Abstract
Accurate strain identification is essential for anyone working with bacteria. For many species, multilocus sequence typing (MLST) is considered the “gold standard” of typing, but it is traditionally performed in an expensive and time-consuming manner. As the costs of whole-genome sequencing (WGS) continue to decline, it becomes increasingly available to scientists and routine diagnostic laboratories. Currently, the cost is below that of traditional MLST. The new challenges will be how to extract the relevant information from the large amount of data so as to allow for comparison over time and between laboratories. Ideally, this information should also allow for comparison to historical data. We developed a Web-based method for MLST of 66 bacterial species based on WGS data. As input, the method uses short sequence reads from four sequencing platforms or preassembled genomes. Updates from the MLST databases are downloaded monthly, and the best-matching MLST alleles of the specified MLST scheme are found using a BLAST-based ranking method. The sequence type is then determined by the combination of alleles identified. The method was tested on preassembled genomes from 336 isolates covering 56 MLST schemes, on short sequence reads from 387 isolates covering 10 schemes, and on a small test set of short sequence reads from 29 isolates for which the sequence type had been determined by traditional methods. The method presented here enables investigators to determine the sequence types of their isolates on the basis of WGS data. This method is publicly available at www.cbs.dtu.dk/services/MLST.
INTRODUCTION
Correct, standardized classification is a basic need for anyone working with bacteria, whether pathogens, commensals, or bacteria used for industrial purposes. Especially in outbreak situations, it is of pivotal importance that the strains of infectious agents be rapidly and accurately identified. A recent example is the outbreak of hemolytic-uremic syndrome and bloody diarrhea caused by an Escherichia coli O104:H4 strain, which in the beginning of May 2011 started spreading in Germany and throughout Europe. Reliable classification, including determination of the multilocus sequence type (MLST), was needed to identify strains related to the outbreak (19, 23). Also, for a range of other species, MLST is used to classify isolates in an understandable and comparable global context (6, 12, 22, 31).
MLST was first developed for Neisseria meningitidis in 1998 to overcome the poor reproducibility between laboratories of older molecular typing schemes (18). The principle behind the MLST scheme is to identify internal nucleotide sequences of approximately 400 to 500 bp in multiple housekeeping genes. Unique sequences (alleles) are assigned a random integer number, and a unique combination of alleles at each locus, an “allelic profile,” specifies the sequence type (ST). Following the introduction of the Neisseria MLST scheme, MLST has been considered the “gold standard” of typing, and additional schemes that cover bacterial and fungal species have been developed. The MLST allele sequences and ST profile tables are stored in curated databases hosted at different sites around the world (1, 14, 15). The PubMLST site collects data from all databases and makes it easily accessible (multilocus sequence typing databases and software, December 2011 [http://pubmlst.org]).
Traditionally, MLST starts with a PCR amplification step using primers that are specific for the loci of the MLST scheme, followed by Sanger sequencing. The procedure is both costly and time-consuming. In this new era of high-throughput sequencing, it may be more rational to use whole-genome sequence (WGS) data for typing. The cost of DNA sequencing has steadily gone down roughly 10-fold every 5 years (25), and the development of next- and third-generation sequencing methods has provided equally great reductions in equipment investments, thus making the technology accessible to individual investigators and routine clinical and microbial laboratories. The challenge, however, is to extract the relevant information from the large amount of data generated by these techniques. To allow comparison with results obtained by other commonly used technologies and with historical data, it is also important to be able to relate the WGS data to typing schemes such as MLST.
We present here the publicly available MLST server (www.cbs.dtu.dk/services/MLST), which uses WGS data for identifying the STs of bacteria.
MATERIALS AND METHODS
MLST databases.
MLST allele sequences and ST profile tables are stored in online databases hosted at five different sites around the world. The University of Oxford collects data from all databases and makes it easily accessible (http://pubmlst.org). In total, 66 bacterial MLST schemes are currently available. Most of them function at the species level, e.g., Escherichia coli and Staphylococcus aureus schemes, while a few function on the genus level, e.g., the Bifidobacterium and Neisseria schemes. Most schemes include 7 housekeeping genes, but schemes with as few as 5 and as many as 10 genes have also been developed. For four bacterial species, two different MLST schemes are available: Acinetobacter baumannii (2; Institut Pasteur, Acinetobacter baumannii MLST database, December 2011 [http://www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html]), Clostridium difficile (10, 17), E. coli (13, 32), and Pasteurella multocida (28; PubMLST, Pasteurella multocida multi-host MLST databases, December 2011 [http://pubmlst.org/pmultocida_multihost/]).
Data sets. (i) Assembled genomes.
In August 2010, 1,212 completely sequenced and assembled bacterial genomes were collected from the NCBI Genome database (http://www.ncbi.nlm.nih.gov/sites/genome). For 336 of these genomes, MLST schemes have been developed and are available through the MLST databases (Table 1).
Table 1.
MLST of preassembled, completely sequenced bacterial isolates
| MLST scheme | No. of loci in scheme | Avg no. of alleles per locusa | No. of STsa | No. of isolatesb | Proportionc of: |
|
|---|---|---|---|---|---|---|
| New alleles | Unknown STs | |||||
| Acinetobacter baumannii_1 | 7 | 82 | 346 | 6 | 0.095 | 0.333 |
| Acinetobacter baumannii_2 | 7 | 34 | 124 | 6 | 0.000 | 0.167 |
| Arcobacter | 7 | 205 | 357 | 2 | 0.214 | 0.500 |
| Bacillus cereus | 7 | 129 | 553 | 10 | 0.043 | 0.100 |
| Bifidobacterium | 7 | 42 | 102 | 11 | 0.221 | 0.273 |
| Bordetella | 7 | 8 | 43 | 5 | 0.400 | 0.400 |
| Borrelia burgdorferi | 8 | 125 | 402 | 2 | 0.000 | 0.000 |
| Brachyspira | 7 | 39 | 36 | 3 | 0.571 | 1.000 |
| Brachyspira hyodysenteriae | 7 | 17 | 66 | 1 | 0.143 | 0.000 |
| Burkholderia pseudomallei | 7 | 46 | 886 | 4 | 0.000 | 0.000 |
| Corynebacterium diphtheriae | 7 | 40 | 227 | 1 | 0.000 | 0.000 |
| Campylobacter fetus | 7 | 10 | 35 | 1 | 0.000 | 0.000 |
| Campylobacter jejuni | 7 | 415 | 5,489 | 6 | 0.000 | 0.000 |
| Campylobacter lari | 7 | 50 | 18 | 1 | 0.000 | 0.000 |
| Campylobacter upsaliensis | 7 | 42 | 138 | 2 | 0.000 | 0.500 |
| Clostridium botulinum | 7 | 10 | 24 | 11 | 0.377 | 0.455 |
| Clostridium difficile_1 | 7 | 18 | 128 | 2 | 0.000 | 0.000 |
| Clostridium difficile_2 | 7 | 14 | 65 | 2 | 0.000 | 0.000 |
| Cronobacter | 7 | 48 | 74 | 2 | 0.000 | 0.000 |
| Enterococcus faecalis | 7 | 61 | 435 | 1 | 0.000 | 0.000 |
| Enterococcus faecium | 7 | 48 | 617 | 26 | 0.011 | 0.038 |
| Escherichia coli_1 | 7 | 228 | 2,333 | 36 | 0.004 | 0.000 |
| Escherichia coli_2 | 8 | 143 | 535 | 36 | 0.031 | 0.250 |
| Flavobacterium psychrophilum | 7 | 15 | 33 | 1 | 0.000 | 0.000 |
| Haemophilus influenzae | 7 | 124 | 939 | 4 | 0.071 | 0.000 |
| Haemophilus parasuis | 7 | 25 | 116 | 1 | 0.000 | 0.000 |
| Helicobacter pylori | 7 | 2,088 | 2,356 | 10 | 0.543 | 0.600 |
| Klebsiella pneumoniae | 7 | 94 | 688 | 3 | 0.000 | 0.000 |
| Lactobacillus casei | 7 | 9 | 40 | 3 | 0.000 | 0.333 |
| Leptospira | 7 | 25 | 117 | 6 | 0.667 | 0.667 |
| Listeria monocytogenes | 7 | 79 | 34 | 6 | 0.000 | 0.000 |
| Mannheimia haemolytica | 7 | 13 | 35 | 3 | 0.000 | 0.000 |
| Moraxella catarrhalis | 8 | 40 | 214 | 1 | 0.000 | 0.000 |
| Neisseria | 7 | 561 | 8,999 | 8 | 0.000 | 0.000 |
| Pasteurella multocida multihost | 7 | 25 | 46 | 1 | 0.000 | 0.000 |
| Pasteurella multocida RIRDC | 7 | 47 | 189 | 1 | 0.000 | 0.000 |
| Porphyromonas gingivalis | 7 | 32 | 138 | 2 | 0.000 | 0.000 |
| Propionibacterium acnes | 7 | 12 | 58 | 2 | 0.000 | 0.000 |
| Pseudomonas aeruginosa | 7 | 116 | 1,070 | 4 | 0.036 | 0.250 |
| Stenotrophomonas maltophilia | 7 | 47 | 56 | 2 | 0.000 | 0.000 |
| Salmonella enterica | 7 | 395 | 1,492 | 18 | 0.008 | 0.167 |
| Sinorhizobium | 10 | 18 | 136 | 2 | 0.000 | 0.000 |
| Staphylococcus aureus | 7 | 244 | 2,107 | 21 | 0.000 | 0.000 |
| Staphylococcus epidermidis | 7 | 34 | 361 | 2 | 0.000 | 0.000 |
| Streptococcus agalactiae | 7 | 58 | 557 | 3 | 0.000 | 0.000 |
| Streptococcus pneumoniae | 7 | 319 | 6,947 | 12 | 0.012 | 0.000 |
| Streptococcus pyogenes | 7 | 89 | 572 | 13 | 0.022 | 0.000 |
| Streptococcus suis | 7 | 87 | 239 | 6 | 0.238 | 0.500 |
| Streptococcus thermophilus | 6 | 22 | 116 | 3 | 0.111 | 0.000 |
| Streptococcus uberis | 7 | 42 | 475 | 1 | 0.000 | 0.000 |
| Streptomyces | 6 | 107 | 135 | 5 | 0.733 | 0.600 |
| Vibrio parahaemolyticus | 7 | 141 | 348 | 1 | 0.000 | 0.000 |
| Vibrio vulnificus | 10 | 40 | 83 | 2 | 0.400 | 1.000 |
| Wolbachia | 5 | 168 | 236 | 4 | 0.000 | 0.000 |
| Xylella fastidiosa | 7 | 17 | 27 | 4 | 0.000 | 0.000 |
| Yersinia pseudotuberculosis | 7 | 11 | 95 | 4 | 0.000 | 0.000 |
Registered in the MLST database.
Number of isolates with completely sequenced genomes tested for this scheme.
Proportion of alleles found in the isolates, or proportion of STs found for the isolates, which were not already registered in the database.
(ii) Sequence reads.
Table 2 shows an overview of the species for which we had short sequence reads, along with the sequencing platforms used. Campylobacter jejuni, E. coli, Klebsiella pneumoniae, Staphylococcus aureus, and Salmonella enterica isolates were sequenced on the Illumina platform generating paired-end reads by TGen (United States). Streptococcus thermophilus, Bifidobacterium animalis, and Lactococcus lactis isolates were sequenced on the Illumina platform generating paired-end reads by Source BioScience (United Kingdom), BaseClear (The Netherlands), and BGI (Hong Kong). Other S. thermophilus, B. animalis, B. longum, and L. lactis isolates were sequenced on the Illumina platform generating single reads by Source BioScience (United Kingdom). P. aeruginosa isolates were sequenced on the Illumina platform generating single reads by Partners HealthCare (Boston, MA) or on the Roche 454 GS platform by the Allegheny-Singer Research Institute (Pittsburgh, PA). WGS data for the 2011 German O104:H4 E. coli outbreak were obtained from publicly available sources. Data from 7 × 314 chips sequenced on the Ion Torrent platform (Life Technologies) for a single isolate were obtained from BGI (23). Data from 8 × 314 chips sequenced on the Ion Torrent platform (Life Technologies) for the outbreak strain LB226692 were obtained from Life Technologies and the University of Münster (19). Illumina MiSeq single-read data for five different isolates of the outbreak strain were obtained from the British Health Protection Agency (HPA). From the Göttingen Genomics Laboratory, we obtained data for two isolates of the outbreak strain sequenced on a Roche 454 GS sequencer. The ABI SOLiD data were the 50 × 50 mate pair data set with 600× coverage of E. coli DH10B, available from the SOLiD software development community.
Table 2.
MLST of completely sequenced bacterial isolates using short sequence reads
| Sequencing platform | Species | No. of isolates | MLST scheme | Proportion of loci with: |
Avg N50 | Log avg N50 | |
|---|---|---|---|---|---|---|---|
| Minor mismatches | Major mismatches | ||||||
| Illumina | |||||||
| Paired-end reads | B.animalis | 5 | Bifidobacterium | 0.000 | 0.029 | 33,113 | 4.52 |
| C. jejuni | 53 | Campylobacter | 0.005 | 0.000 | 131,571 | 5.12 | |
| E. coli | 15 | E. coli scheme 1 | 0.010 | 0.000 | 195,822 | 5.29 | |
| E. coli | 15 | E. coli scheme 2 | 0.075 | 0.000 | 196,463 | 5.29 | |
| K. pneumoniae | 4 | K. pneumoniae | 0.000 | 0.000 | 207,167 | 5.32 | |
| L. lactis | 34 | L. lactis | 0.564 | 0.039 | 44,525 | 4.65 | |
| S. aureus | 83 | S. aureus | 0.017 | 0.009 | 196,736 | 5.29 | |
| S. enterica | 50 | S. enterica | 0.000 | 0.009 | 249,501 | 5.40 | |
| S. thermophilus | 13 | S. thermophilus | 0.090 | 0.000 | 47,686 | 4.68 | |
| Single reads | E. coli | 6 | E. coli scheme 1 | 0.000 | 0.000 | 46,479 | 4.67 |
| B. animalis | 2 | Bifidobacterium | 0.000 | 0.071 | 24,979 | 4.40 | |
| B.longum | 2 | Bifidobacterium | 0.000 | 0.000 | 22,548 | 4.35 | |
| L. lactis | 7 | L. lactis | 0.238 | 0.119 | 14,114 | 4.15 | |
| P. aeruginosa | 81 | P. aeruginosa | 0.019 | 0.125 | 9,380 | 3.97 | |
| S. thermophilus | 9 | S. thermophilus | 0.000 | 0.000 | 28,823 | 4.46 | |
| Roche 454 | E. coli | 3 | E. coli scheme 1 | 0.000 | 0.000 | 92,131 | 4.96 |
| P. aeruginosa | 2 | P. aeruginosa | 0.000 | 0.000 | 60,477 | 4.78 | |
| Ion Torrent | E. coli | 2 | E. coli scheme 1 | 0.000 | 0.500 | 13,779 | 4.14 |
| SOLiD | E. coli | 1 | E. coli scheme 1 | 0.286 | 0.000 | 165,835 | 5.22 |
Draft assembly of short sequence reads.
If sequence reads are given as input to the MLST server, the reads are assembled de novo prior to ST prediction. Short-read data produced from all major next- and third-generation sequencing platforms, such as the Illumina, Roche 454 GS, and Applied Biosystems SOLiD platforms and the Life Science Ion Torrent personal genome machine (PGM), are supported (8, 18a, 24, 26, 30). The de novo assembly creates contiguous sequences without gaps from the DNA sequence reads, termed contigs, and when paired-end or mate-paired reads are available, these are used to combine the contigs into scaffolds. As a measure of the quality of the draft assembly, the N50 value is calculated for the assembled genomes. The N50 value for contigs or scaffolds is defined as the length of the shortest contig or scaffold in the set of the largest contigs or scaffolds that represents at least 50% of the assembly (20). The assembly is available for download from the MLST server.
Illumina sequence data are assembled using Velvet, version 1.1.04 (34). Prior to assembly, paired-end data are filtered and trimmed using the following steps. (i) All reads containing the character N are removed. (ii) If a read matches at least 15 nucleotides (nt) of a sequencing primer/adaptor, the read is trimmed at the 5′ coordinate of the match. (iii) The 3′ tail is trimmed up to a quality score of 15 (phred scale). (iv) The minimum average quality of the read after trimming is 20. (v) The length of the read after trimming is at least 15 nt. We do not trim Illumina single-end data, since benchmarking showed that this reduced the overall quality of the assemblies and of MLST prediction for the data sets used in the study. Then, in parallel, several assemblies using k-mer sizes from 33% to 80% of the average read length are run, and the assembly with the best cumulative rank for N50, number of contigs, and length of the largest contig is selected as the best assembly.
Both Roche 454 GS and Ion Torrent PGM sequence data are assembled using the Roche proprietary GS De Novo Assembler software, version 2.6 (Newbler 2.6). If given standard flowgram files (.sff), the assembler clips and trims the data prior to assembly.
For Applied Biosystems SOLiD sequence data, assembly is performed using the SOLiD System de novo Accessory Tools, version 2.2. The assembly pipeline uses colorspace Velvet 0.7.55 (34) for the assembly and is run without read error correction (with SAET) or postassembly analysis in order to decrease run time. For all sequencing technologies, single-end, paired-end, or mate-paired reads can be used for assembly.
After uploading of short read data, the assembly is available for download from the MLST server.
Implementation of MLST on completely sequenced bacteria.
An automatic weekly download script was set up for all allele sequences and ST profiles from the MLST databases. Via a script written in Perl, the assembled bacterial genome was converted into a BLAST database. Using the specified MLST scheme, the genome was searched by BLAST for all MLST alleles for all genes. Statistically significant alignments between the query sequence (the MLST alleles) and sequences in the BLAST genome database are called high-scoring segment pairs (HSP) according to BLAST terminology. As the Expect threshold, we use the default value, which is 10.
The best-matching MLST allele is found by calculating the length score (LS) as QL − HL + G, where QL is the length of the MLST allele, HL is the length of the HSP, and G is the number of gaps in the HSP. The allele with the lowest LS and, secondly, with the highest percentage of identity (ID) is selected as the best-matching MLST allele. A perfectly matching MLST allele will have an LS of zero and 100% ID, meaning that all the nucleotides of the MLST allele match with the nucleotides in the genome across the entire length of the allele. Note that the BLAST HSP E value or score cannot be used for selecting the correct allele, since a long allele with a percentage of ID below 100% can have a lower E value (i.e., a higher score) than a shorter allele with 100% ID. Per definition, the shorter allele with 100% ID over the whole length is the correct allele.
After identification of the MLST allele for all genes of the MLST scheme, the ST is determined on the basis of the combination of identified alleles.
RESULTS
MLST implementation.
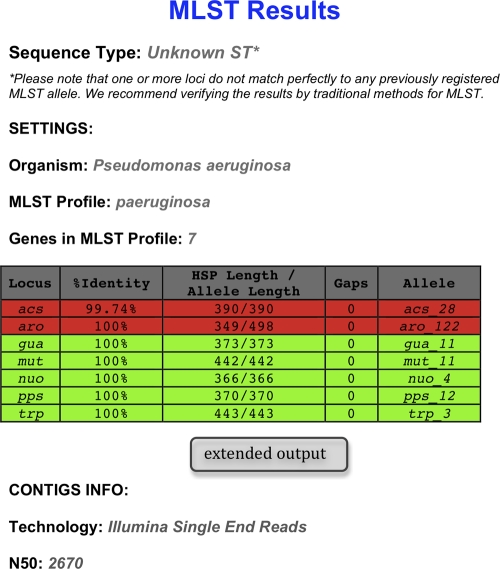
For MLST of completely sequenced bacterial genomes, short sequence reads are, in a first step, assembled to draft genomes as described in Materials and Methods. It is also possible to bypass the assembly step and to input a complete or partial preassembled genome. The minimum requirement for a partial genome is that it contain all the loci necessary for MLST. For a specific MLST scheme, the MLST alleles of each locus are aligned to the genome by using BLAST. The closest-matching MLST allele is selected, and the ST is determined based on the combination of MLST alleles. Two different output formats are available. The short output format includes the identified ST and details about the concordance of each locus with the best-matching MLST allele in the database. Figure 1 shows an example of the short output format from the typing of a P. aeruginosa isolate. The extended output format additionally includes the nucleotide sequences of the MLST alleles identified (see Fig. S1 in the supplemental material). This format can be useful for drawing phylogenetic trees.
Fig 1.
MLST results for a P. aeruginosa isolate in the short output format. By use of the MLST Web server, a P. aeruginosa strain that had been sequenced on the Illumina platform generating single reads was typed. For the purpose of the example, we have chosen to show the results obtained by using short sequence reads that assemble into a draft genome with a low N50. Shown are the name of the loci in the MLST scheme, the percentage of nucleotides that are identical in the best-matching MLST allele in the database and the corresponding sequence in the genome (% identity), the length of the alignment between the best-matching MLST allele in the database and the corresponding sequence in the genome (also called the high-scoring segment pair [HSP]), the length of the best-matching MLST allele in the database, the number of gaps in the HSP, and the name of the best-matching MSLT allele. Note that for a perfectly matching allele, the percentage of identity will be 100%, the allele length will equal the HSP length, and the number of gaps will be zero. Green indicates a perfect match, while red indicates an imperfect match.
MLST of 336 assembled bacterial genomes.
To evaluate our method, we used it for identification of the STs of 336 completely sequenced and preassembled bacterial genomes. These bacteria cover 56 MLST schemes. Table 1 shows the results with regard to the proportion of the MLST alleles in the tested genomes that were previously unseen and hence were not registered in the MLST databases. For 34 MLST schemes, all alleles in the MLST loci in the tested genomes matched perfectly to an allele already registered in the MLST databases (the proportion of new alleles equaled zero), while for the remaining 22 MLST schemes, 0.4% to 73.3% of the MLST alleles in the genomes were not in the MLST databases.
Two MLST schemes exist for E. coli: E. coli scheme 1, which employs seven genes (adk, fumC, gyrB, icd, mdh, purA, recA) (32), and E. coli scheme 2, which employs eight genes (dinB, icdA, pabB, polB, putP, trpA, trpB, uidA) (13). When the 36 completely sequenced E. coli isolates were typed using E. coli scheme 1, only one allele (0.4%) was not in the database. When E. coli scheme 2 was used, 10 alleles (3.1%) were not in the database. This difference in the proportion of previously unseen alleles may reflect either the coverage of the MLST databases (that is, how large a fraction of the total number of alleles they contain) or the rates of evolution of the genes used by the two schemes. The database for E. coli scheme 1 contains on average 228 alleles per locus, while that for E. coli scheme 2 contains on average 143 alleles per locus. Accordingly, the higher number of previously unseen alleles found by using E. coli scheme 2 seems mostly to reflect the fact that this database is less complete than the database for scheme 1.
The three MLST schemes that resulted in the highest number of previously unseen MLST alleles were the Brachyspira (57.1% new alleles), Leptospira (66.7% new alleles), and Streptomyces (73.3% new alleles) schemes. These schemes are meant to cover a whole genus rather than a specific species. As a consequence, these databases are expected to contain far more alleles than databases that aim at covering only a single species. However, this was not the case, as can be seen from Table 1. The Neisseria scheme also aims at covering a whole genus, but here no new alleles were found in the eight Neisseria genomes tested (two Neisseria gonorrhoeae and six Neisseria meningitidis genomes). Indeed, the Neisseria database is the second largest database, containing 561 alleles per locus, in accord with the early establishment of the database in 1998 (18). Of interest, the Helicobacter pylori database contains an average of 2,088 alleles per locus and as such is by far the largest database. Apparently, this does not mean that the database is in any way complete, since more than half of the alleles in the 10 H. pylori genomes tested are not in the database. This observation indicates that the genes selected for the H. pylori MLST scheme (33) are evolving faster than the genes that are generally used in the MLST schemes. This idea is in line with studies showing that in general, H. pylori has high rates of recombination and mutation (5, 7, 29).
Eight bacterial MLST schemes were not tested in this analysis, since we did not have access to complete genomes from these species. However, it is possible to use the MLST Web server with these species as well (Brachyspira intermedia, Burkholderia cepacia complex, Campylobacter helveticus, Campylobacter insulaenigrae, Streptococcus oralis, Streptococcus equi subsp. zooepidemicus, Clostridium septicum, and Chlamydiales spp.).
From short sequence reads to MLST.
MLST implementation was then tested on short sequence reads from 387 bacterial isolates covering 10 MLST schemes and four sequencing platforms. Table 2 shows the results. We have divided the genomic MLST loci that did not perfectly match an MLST allele in the databases into major and minor mismatches. The major mismatches occur when the MLST allele from the MLST database exceeds the length of the contig, meaning that the MLST locus is only partly contained in the contig. In Fig. S1 in the supplemental material, the aro gene represents a major mismatch in a P. aeruginosa genome. The minor mismatches are all other types of mismatches and are equivalent to the “new alleles” of Table 1. In Fig. S1, the acs gene represents a minor mismatch.
For 11 of the 15 sets of isolates sequenced by the Illumina technology for paired-end or single reads, and for all of the isolates sequenced on the Roche 454 GS platform, the frequency of alleles with minor mismatches was below 2%. For the remaining four sets of isolates, where the frequency of minor mismatches was above 2% (E. coli, Illumina paired-end reads, E. coli MLST scheme 2; L. lactis, Illumina paired-end and single reads; S. thermophilus, Illumina paired-end reads), this is likely to reflect the small size of the MLST database.
Whereas the proportion of alleles with minor mismatches reflects the coverage of the database for the selected scheme, the proportion of alleles with major mismatches reflects how well the short sequence reads have been assembled into a draft genome. The N50 value is a measure of the quality of the draft assembly: the higher the N50 value, the better the quality of the assembly. In general, Illumina paired-end reads were assembled into draft genomes with higher N50 values (average N50, 165,149; 95% confidence interval [95% CI], 150,491 to 179,807) than Illumina single reads (average N50, 13,943; 95% CI, 11,824 to 16,062). For the remaining sequencing platforms, we have too little data to draw conclusions on the general quality of the assembled draft genomes. Furthermore, the variability can be very large, as evidenced by the two E. coli isolates that were sequenced on the Life Sciences Ion Torrent PGM platform. While the isolate sequenced by Life Technologies and the University of Münster had an N50 value of 28,537, the isolate sequenced by BGI was assembled into a draft genome with an N50 of 666. As a comment on this poor N50 value, it should be noted that only the FASTQ files from the sequencing, not the flowgram files, were available to us.
For the assembled P. aeruginosa genomes, 13.2% of the alleles contained major mismatches. However, more than 40% of the alleles with major mismatches were found in the assembled genomes of only five isolates. The average N50 of these five draft genomes was as low as 503 (95% CI, 175 to 831).
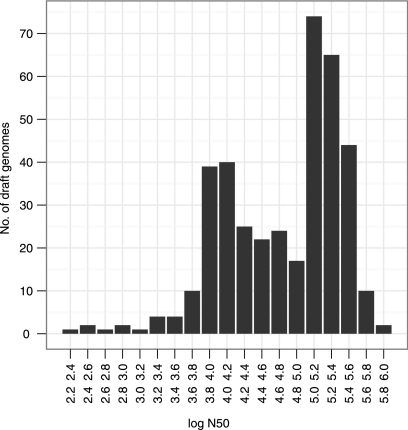
Figure 2 shows the distribution of the log N50 for all assembled draft genomes. Fifteen draft genomes had a log N50 below 3.6 (N50 below 4,000). The remaining draft assemblies are contained within two peaks, roughly separating the draft genomes based on single reads from those based on paired-end reads.
Fig 2.
Distribution of log N50 values for 387 assembled draft genomes.
For a small subset of the P. aeruginosa and S. aureus isolates, and for all K. pneumoniae isolates, the ST had been determined previously by traditional methods. For 10 of the E. coli isolates, the WGS data were obtained from publicly available sources. These isolates were all from the 2011 German E. coli O104:H4 outbreak, the causative agent of which has been found to belong to ST-678 (4, 19, 23). Table 3 shows that 25 of the 29 isolates with known STs were assigned the correct ST on the basis of our method for MLST. Three of the P. aeruginosa isolates were not assigned the correct ST. Instead, they all contained major mismatches and were assigned the ST “unknown” (N50 values, 371, 453, and 1,154). For the E. coli isolate sequenced by BGI using the Life Sciences Ion Torrent PGM, the MLST loci likewise contained major mismatches and the ST “unknown” was assigned.
Table 3.
Isolates with known STs
| Sequencing platform | Species | No. of isolates with known STsa | No. of correctly identified STsb |
|---|---|---|---|
| Illumina | |||
| Paired-end reads | S. aureus | 6 | 6 |
| K. pneumoniae | 4 | 4 | |
| Single reads | E. coli | 6 | 6 |
| P. aeruginosa | 7 | 4 | |
| Roche 454 | E. coli | 2 | 2 |
| P. aeruginosa | 2 | 2 | |
| Ion Torrent | E. coli | 2 | 1 |
Determined by traditional methods.
Predicted by using WGS data.
DISCUSSION
WGS of bacterial pathogens has become an option for more scientists than formerly and even for routine laboratories due to the declining costs of sequencing and the increasing number of analytic methods available. WGS may be useful in trend studies, in diagnostics, and for surveillance. Depending on the technology, WGS can be performed in a couple of hours. By combining this speed with low costs and the right tools, real-time surveillance and quick detection of outbreaks will become possible. As both the costs of technology and the run times continue to decline, WGS will become increasingly available to routine diagnostic laboratories. The challenges will thus be not to produce the sequence data but to extract the relevant information so as to allow for comparisons over time and between laboratories. Ideally, this information should also allow for comparison to historical data.
We have developed, implemented, and evaluated an MLST predictor based on WGS data. The method is publicly available at www.cbs.dtu.dk/services/MLST. The user can upload either a preassembled complete or partial bacterial genome or short sequence reads from one of four sequencing platforms. Currently, 70 different MLST schemes for 66 species are available.
The MLST Web server was specifically designed for ease of use, for the benefit of investigators with limited bioinformatics experience. The first step is to upload the preassembled genome or short sequence reads. In the case of short sequence reads, the sequence platform also needs to be specified. After one selects the MLST scheme to be used, the job can be submitted.
Jolley and Maiden have developed a Web-accessible database system, BIGSdb, that can also use WGS for MLST (16). This system, however, works only on UNIX/Linux systems and requires the installation of a whole range of programs and databases. The MLST Web server presented here can be used by anyone with a computer and a reasonably fast Internet connection.
Although new typing methods are expected to emerge in the wake of complete genome sequencing, e.g., single nucleotide polymorphism (SNP) typing (9, 11) and pangenome family trees (27), these methods lack standardized implementation and general acceptance in the scientific community. We therefore believe that MLST will still be considered the “gold standard” for typing for some time. In addition, for many years, knowledge of the ST will be crucial for comparison to data from isolates that were characterized before complete genome data became easily available.
The MLST server will continue to be improved, e.g., by addition of an option for the automatic detection of species, and hence the selection of the MLST scheme to be used, based on 16S rRNA typing. Furthermore, it will become possible to obtain a phylogenetic tree as output, which will enable the user to see how the ST of the query isolate relates to other STs.
Additional features for analyzing WGS data are also under development. These include the identification of antimicrobial resistance and virulence genes, as in a study described recently (3). Furthermore, we are developing methods for species identification and phylogenetic analysis based on SNP and pangenome analysis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Center for Genomic Epidemiology at the Technical University of Denmark and was funded by grant 09-067103/DSF from the Danish Council for Strategic Research.
We are grateful to Mads Bennedsen, Birgitte Stuer-Lauridsen, and colleagues at Chr Hansen A/S (Hørsholm, Denmark) for sharing unpublished genome sequence data. We are grateful to Hans-Henrik Stærfeldt and John Damm Sørensen for excellent technical assistance.
Footnotes
Published ahead of print 11 January 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Aanensen DM, Spratt BG. 2005. The multilocus sequence typing network: mlst.net. Nucleic Acids Res. 33:W728–W733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartual SG, et al. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bennedsen M, Stuer-Lauridsen B, Danielsen M, Johansen E. 2011. Screening for antimicrobial resistance genes and virulence factors via genome sequencing. Appl. Environ. Microbiol. 77:2785–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bielaszewska M, et al. 2011. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 11:671–676 [DOI] [PubMed] [Google Scholar]
- 5. Bjorkholm B, et al. 2001. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 98:14607–14612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. David MD, Kearns AM, Gossain S, Ganner M, Holmes A. 2006. Community-associated meticillin-resistant Staphylococcus aureus: nosocomial transmission in a neonatal unit. J. Hosp. Infect. 64:244–250 [DOI] [PubMed] [Google Scholar]
- 7. Falush D, et al. 2001. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl. Acad. Sci. U. S. A. 98:15056–15061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fedurco M, Romieu A, Williams S, Lawrence I, Turcatti G. 2006. BTA, a novel reagent for DNA attachment on glass and efficient generation of solid-phase amplified DNA colonies. Nucleic Acids Res. 34:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gardy JL, et al. 2011. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N. Engl. J. Med. 364:730–739 [DOI] [PubMed] [Google Scholar]
- 10. Griffiths D, et al. 2010. Multilocus sequence typing of Clostridium difficile. J. Clin. Microbiol. 48:770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hendriksen RS, et al. 2011. Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. mBio 2(4):e00157–11 doi:10.1128/mBio.00157-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heym B, Le Moal M, Armand-Lefevre L, Nicolas-Chanoine MH. 2002. Multilocus sequence typing (MLST) shows that the ‘Iberian’ clone of methicillin-resistant Staphylococcus aureus has spread to France and acquired reduced susceptibility to teicoplanin. J. Antimicrob. Chemother. 50:323–329 [DOI] [PubMed] [Google Scholar]
- 13. Jaureguy F, et al. 2008. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics 9:560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jolley KA, Chan MS, Maiden MC. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jolley KA, Maiden MC. 2006. AgdbNet—antigen sequence database software for bacterial typing. BMC Bioinformatics 7:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lemee L, Dhalluin A, Pestel-Caron M, Lemeland JF, Pons JL. 2004. Multilocus sequence typing analysis of human and animal Clostridium difficile isolates of various toxigenic types. J. Clin. Microbiol. 42:2609–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maiden MC, et al. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a. Margulies M, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mellmann A, et al. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller JR, Koren S, Sutton G. 2010. Assembly algorithms for next-generation sequencing data. Genomics 95:315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reference deleted.
- 22. Oliveira DC, Tomasz A, de Lencastre H. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of meticillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180–189 [DOI] [PubMed] [Google Scholar]
- 23. Rohde H, et al. 2011. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N. Engl. J. Med. 365:718–724 [DOI] [PubMed] [Google Scholar]
- 24. Rothberg JM, et al. 2011. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475:348–352 [DOI] [PubMed] [Google Scholar]
- 25. Service RF. 2006. Gene sequencing. The race for the $1000 genome. Science 311:1544–1546 [DOI] [PubMed] [Google Scholar]
- 26. Shendure J, et al. 2005. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309:1728–1732 [DOI] [PubMed] [Google Scholar]
- 27. Snipen L, Ussery DW. 2010. Standard operating procedure for computing pangenome trees. Stand. Genomic Sci. 2:135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Subaaharan S, Blackall LL, Blackall PJ. 2010. Development of a multi-locus sequence typing scheme for avian isolates of Pasteurella multocida. Vet. Microbiol. 141:354–361 [DOI] [PubMed] [Google Scholar]
- 29. Suerbaum S, et al. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 95:12619–12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Turcatti G, Romieu A, Fedurco M, Tairi AP. 2008. A new class of cleavable fluorescent nucleotides: synthesis and optimization as reversible terminators for DNA sequencing by synthesis. Nucleic Acids Res. 36:e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wagenlehner FM, et al. 2007. Management of a large healthcare-associated outbreak of Panton-Valentine leucocidin-positive meticillin-resistant Staphylococcus aureus in Germany. J. Hosp. Infect. 67:114–120 [DOI] [PubMed] [Google Scholar]
- 32. Wirth T, et al. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wirth T, et al. 2004. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: lessons from Ladakh. Proc. Natl. Acad. Sci. U. S. A. 101:4746–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.