lrp are involved in small-molecule transport and amino acid metabolism (13, 60). According to our genetic and molecular analyses, in the absence of l-leucine, Lrp binds the promoter region of the tonB3 operon in at least four very distinct locations. An essential binding site for the Lrp protein, resulting in full activation of the tonB3 operon, is located in the far-upstream positions −509 to −443 (with a second small region at positions −415 to −398). The footprints showed phased hypersensitivity (26) that is consistent with Lrp bending or wrapping of large regions of DNA. It has been described that this protein can form multimers (dimers, octamers, and hexadecamers) (12, 19). The activation at long distance observed for Lrp in this work is not uncommon for this protein; however, the location of this essential binding site at those positions in the tonB3 promoter makes this regulatory region one of the farthest described so far for Lrp (48, 64). A third binding site, located in the middle region of the promoter comprising positions −265 to −208, presented a large footprint that is also characteristic for this protein. It is worth noting that in some regions we could not determine the exact boundaries of the binding sites for this protein due to the intrinsic resistance to DNase I observed. A partial deletion of this binding site demonstrated that it was also essential for full activation of the tonB3 operon. Finally, a fourth binding site for Lrp was located upstream of the putative binding site for the RNA polymerase and just downstream of the binding site for CRP.
While in class I and class II promoters CRP interacts directly with the RNA polymerase and other activators are not needed, complex promoters require a regulator in addition to CRP and have been designated class III promoters (7, 8). In those cases, CRP-induced DNA bending can reposition an activator into a productive complex with RNA polymerase (e.g., MalT), or CRP can modulate the binding of an activator to the DNA (e.g., AraC) (5, 52, 68). Since Lrp can induce bending of the DNA (62), according to our results, in the tonB3 promoter region Lrp might help to form a nucleoprotein complex that facilitates interaction between cAMP-CRP and the RNA polymerase. A similar situation has also been described for E. coli in the papBA operon, where Lrp interacts with the DNA region located between the CRP binding site and the RNA polymerase (63). The location of Lrp between those two proteins helps the CRP to interact with the alpha C-terminal domain (α-CTD) region of the RNA polymerase (63). In another example, CRP binds at bp −121.5 upstream of the transcription start site of the proP P2 promoter and coactivation is achieved by Fis and CRP independently contacting each of the two α-CTDs (36). Further work is needed to establish whether in the tonB3 promoter CRP interacts directly with the RNA polymerase (with or without Lrp help) or whether CRP helps in the interaction between the RNA polymerase and the far-located Lrp proteins or another unrecognized activator. Although our genetic and DNase I footprinting experiments support the hypothesis of these two proteins acting in concert to drive transcription, it is also possible that under some growth conditions Lrp might displace CRP from the promoter region.
Thus, we were able to identify not only two direct activators of the TonB3 system but also growth conditions where it is expressed at high levels in addition to the previously recognized HS-FAC condition. The interesting nutritional parallelism between human serum and M9 with a low CAA concentration and glycerol as the sole carbon source suggests that when V. vulnificus is growing under the former condition it is deprived of amino acids (a signal sensed as low levels of l-leucine) and rapidly utilizes glucose (the second signal). In addition to l-leucine, other amino acids, such as l-alanine, l-isoleucine, l-valine, and l-methionine, repressed the expression of the TonB3 system when added to the minimal medium (data not shown). This result is consistent with a recent discovery that E. coli Lrp responds to several of these amino acids (25). In line with those observations, when we performed a microarray analysis with total RNA obtained from V. vulnificus cells growing in HS-FAC, we found that the genes that encode enzymes involved in the glyoxylate shunt as well as glycerol metabolism were clearly induced after 4 to 6 h of growth in serum (Alice and Crosa, unpublished data), indicating the utilization of glycerol and/or lipids as carbon sources after the glucose depletion. According to the Human Serum Metabolome Database (HMDB), the concentration of l-leucine in normal human serum is 123 μmol liter−1 (range, 98 to 148 μmol liter−1), while the concentration of glucose is 4,440.0 ± 370.0 μmol liter−1 (51). These values indicate that Lrp could bind the promoter region of the tonB3 operon when V. vulnificus grows in human serum, and if glucose is depleted, cAMP-CRP could bind and allow full activation of this system. In agreement with this hypothesis, when we added glucose or 10 mM l-leucine to the HS-FAC used throughout this work, we observed a reduction in the expression of the tonB3-lacZ fusion without the growth being notably affected (Fig. 6A). This TonB3 complex appears to play a role during V. vulnificus invasion, suggesting that the substrate(s) being transported for this system inside the host is relevant for that process. It is tempting to speculate that the transition between “feast” and “famine” (9, 43), sensed as l-leucine starvation (and maybe the stringent response), constitutes an important signal when this pathogen grows in serum or inside the host. The study of the physiology of V. vulnificus growing in human serum could lead to the discovery of new mechanisms used by this pathogen during the rapid invasion observed in infected susceptible individuals.
As mentioned above, Lrp and CRP appear to act in concert in the activation of the TonB3 operon, sensing at least two different environmental signals: low concentrations of l-leucine and glucose. In E. coli, a number of genes are controlled by both Lrp and Crp (15, 18, 35, 49, 65, 67, 70). A question that remains to be answered is which other protein(s) binds this DNA when Lrp and CRP are not bound (i.e., when the operon is not actively transcribed). Recently, Lee et al. (30), in a search of binding sites for the quorum-sensing regulator SmcR in the V. vulnificus genome, identified and confirmed a binding site for this protein in front of the tonB3 operon. More interestingly, the binding site sequence identified by DNase I footprinting for SmcR lies immediately upstream of the CRP binding region identified in this work. Since the bindings of these two proteins to the promoter region seem to be mutually exclusive (Alice and Crosa, unpublished data) we are currently exploring the hypothesis that this operon responds to different environmental signals in addition to or together with those described here.
Since the sequence of the tonB3 promoter region is longer than usual and Lrp and CRP binding might occur at defined moments of growth, the most plausible situation when the cells are growing rapidly (i.e., when glucose and/or high CAA concentrations are present) is that other transcriptional factors and/or nucleoid-associated proteins (NAPs), such as Fis, H-NS, or HU, could bind this region, affecting the conformation and/or the supercoiling of the chromosomal DNA and consequently the expression of the tonB3 operon. Of course, some of those NAPs could also work in concert with Lrp and/or CRP for the activation of this operon. We are currently pursuing these hypotheses in what we believe is an interesting model to study the timely expression of an orphan TonB system that is induced mainly under conditions of nutritional deprivation together with possible changes in the organization of the chromosome.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant AI065981 from the National Institutes of Health to J.H.C. and by a Beginning Grant-in-Aid Program Award (10BGIA2560022) from the American Heart Association to A.F.A.
We thank David Lewinshon and Gwendolyn Swarbrick for the generous provision of pooled healthy human serum and Larry David and John Klimek at the Proteomic Shared Resource at OHSU.
Footnotes
Published ahead of print 3 February 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alice AF, Naka H, Crosa JH. 2008. Global gene expression as a function of the iron status of the bacterial cell: influence of differentially expressed genes in the virulence of the human pathogen Vibrio vulnificus. Infect. Immun. 76: 4019– 4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blanvillain S, et al. 2007. Plant carbohydrate scavenging through tonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2: e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brennt CE, Wright AC, Dutta SK, Morris JGJ. 1991. Growth of Vibrio vulnificus in serum from alcoholics: association with high transferrin iron saturation. J. Infect. Dis. 164: 1030– 1032 [DOI] [PubMed] [Google Scholar]
- 4. Browning DF, et al. 2004. Modulation of CRP-dependent transcription at the Escherichia coli acsP2 promoter by nucleoprotein complexes: anti-activation by the nucleoid proteins FIS and IHF. Mol. Microbiol. 51: 241– 254 [DOI] [PubMed] [Google Scholar]
- 5. Browning DF, Busby SJ. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2: 57– 65 [DOI] [PubMed] [Google Scholar]
- 6. Busby S, Ebright RH. 1997. Transcription activation at class II CAP-dependent promoters. Mol. Microbiol. 23: 853– 859 [DOI] [PubMed] [Google Scholar]
- 7. Busby S, Ebright RH. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293: 199– 213 [DOI] [PubMed] [Google Scholar]
- 8. Busby S, Kolb A, Buc H. 1995. The E. coli cyclic AMP receptor protein, p 177– 191 In Eckstein F, Lilley DMJ. (ed), Nucleic acids and molecular biology, vol 9 Springer, Berlin, Germany [Google Scholar]
- 9. Calvo JM, Matthews RG. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58: 466– 490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cardew AS, Fox KR. 2010. DNase I footprinting. Methods Mol. Biol. 613: 153– 172 [DOI] [PubMed] [Google Scholar]
- 11. Chen CF, et al. 1997. Metabolic regulation of lrp gene expression in Escherichia coli K-12. Microbiology 143: 2079– 2084 [DOI] [PubMed] [Google Scholar]
- 12. Chen S, Calvo JM. 2002. Leucine-induced dissociation of Escherichia coli Lrp hexadecamers to octamers. J. Mol. Biol. 318: 1031– 1042 [DOI] [PubMed] [Google Scholar]
- 13. Cho BK, Barrett CL, Knight EM, Park YS, Palsson BO. 2008. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105: 19462– 19467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi MH, et al. 2006. Effect of the crp mutation on the utilization of transferrin-bound iron by Vibrio vulnificus. FEMS Microbiol. Lett. 257: 285– 292 [DOI] [PubMed] [Google Scholar]
- 15. Colland F, Barth M, Hengge-Aronis R, Kolb A. 2000. Sigma factor selectivity of Escherichia coli RNA polymerase: role for CRP, IHF and lrp transcription factors. EMBO J. 19: 3028– 3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crosa JH. 1980. A plasmid associated with virulence in the marine fish pathogen Vibrio anguillarum specifies an iron-sequestering system. Nature 284: 566– 568 [DOI] [PubMed] [Google Scholar]
- 17. Cui Y, Wang Q, Stormo GD, Calvo JM. 1995. A consensus sequence for binding of Lrp to DNA. J. Bacteriol. 177: 4872– 4880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De la Cruz MA, Calva E. 2010. The complexities of porin genetic regulation. J. Mol. Microbiol. Biotechnol. 18: 24– 36 [DOI] [PubMed] [Google Scholar]
- 19. de los Rios S, Perona JJ. 2007. Structure of the Escherichia coli leucine-responsive regulatory protein Lrp reveals a novel octameric assembly. J. Mol. Biol. 366: 1589– 1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70: 939– 1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8: 185– 195 [DOI] [PubMed] [Google Scholar]
- 22. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76: 1648– 1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gulig PA, Bourdage KL, Starks AM. 2005. Molecular pathogenesis of Vibrio vulnificus. J. Microbiol. 43: (Spec No) 118– 131 [PubMed] [Google Scholar]
- 24. Hagen TA, Cornelissen CN. 2006. Neisseria gonorrhoeae requires expression of TonB and the putative transporter TdfF to replicate within cervical epithelial cells. Mol. Microbiol. 62: 1144– 1157 [DOI] [PubMed] [Google Scholar]
- 25. Hart BR, Blumenthal RM. 2011. Unexpected coregulator range for the global regulator Lrp of Escherichia coli and Proteus mirabilis. J. Bacteriol. 193: 1054– 1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hochschild A, Ptashne M. 1986. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell 44: 681– 687 [DOI] [PubMed] [Google Scholar]
- 27. Jones MK, Oliver JD. 2009. Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77: 1723– 1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kolb A, Busby S, Buc H, Garges S, Adhya S. 1993. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62: 749– 795 [DOI] [PubMed] [Google Scholar]
- 29. Landgraf JR, Wu J, Calvo JM. 1996. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J. Bacteriol. 178: 6930– 6936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee DH, et al. 2008. A consensus sequence for binding of SmcR, a Vibrio vulnificus LuxR homologue, and genome-wide identification of the SmcR regulon. J. Biol. Chem. 283: 23610– 23618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Linn T, St Pierre R. 1990. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J. Bacteriol. 172: 1077– 1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lintner RE, et al. 2008. Limited functional conservation of a global regulator among related bacterial genera: Lrp in Escherichia, Proteus and Vibrio. BMC Microbiol. 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu M, Alice AF, Naka H, Crosa JH. 2007. The HlyU protein is a positive regulator of rtxA1, a gene responsible for cytotoxicity and virulence in the human pathogen Vibrio vulnificus. Infect. Immun. 75: 3282– 3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lohmiller S, Hantke K, Patzer SI, Braun V. 2008. TonB-dependent maltose transport by Caulobacter crescentus. Microbiology 154: 1748– 1754 [DOI] [PubMed] [Google Scholar]
- 35. Man TK, Pease AJ, Winkler ME. 1997. Maximization of transcription of the serC (pdxF)-aroA multifunctional operon by antagonistic effects of the cyclic AMP (cAMP) receptor protein-cAMP complex and Lrp global regulators of Escherichia coli K-12. J. Bacteriol. 179: 3458– 3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McLeod SM, Xu J, Johnson RC. 2000. Coactivation of the RpoS-dependent proP P2 promoter by fis and cyclic AMP receptor protein. J. Bacteriol. 182: 4180– 4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meselson M, Yuan R. 1968. DNA restriction enzyme from E. coli. Nature 217: 1110– 1114 [DOI] [PubMed] [Google Scholar]
- 38. Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39. Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178: 1310– 1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morales VM, Backman A, Bagdasarian M. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97: 39– 47 [DOI] [PubMed] [Google Scholar]
- 41. Muller CM, et al. 2009. Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP-cAMP. PLoS Pathog. 5: e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Neugebauer H, et al. 2005. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J. Bacteriol. 187: 8300– 8311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Newman EB, D'Ari R, Lin RT. 1992. The leucine-Lrp regulon in E. coli: a global response in search of a raison d'etre. Cell 68: 617– 619 [DOI] [PubMed] [Google Scholar]
- 44. Newman EB, Lin R. 1995. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annu. Rev. Microbiol. 49: 747– 775 [DOI] [PubMed] [Google Scholar]
- 45. Noinaj N, Guillier M, Barnard TJ, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64: 43– 60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nordhoff E, et al. 1999. Rapid identification of DNA-binding proteins by mass spectrometry. Nat. Biotechnol. 17: 884– 888 [DOI] [PubMed] [Google Scholar]
- 47. Oh MH, Lee SM, Lee DH, Choi SH. 2009. Regulation of the Vibrio vulnificus hupA gene by temperature alteration and cyclic AMP receptor protein and evaluation of its role in virulence. Infect. Immun. 77: 1208– 1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Paul L, Blumenthal RM, Matthews RG. 2001. Activation from a distance: roles of Lrp and integration host factor in transcriptional activation of gltBDF. J. Bacteriol. 183: 3910– 3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Paul L, Mishra PK, Blumenthal RM, Matthews RG. 2007. Integration of regulatory signals through involvement of multiple global regulators: control of the Escherichia coli gltBDF operon by Lrp, IHF, Crp, and ArgR. BMC Microbiol. 7: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Postle K, Kadner RJ. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49: 869– 882 [DOI] [PubMed] [Google Scholar]
- 51. Psychogios N, et al. 2011. The human serum metabolome. PLoS One 6: e16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Richet E, Sogaard-Andersen L. 1994. CRP induces the repositioning of MalT at the Escherichia coli malKp promoter primarily through DNA bending. EMBO J. 13: 4558– 4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schauer K, Gouget B, Carriere M, Labigne A, de Reuse H. 2007. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol. Microbiol. 63: 1054– 1068 [DOI] [PubMed] [Google Scholar]
- 54. Schauer K, Rodionov DA, de Reuse H. 2008. New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 33: 330– 338 [DOI] [PubMed] [Google Scholar]
- 55. Senanayake SD, Brian DA. 1995. Precise large deletions by the PCR-based overlap extension method. Mol. Biotechnol. 4: 13– 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shimada T, Fujita N, Yamamoto K, Ishihama A. 2011. Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS One 6: e20081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering transposon mutagenesis in gram negative bacteria. Biotechnology (NY) 1: 787– 796 [Google Scholar]
- 58. Stork M, et al. 2010. An outer membrane receptor of Neisseria meningitidis involved in zinc acquisition with vaccine potential. PLoS Pathog. 6: e1000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stork M, Otto BR, Crosa JH. 2007. A novel protein, TtpC, is a required component of the TonB2 complex for specific iron transport in the pathogens Vibrio anguillarum and Vibrio cholerae. J. Bacteriol. 189: 1803– 1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tani TH, Khodursky A, Blumenthal RM, Brown PO, Matthews RG. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. U. S. A. 99: 13471– 13476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. U. S. A. 84: 2833– 2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang Q, Calvo JM. 1993. Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of a higher-order nucleoprotein structure. EMBO J. 12: 2495– 2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weyand NJ, Braaten BA, van der Woude M, Tucker J, Low DA. 2001. The essential role of the promoter-proximal subunit of CAP in pap phase variation: Lrp- and helical phase-dependent activation of papBA transcription by CAP from −215. Mol. Microbiol. 39: 1504– 1522 [DOI] [PubMed] [Google Scholar]
- 64. Wiese DE, II, Ernsting BR, Blumenthal RM, Matthews RG. 1997. A nucleoprotein activation complex between the leucine-responsive regulatory protein and DNA upstream of the gltBDF operon in Escherichia coli. J. Mol. Biol. 270: 152– 168 [DOI] [PubMed] [Google Scholar]
- 65. Wonderling LD, Stauffer GV. 1999. The cyclic AMP receptor protein is dependent on GcvA for regulation of the gcv operon. J. Bacteriol. 181: 1912– 1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wright AC, Simpson LM, Oliver JD. 1981. Role of iron in the pathogenesis of Vibrio vulnificus infections. Infect. Immun. 34: 503– 507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yang L, Lin RT, Newman EB. 2002. Structure of the Lrp-regulated serA promoter of Escherichia coli K-12. Mol. Microbiol. 43: 323– 333 [DOI] [PubMed] [Google Scholar]
- 68. Zhang X, Schleif R. 1998. Catabolite gene activator protein mutations affecting activity of the araBAD promoter. J. Bacteriol. 180: 195– 200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang Z, et al. 2005. Functional interactions between the carbon and iron utilization regulators, Crp and Fur, in Escherichia coli. J. Bacteriol. 187: 980– 990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhi J, Mathew E, Freundlich M. 1998. In vitro and in vivo characterization of three major dadAX promoters in Escherichia coli that are regulated by cyclic AMP-CRP and Lrp. Mol. Gen. Genet. 258: 442– 447 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.