Abstract
Previous reports indicate that the expression and/or activity of the protein-tyrosine phosphatase (PTP) LAR are increased in insulin-responsive tissues of obese, insulin-resistant humans and rodents, but it is not known whether these alterations contribute to the pathogenesis of insulin resistance. To address this question, we generated transgenic mice that overexpress human LAR, specifically in muscle, to levels comparable to those reported in insulin-resistant humans. In LAR-transgenic mice, fasting plasma insulin was increased 2.5-fold compared with wild-type controls, whereas fasting glucose was normal. Whole-body glucose disposal and glucose uptake into muscle in vivo were reduced by 39–50%. Insulin injection resulted in normal tyrosyl phosphorylation of the insulin receptor and insulin receptor substrate 1 (IRS-1) in muscle of transgenic mice. However, phosphorylation of IRS-2 was reduced by 62%, PI3′ kinase activity associated with phosphotyrosine, IRS-1, or IRS-2 was reduced by 34–57%, and association of p85α with both IRS proteins was reduced by 39–52%. Thus, overexpression of LAR in muscle causes whole-body insulin resistance, most likely due to dephosphorylation of specific regulatory phosphotyrosines on IRS proteins. Our data suggest that increased expression and/or activity of LAR or related PTPs in insulin target tissues of obese humans may contribute to the pathogenesis of insulin resistance.
Glucose homeostasis is essential for normal mammalian function. Accordingly, blood glucose is maintained in a tight range by the actions of insulin and counterregulatory hormones. Insulin promotes glucose uptake into muscle and adipose tissue and inhibits glucose production by the liver. Impairment of the normal response to insulin (insulin resistance) is a common disorder of substantial medical importance: insulin resistance is a major risk factor for hypertension, dyslipidemia, cardiovascular disease and cardiac death, and polycystic ovarian disease, as well as diabetes (1). The mechanisms underlying insulin resistance remain largely unknown.
Insulin action is mediated by a cascade of tyrosyl phosphorylation events, initiated by binding of insulin to the insulin receptor (IR) (2, 3). Binding increases the kinase activity of the IR, which then phosphorylates insulin receptor substrates (IRSs) on multiple tyrosyl residues. Phosphotyrosyl residues on IRSs act as docking sites for many SH2 domain-containing proteins, including the p85 regulatory subunit of PI3′ kinase (PI3K). On binding to IRS proteins, PI3K is activated and promotes glucose uptake. Skeletal muscle is the major site of insulin-stimulated glucose uptake in vivo. Both IRS-1 and IRS-2 are expressed in skeletal muscle; data are conflicting regarding the relative role of each IRS in mediating insulin-stimulated glucose uptake in muscle (4–8).
A critical regulatory step in insulin signal transduction is the dephosphorylation of signaling molecules by protein tyrosine phosphatases (PTPs), which helps to terminate insulin signaling (9, 10). In insulin-resistant states such as obesity and type 2 diabetes, insulin signaling is impaired. Several studies of obese humans and rodents report that the expression and/or activity of specific PTPs, including leukocyte antigen-related phosphatase (LAR), protein-tyrosine phosphatase 1B (PTP1B), and src-homology phosphatase-2 (SHP2), are increased in muscle and adipose tissue (11–15). We have also observed increased levels of LAR expression in immunoblots of liver extracts from Zucker rats (J.M.Z., unpublished observations). Immunodepletion experiments suggest that a 2–3-fold elevation of LAR accounts for most of the enhanced PTP activity in both adipose tissue (12) and muscle (13) of obese humans.
In cultured cells, LAR can modulate insulin signaling, at least when overexpressed and/or ectopically expressed (16, 17). LAR overexpression reportedly inhibits IR tyrosyl phosphorylation (16, 17). In vitro studies suggest that LAR preferentially dephosphorylates insulin receptor Tyr-1150 (18), one of three tyrosyl residues that are critical for receptor activity (2). IRS-1 also is a substrate of LAR in vitro (19, 20).
Although these studies suggest that LAR could play a role in insulin action, data from in vivo experiments are inconclusive. Two independent lines of LAR knockout mice give conflicting results about the role of LAR in normal glucose homeostasis (ref. 21 and W. J. A. J. Hendriks and M. P. H. Møller, personal communication). The absence of LAR in the brain, where it may be important for establishing and maintaining neuronal networks (22, 23), might contribute to the complex phenotype seen in LAR knockout mice (21) and obscure a role of LAR in glucose homeostasis. Even if LAR has no role in regulating insulin signaling under normal conditions in vivo, it is possible that overexpressed LAR (as has been observed in insulin-resistant states), might pathologically impair insulin signaling. However, it remains unclear whether overexpression of LAR contributes to the pathogenesis of insulin resistance. To address this issue, we overexpressed LAR selectively in muscle of transgenic mice to levels comparable to those reported in insulin resistant humans. These mice have defective insulin action specifically in muscle, which causes whole body insulin resistance.
Materials and Methods
Transgenic MCK-hLAR (Muscle Creatine Kinase–Human LAR) Mice.
A 7.7-kb EcoRI fragment of human LAR cDNA from plasmid pSP6-LAR (24) was inserted into a BstEII site in p3300MCK-CAT (25), downstream of the muscle creatine kinase promoter/enhancer and start site, and upstream of intron and SV40 polyadenylation sequences (Fig. 1a). The 12.6-kb BssHII partial digestion fragment of the resultant plasmid was injected into the pronuclei of fertilized zygotes from FVB mice. Two founders overexpressing human LAR were obtained. Mice heterozygotic for the transgene were studied. Mice were housed at 22°C with a 12-h light/dark cycle and were fed a standard diet (Purina Autoclavable Rodent Diet 5010) ad libitum.
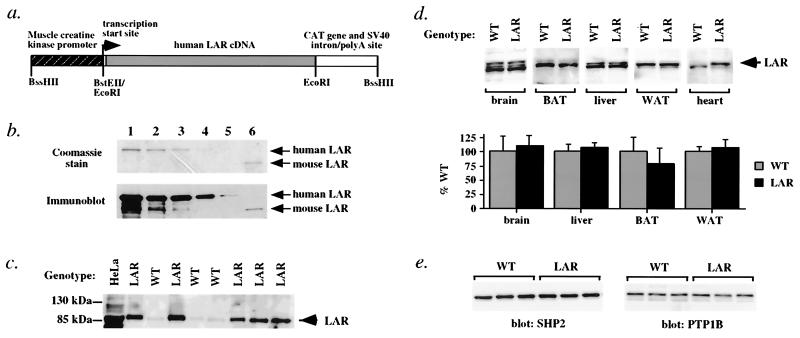
Figure 1.
Transgenic overexpression of human LAR in MCK-hLAR mice is muscle-specific. (a) MCK-hLAR transgene. A schematic diagram of the human LAR transgene used to generate muscle-specific human LAR overexpressing mice is shown. Sequences corresponding to the muscle creatine kinase gene enhancer/promoter, and CAT gene and the intron/polyadenylation site of SV40 are shown as stippled and open boxes, respectively. Sequences corresponding to the LAR cDNA are represented as a gray box. Relevant restriction sites and the transcription start site are indicated. (b) Reactivity of anti-human LAR antibody to human and mouse LAR. Human and mouse GST-LAR fusions proteins were isolated from bacterial lysates, separated by SDS/PAGE, and stained with Coomassie brilliant blue R-250 (Upper) or blotted onto nitrocellulose and probed with anti-human LAR antibody (Lower). Lanes 1–5 contain serial dilutions of human GST-LAR and lane 6 contains mouse GST-LAR. The positions of the GST-LAR proteins are indicated. (c) Overexpression of LAR in MCK-hLAR transgenic mice. Muscle glycoproteins were separated by SDS/PAGE and immunoblotted for LAR. Molecular weight markers are shown, and the position of the 85-kDa subunit of LAR is indicated with an arrow. Results are representative of seven WT and eleven transgenic mice from two different founders. HeLa cell lysates are shown as a positive control. (d) Tissue distribution of LAR overexpression. LAR levels in brain, liver, heart, interscapular brown adipose tissue (BAT), and perigonadal WAT of male mice were determined as in c. Representative blots and quantification of three WT and three transgenic mice are shown. (e) Expression of SHP2 and PTP1B. Muscle lysates from female mice were immunoblotted with polyclonal antisera against SHP2 or PTP1B. Each lane represents one animal. Results are representative of immunoblots on a total of 7–10 male and 12–13 female animals per genotype.
PCR Genotyping of hLAR Transgenic Mice.
Toe DNA was prepared by proteinase K digestion (26). Human LAR was amplified by PCR. Primers and reaction conditions are available from J.M.Z.
Preparation of LAR-GST Fusion Proteins.
Human and murine GST-LAR fusion proteins were isolated and purified on glutathione agarose beads by using standard procedures. Duplicate gels were Coomassie-stained or transferred to nitrocellulose and immunoblotted with polyclonal antibodies against the human LAR cytoplasmic domain (a gift of M. Streuli, Dana–Farber Cancer Institute, Boston) (27), followed by enhanced chemiluminesence (ECL from Amersham Pharmacia). Plasmid pGEX-hLAR (gift of M. Streuli) contains human LAR amino acids 1275–1881 (28), whereas pGEX-mLAR cDNA#1 (gift of W. Hendriks, University of Nijmegen, Nijmegen, The Netherlands) encodes nucleotides 421-2202 of murine LAR (29).
Detection of hLAR Expression in Transgenic Mice.
Tissue lysates [from liver, heart, interscapular brown adipose tissue (BAT), perigonadal white adipose tissue (WAT), and hindlimb muscle] were prepared as described (30). To detect LAR, 1 mg of protein was incubated with wheat germ agarose beads (Sigma) overnight and washed twice in PBS with 1 mM sodium vanadate and twice with PBS alone, and bound proteins were separated by SDS/PAGE on 10% gels and subjected to anti-LAR immunoblotting. Muscle lysates also were immunoblotted with polyclonal antibodies against the IRβ subunit (Santa Cruz Biotechnology), SHP2 (Santa Cruz Biotechnology), PTP1B (31), IRS-1, and IRS-2 (gifts of M. White, Joslin Diabetes Center, Boston; ref. 32), and a monoclonal antibody to p85α (Upstate Biotechnology, Lake Placid, NY), following the manufacturer's directions or as described in the cited publications.
Tyrosyl Phosphorylation of IR, IRS-1, and IRS-2.
Fasted (15–18 h) mice were injected i.v. with 10 milliunits of insulin per gram of body weight (Humulin from Eli Lilly) and killed 3 min later; various tissues were then dissected and frozen in liquid N2. Lysates (0.5–1 mg protein) were subjected to serial immunoprecipitations with anti-IRS-1 or -IRS-2 polyclonal antisera (3 μl) followed by anti-IR polyclonal antisera (1 μl), bound to protein A-Sepharose (Sigma), essentially as described (30). Immune complexes were resolved by SDS/PAGE, subjected to antiphosphotyrosine immunoblotting (PY20 or PY99, Santa Cruz Biotechnology), and visualized by using ECL. The efficiency of immunoprecipitation of IR was ≥70%, of IRS-1 was ≥95%, and of IRS-2 was ≥90% (data not shown).
Ex Vivo Insulin Stimulation of Muscle.
Extensor digitorum longus (EDL) or soleus muscles were rapidly dissected from fasted female mice, and incubated for 10 min in pregassed (95% O2, 5% CO2) Krebs–Henseleit buffer [KHB: 118.5 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 25 mM NaHCO3, 2.5 mM CaCl2, 1.2 mM Mg SO4, 0.1% BSA, 5 mM Hepes (pH 7.4)] containing 10 mM glucose at 35°C, followed by 29°C for 10 min, and then stimulated with or without insulin (33 mM) for 3 or 6 min. Muscle was rapidly removed, blotted dry, and frozen in liquid N2, and lysates were prepared as described above.
PI3 Kinase Assays.
PI3K activity associated with IRS-1, IRS-2, or antiphosphotyrosine immune complexes (from 0.25–1 mg muscle protein) was determined as reported (30).
Measurement of Glucose, Insulin, and Free Fatty Acid Levels.
For hyperinsulinemic–euglycemic clamp studies, plasma glucose was measured by glucose oxidase reaction using a Beckman glucose analyzer II (Beckman Coulter). For all other studies, glucose levels were measured by using a One Touch II glucometer (Lifescan from Johnson & Johnson, Milpitas, CA). For hyperinsulinemic–euglycemic clamp studies, plasma insulin levels were determined by RIA (Linco Research Immunoassay, St. Charles, MO). For all other experiments, serum insulin levels were determined with a rat insulin ELISA (Crystal Chem, Chicago, IL) by using mouse insulin standards. Serum free fatty acid levels were measured by using a colorometric diagnostic assay (Wako Biochemicals, Osaka).
Hyperinsulinemic–Euglycemic Clamps.
Hyperinsulinemic–euglycemic (111 ± 3 mg/dl) clamps were performed as described (33) on male mice 5–8 months of age. Insulin was infused continuously for 120 min at 2.5 milliunits/kg/min.
Body Composition.
Carcasses (with their gastrointestinal tracts removed) were weighed, dried at 60°C, reweighed to determine their water content, and hydrolyzed in ethanolic potassium hydroxide (34). Body triglyceride content was determined by enzymatic measurement of glycerol (product no. 337–40A; Sigma). All other values were derived as described (34).
Statistical Analyses.
Data are presented as means ± SEM. Statistical analyses were performed by using statview software (Abacus Concepts, Berkeley, CA). Statistical significance was tested with unpaired Student's t tests (two-tailed unless otherwise indicated).
Results
Generation of Mice Overexpressing LAR Selectively in Muscle.
Transgenic mice overexpressing LAR in muscle were generated by ligating the hLAR cDNA downstream of the 3.3-kb MCK promoter/enhancer (see Materials and Methods and Fig. 1a), which directs high-level expression in skeletal muscle and lower levels in cardiac muscle (35). Two founders expressing the LAR transgene were generated. MCK-hLAR mice were born at the expected Mendelian frequency and showed normal growth curves and development (data not shown). Carcass analysis showed a 19% increase in body lipid per mouse (Table 1), which was not seen by dual-energy x-ray absorptiometry (DEXA) analysis (Lunar Piximus Densitometer, Lunar, Madison, WI; data not shown), or when body lipid was expressed as a percent of body weight. Fat-free dry mass was unchanged (Table 1).
Table 1.
Body composition of MCK-hLAR mice
| Genotype (n) | BW, g | Water, g | Lipid
|
Fat-free dry mass
|
||
|---|---|---|---|---|---|---|
| g | % BW | g | % BW | |||
| WT (5) | 27.78 ± 0.50 | 17.48 ± 0.43 | 2.62 ± 0.11 | 9.43 ± 0.45 | 7.68 ± 0.13 | 27.67 ± 0.29 |
| MCK-hLAR (5) | 28.58 ± 0.21 | 17.80 ± 0.23 | 3.13 ± 0.16* | 10.94 ± 0.57 | 7.65 ± 0.05 | 26.78 ± 0.25 |
Body composition was performed on 5-month-old male mice. Data are expressed as mean ± SEM. BW, body weight. *, P < 0.04.
The level of overexpression of the human LAR transgene was assessed by using antibody generated against the human 85-kDa cytoplasmic domain (27). The reactivity of the antibody to human GST-LAR was ≈20 times greater than for mouse GST-LAR (Fig. 1b, compare lanes 3 and 6). After correcting for differences in antibody cross-reactivity, both lines of mice overexpressed LAR ≈2–4-fold in skeletal muscle (Fig. 1c) and at a lower level in heart (Fig. 1c) and expression was stable over multiple generations. Other tissues, including liver, brain, WAT, and brown adipose tissue (BAT), showed no overexpression of LAR (Fig. 1d). Two other PTPs implicated in regulating insulin signaling, SHP2 and PTP1B (9, 10), were expressed at similar levels in muscle of MCK-hLAR transgenic and wild-type (WT) mice (Fig. 1e). Both MCK-hLAR lines exhibited the same phenotype, which is described below.
MCK-hLAR Mice Are Insulin-Resistant.
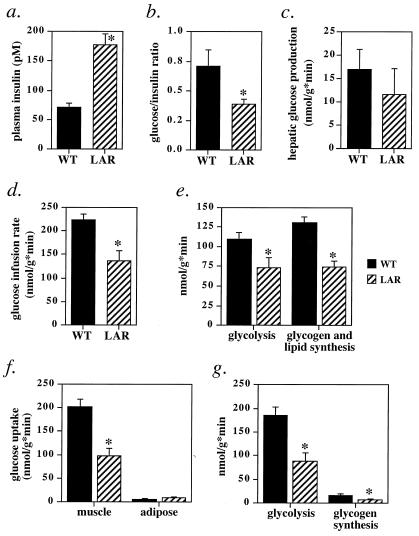
Insulin resistance in the ambient state was evident in male MCK-hLAR mice by a 2.5-fold elevation in fasting plasma insulin levels (Fig. 2a) with normal blood glucose (see Table 2, which is published as supplemental data on the PNAS web site, www.pnas.org) and by a reduced ratio of glucose to insulin 15 min after glucose challenge (Fig. 2b). Fed blood glucose and insulin levels were normal (see Table 2). There also was a tendency toward increased insulin levels and decreased glucose/insulin ratios in fed females (data not shown). MCK-hLAR mice were not overtly diabetic, and fasted and fed serum free fatty acid levels were normal (see Table 2). To investigate the effects of LAR overexpression on metabolic pathways in vivo, mice were subjected to hyperinsulinemic (2.5 milliunits/kg/min)–euglycemic clamps. This resulted in submaximal plasma insulin levels, which suppressed hepatic glucose production similarly in WT and MCK-hLAR mice (Fig. 2c), indicating that liver remains insulin-responsive in MCK-hLAR mice. However, the rate of glucose infusion needed to maintain euglycemia was decreased by 39% (Fig. 2d), indicating insulin resistance at the whole-body level. Glucose can be metabolized via glycolysis or used for glycogen and lipid synthesis. Both pathways were suppressed to comparable extents in MCK-hLAR mice (by 33% and 43%, respectively; Fig. 2e).
Figure 2.
Insulin resistance in MCK-hLAR mice. Fasting plasma insulin (a), glucose/insulin ratio 15 min after glucose injection (b), hepatic glucose production (c), glucose infusion rate (d), whole body glycolysis and glycogen and lipid synthesis (e), glucose uptake into gastrocnemius muscle and epididymal white adipose tissue (WAT) (f), and muscle glycolysis and glycogen synthesis (g) in MCK-hLAR mice. (a) Male mice 5–8 months old were fasted for 16 h overnight and blood was obtained from tail vein. (b) Male mice 6–8 months old were fasted 14–16 h. Glucose (1 mg/g) was injected i.p. and blood was obtained 15 min later. The glucose/insulin ratio was determined by dividing glucose (mg/dl) at 15 min by insulin (ng/ml) at 15 min for each animal. (c–g) Male mice, aged 5–8 months, were subjected to hyperinsulinemic–euglycemic clamp studies with an insulin infusion rate of 2.5 milliunits/kg/min for 120 min, resulting in insulin levels of 741 ± 66 pM in WT mice and 788 ± 109 pM in MCK-hLAR mice (p = NS). Results are mean ± SEM of 5–6 mice (a and c–g) or 10–12 mice (b) per genotype. *, P < 0.05 one-tailed t test.
We also measured glucose uptake into skeletal muscle and WAT during hyperinsulinemic–euglycemic clamps to determine whether the insulin resistance in the MCK-hLAR mice is due to impaired glucose uptake only in muscle or whether adipose tissue is affected indirectly. Glucose uptake specifically into muscle was decreased by 50% in MCK-hLAR mice compared with WT mice (Fig. 2f). Glycolysis and glycogen synthesis were impaired to similar extents in muscle (49% and 41%, respectively; Fig. 2g). In contrast, in WAT of MCK-hLAR mice, glucose uptake was unchanged (Fig. 2f). Thus, insulin action is preserved in adipose tissue and liver, whereas it is impaired in muscle, and the defect in muscle is sufficient to reduce whole-body glucose disposal.
Effects of hLAR Overexpression on Insulin Signal Transduction.
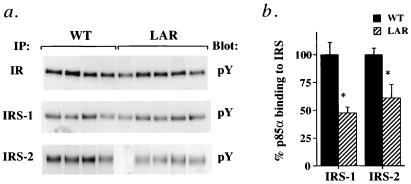
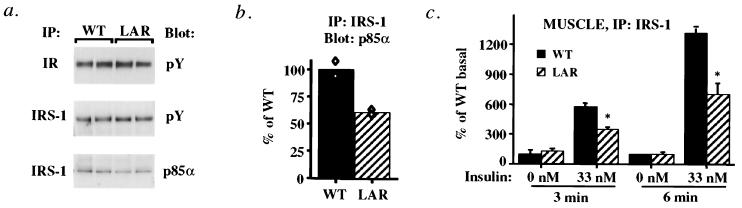
Insulin injection stimulated tyrosyl phosphorylation of the IR by 15–20-fold and of IRS-1 by 5–7-fold in muscle of both control and MCK-hLAR mice (data not shown). There were no differences in tyrosyl phosphorylation of IR or IRS-1 between genotypes (Fig. 3a), but insulin-stimulated IRS-2 tyrosyl phosphorylation was reduced by 62% in muscle of MCK-hLAR mice (Fig. 3a). Although we did not detect changes in tyrosyl phosphorylation of IRS-1 by immunoblotting for phosphotyrosine, the amount of p85α coimmunoprecipitating with either IRS-1 or IRS-2 was reduced by 39–52% (Fig. 3b). There were no differences in the amounts of IR, IRS-1, IRS-2, or p85α in muscle of MCK-hLAR mice compared with controls (data not shown). Morever, consistent with the effect of LAR overexpression on tyrosyl phosphorylation of only some components of the insulin signaling pathway, the pattern of tyrosyl phosphorylation in total muscle lysates was largely the same in WT and MCK-hLAR mice (data not shown). These data indicate that LAR overexpression has highly specific and selective effects on muscle phosphotyrosyl proteins.
Figure 3.
In vivo insulin-stimulated IR, IRS-1, and IRS-2 phosphorylation (a) and p85α association (b). Male mice were fasted for 16–18 h, injected i.v. with saline or insulin (10 units/kg), and killed 3 min postinjection. (a) Gastrocnemius muscle lysates were subjected to immunoprecipitation with anti-IR, anti-IRS-1, or anti-IRS-2 polyclonal antisera and immunoblotted with antiphosphotyrosine antibodies. (b) IRS-1 and IRS-2 immunoprecipitates were also blotted with anti-p85α monoclonal antibody. Each lane represents one animal. Immunoblots and bars (mean ± SEM) are representative of three separate experiments each on a total of 11–12 mice in each group. Similar results were seen in male and female mice. *, P < 0.05.
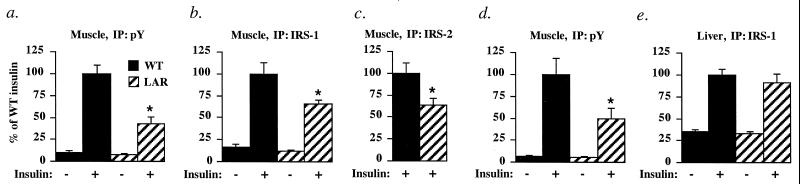
Insulin increased phosphotyrosine-associated PI3K activity 9–17-fold (Fig. 4 a and d) and IRS-1-associated PI3K 5-fold (Fig. 4b) in muscle of WT mice. Insulin-stimulated phosphotyrosine-associated PI3K activity was reduced by 51–57%, and IRS-1- and IRS-2-associated PI3K activities were reduced by 34–37% in muscle of MCK-hLAR mice (Figs. 4 a–d). In liver of MCK-hLAR mice, insulin-stimulated phosphotyrosine-associated (data not shown) and IRS-1-associated (Fig. 4e) PI3K activities were normal, indicating that PI3K activity is impaired only where LAR is overexpressed.
Figure 4.
PI3K activity in MCK-hLAR mice. Phosphotyrosine- (a), IRS-1- (b), or IRS-2-associated (c) PI3K activity in muscle of male mice, phosphotyrosine-associated PI3K activity in muscle of female mice (d), and IRS-1-associated PI3K activity in liver of female mice (e). Mice were fasted for 16–18 h, injected i.v. with saline or insulin (10 units/kg), and killed 3 min postinjection. PI3K activity was measured in phosphotyrosine, IRS-1, or IRS-2 immunoprecipitates of hind-limb muscles or liver as described in Materials and Methods. Results are mean ± SEM for three to five mice per group. *, difference from nontransgenic insulin-stimulated is P < 0.05. Similar results were seen in male and female mice.
To test whether these insulin signaling defects are intrinsic to muscle or due to the metabolic milieu, we measured PI3K activity in muscles ex vivo. In parallel with the results in vivo, in muscles from MCK-hLAR mice stimulated ex vivo with insulin, IR, and IRS-1 tyrosyl phosphorylation were normal (Fig. 5a). However, IRS-1-associated p85α was reduced by 50% (Fig. 5 a and b), and IRS-1-associated PI3K activity was reduced by 39–46% compared with WT controls (Fig. 5c). Thus, overexpression of LAR in muscle impairs insulin-stimulated PI3K activity ex vivo and in vivo.
Figure 5.
Ex vivo insulin signaling in muscle of MCK-hLAR mice. Phosphorylation of IR and IRS-1 (a) and p85 association with IRS-1 (b). Soleus or extensor digitorum longus (EDL) muscles from female mice fasted for 16–18 h were incubated ex vivo without or with 33 nM insulin for 3 or 6 min. Muscle lysates were subjected to immunoprecipitation with anti-IR or anti-IRS-1 polyclonal antisera and immunoblotted with anti-phosphotyrosine antibodies as described in Materials and Methods. IRS-1 immunoprecipitates were also blotted with anti-p85α monoclonal antibody. Blots are representative of 4–12 mice per genotype. Each lane of immunoblots represents two animals. Bars are averages of two determinations each on two mice per group. Each symbol represents the value for muscle pooled from two mice. (c) IRS-1-associated PI3K activity. Bars are mean ± SEM of muscles from four different animals. *, P < 0.05 compared with WT.
Discussion
Recent work has begun to uncover the complexities of insulin resistance, including the fact that resistance in different insulin target tissues or at different steps in the insulin action cascade in a single tissue leads to markedly different phenotypes (36–38). The current study demonstrates that muscle-specific overexpression of a single tyrosine phosphatase, LAR, in transgenic mice reduces insulin signaling in muscle and impairs whole body glucose disposal. Because LAR is one of the major PTPs that is reportedly increased in expression and/or activity in muscle and adipose tissue of insulin-resistant, obese humans, our findings have potentially important implications for understanding the pathogenesis of insulin resistance.
Although multiple studies show that PTP overexpression accompanies insulin-resistant states such as obesity, it has not been clear whether these changes are causal or merely correlative. Our data show that increased LAR expression in muscle produces insulin resistance, as manifested by increased fasting plasma insulin levels and decreased whole-body glucose disposal. Glucose uptake in muscle is impaired, resulting in defects in both major pathways of glucose metabolism (i.e., glycolysis and glycogen synthesis). This defect appears to result from impaired signaling via PI3K, and possibly other insulin-stimulated pathways. Defective insulin action is restricted to muscle, the only tissue in which the transgene is expressed.
Recently, the role of the insulin receptor in muscle in whole-body insulin responsiveness has been questioned (36, 39, 40). Our study shows that attenuating insulin signaling selectively in muscle is sufficient to cause insulin resistance. Although relatively mild, this resistance is at least as great as the insulin resistance reported in mice with total ablation of the insulin receptor from muscle (MIRKO mice) (36, 41). The phenotype of MCK-hLAR mice differs from that of MIRKO mice, because MIRKO mice have a much more significant increase in fat pad mass and have dyslipidemia (36). These differences may reflect the fact that LAR may act at certain specific sites in the insulin signaling cascade, as suggested by our biochemical analyses, and thereby alter only some of the biological actions of insulin.
In muscle of MCK-hLAR mice, insulin-stimulated PI3K activity associated with phosphotyrosine, IRS-1, or IRS-2 is impaired, but no reduction in total cellular tyrosyl phosphorylation or tyrosyl phosphorylation of IR or IRS-1 is detected. Thus, although previous studies in cell lines indicate that LAR can dephosphorylate IR (16–18), our data suggest that in vivo, LAR acts more distally, perhaps by directly dephosphorylating specific sites on IRS proteins. This finding is consistent with the demonstration that IRS-1 is a substrate for LAR in vitro (19, 20). However, we cannot exclude the possibility that LAR may dephosphorylate specific tyrosyl residues on IR that are not detected when using an assay of total IR tyrosyl phosphorylation. Whereas the binding of insulin to its receptor elicits rapid autophosphorylation of six tyrosine residues in the receptor β subunit, the kinase activity of IR is largely determined by phosphorylation of three C-terminal tyrosines (positions 1146, 1150, and 1151; ref. 2). Mutation or dephosphorylation of one or more of these reduces insulin-stimulated receptor kinase activity (2). Dephosphorylation of a single IR tyrosyl residue could contribute to the defects in MCK-hLAR mice. We also cannot exclude the possibility that LAR overexpression down-regulates insulin signaling by an indirect mechanism such as activation of serine phosphorylation. Although we have demonstrated resistance to insulin-stimulated glucose uptake and impaired p85 binding to IRS-1 and IRS-2 in muscle of MCK-LAR overexpressing mice, this does not prove that impaired IRS function is the major cause of insulin resistance; other pathways also may be involved.
Some early work on PTPs suggested that these enzymes might have broad specificity. However, many recent studies (reviewed in ref. 42) emphasize the exquisite in vivo specificity of these enzymes for particular phosphotyrosyl proteins—and in some cases even particular tyrosyl phosphorylation sites within the same protein. Although our results suggest that the observed effects of LAR overexpression on glucose homeostasis do not reflect global perturbation of cellular tyrosyl phosphorylation pathways, it remains possible that other proteins are affected, especially because the effect on IRS-1 was not detected with antiphosphotyrosyl immunoblotting.
The potential for PTPs to regulate insulin action and metabolism in vivo was recently demonstrated by the PTP1B knockout mouse, which has increased insulin sensitivity as well as resistance to diet-induced obesity. The MCK-hLAR mice provide proof that overexpression of a specific PTP in muscle can cause insulin resistance. Several PTPs are overexpressed simultaneously in muscle and adipose tissue of insulin-resistant obese humans and rodents. There may be an additive effect resulting in more severe insulin resistance if additional PTPs are also overexpressed, and/or additional insulin target tissues are directly affected. Most forms of obesity and diabetes are polygenic; the concurrence of increased PTP expression with other genetic abnormalities also could result in more severe insulin resistance. Thus, our data support an important pathogenic role for the overexpression of LAR or PTPs with similar substrate specificity in the development of insulin resistance and suggest that LAR may be a target for antidiabetic therapy.
Supplementary Material
Acknowledgments
We thank Joel Lawitts for embryo injections, Zhong Li for technical assistance, and Dr. Christos Mantzoros for help with statistical analysis. J.M.Z. and L.D.K. were supported by National Institutes of Health National Research Service Awards DK09903 and AI09815, respectively, Y.-B.K. by an Uehara Memorial Foundation Research Fellowship and an American Diabetes Association mentor-based fellowship, O.D.P. by Association de Langue Francaise d'Etudes du Diabetes et des Anomalies du Metabolisme and the Nestle Society, and O.B. by the Human Frontier Science Program (LT 0020/1999). This work was supported by National Institutes of Health Grants R01 DK43051, P30 DK46200, P30 DK57521, and P01 DK56116 (to B.B.K), R01 CA49152, P01 DK50654, and P50 HL56993 (to B.G.N.), and R01 DK40936 and P30 DK45735 (to G.I.S.). G.I.S. is an investigator and J.K.K. is an associate of the Howard Hughes Medical Institute.
Abbreviations
- LAR
leukocyte antigen-related
- PTP
protein-tyrosine phosphatase
- IR
insulin receptor
- IRS
IR substrate
- PI3K
PI3′ kinase
- MCK
muscle creatine kinase
- hLAR
human LAR
- WAT
white adipose tissue
- SHP2
src-homology phosphatase-2
- PTP1B
protein-tyrosine phosphatase 1B
References
- 1.Mantzoros C S, Flier J S. Adv Endocrinol Metab. 1995;6:193–232. [PubMed] [Google Scholar]
- 2.White M F. Diabetologia. 1997;40, Suppl. 2:S2–S17. doi: 10.1007/s001250051387. [DOI] [PubMed] [Google Scholar]
- 3.White M F, Yenush L. Curr Top Microbiol Immunol. 1998;228:179–208. doi: 10.1007/978-3-642-80481-6_8. [DOI] [PubMed] [Google Scholar]
- 4.Withers D J, Burks D J, Towery H H, Altamuro S L, Flint C L, White M F. Nat Genet. 1999;23:32–40. doi: 10.1038/12631. [DOI] [PubMed] [Google Scholar]
- 5.Higaki Y, Wojtaszewski J F, Hirshman M F, Withers D J, Towery H, White M F, Goodyear L J. J Biol Chem. 1999;274:20791–20795. doi: 10.1074/jbc.274.30.20791. [DOI] [PubMed] [Google Scholar]
- 6.Kido Y, Burks D J, Withers D, Bruning J C, Kahn C R, White M F, Accili D. J Clin Invest. 2000;105:199–205. doi: 10.1172/JCI7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gazdag A C, Dumke C L, Kahn C R, Cartee G D. Diabetes. 1999;48:1930–1936. doi: 10.2337/diabetes.48.10.1930. [DOI] [PubMed] [Google Scholar]
- 8.Yamauchi T, Tobe K, Tamemoto H, Ueki K, Kaburagi Y, Yamamoto-Honda R, Takahashi Y, Yoshizawa F, Aizawa S, Akanuma Y, et al. Mol Cell Biol. 1996;16:3074–3084. doi: 10.1128/mcb.16.6.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein B J, Li P M, Ding W, Ahmad F, Zhang W R. Vitam Horm (San Francisco) 1998;54:67–96. doi: 10.1016/s0083-6729(08)60922-x. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein B J, Ahmad F, Ding W, Li P M, Zhang W R. Mol Cell Biochem. 1998;182:91–99. [PubMed] [Google Scholar]
- 11.Ahmad F, Goldstein B J. Metabolism. 1995;44:1175–1184. doi: 10.1016/0026-0495(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad F, Considine R V, Goldstein B J. J Clin Invest. 1995;95:2806–2812. doi: 10.1172/JCI117985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmad F, Azevedo J L, Cortright R, Dohm G L, Goldstein B J. J Clin Invest. 1997;100:449–458. doi: 10.1172/JCI119552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad F, Considine R V, Bauer T L, Ohannesian J P, Marco C C, Goldstein B J. Metabolism. 1997;46:1140–1145. doi: 10.1016/s0026-0495(97)90206-7. [DOI] [PubMed] [Google Scholar]
- 15.McGuire M C, Fields R M, Nyomba B L, Raz I, Bogardus C, Tonks N K, Sommercorn J. Diabetes. 1991;40:939–942. doi: 10.2337/diab.40.7.939. [DOI] [PubMed] [Google Scholar]
- 16.Zhang W R, Li P M, Oswald M A, Goldstein B J. Mol Endocrinol. 1996;10:575–584. doi: 10.1210/mend.10.5.8732688. [DOI] [PubMed] [Google Scholar]
- 17.Li P M, Zhang W R, Goldstein B J. Cell Signalling. 1996;8:467–473. doi: 10.1016/s0898-6568(96)00101-5. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto N, Feener E P, Zhang W R, Goldstein B J. J Biol Chem. 1992;267:13811–13814. [PubMed] [Google Scholar]
- 19.Goldstein B J, Bittner-Kowalczyk A, White M F, Harbeck M. J Biol Chem. 2000;275:4283–4289. doi: 10.1074/jbc.275.6.4283. [DOI] [PubMed] [Google Scholar]
- 20.Calera M R, Vallega G, Pilch P F. J Biol Chem. 2000;275:6308–6312. doi: 10.1074/jbc.275.9.6308. [DOI] [PubMed] [Google Scholar]
- 21.Ren J M, Li P M, Zhang W R, Sweet L J, Cline G, Shulman G I, Livingston J N, Goldstein B J. Diabetes. 1998;47:493–497. doi: 10.2337/diabetes.47.3.493. [DOI] [PubMed] [Google Scholar]
- 22.Yeo T T, Yang T, Massa S M, Zhang J S, Honkaniemi J, Butcher L L, Longo F M. J Neurosci Res. 1997;47:348–360. doi: 10.1002/(sici)1097-4547(19970201)47:3<348::aid-jnr13>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 23.Van Vactor D. Curr Opin Cell Biol. 1998;10:174–181. doi: 10.1016/s0955-0674(98)80139-7. [DOI] [PubMed] [Google Scholar]
- 24.Streuli M, Krueger N X, Hall L R, Schlossman S F, Saito H. J Exp Med. 1988;168:1523–1530. doi: 10.1084/jem.168.5.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaynes J B, Johnson J E, Buskin J N, Gartside C L, Hauschka S D. Mol Cell Biol. 1988;8:62–70. doi: 10.1128/mcb.8.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kogan S C, Doherty M, Gitschier J. N Engl J Med. 1987;317:985–990. doi: 10.1056/NEJM198710153171603. [DOI] [PubMed] [Google Scholar]
- 27.Debant A, Serra-Pages C, Seipel K, O'Brien S, Tang M, Park S H, Streuli M. Proc Natl Acad Sci USA. 1996;93:5466–5471. doi: 10.1073/pnas.93.11.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pulido R, Serra-Pages C, Tang M, Streuli M. Proc Natl Acad Sci USA. 1995;92:11686–11690. doi: 10.1073/pnas.92.25.11686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaapveld R Q, van den Maagdenberg A M, Schepens J T, Weghuis D O, Geurts van Kessel A, Wieringa B, Hendriks W J. Genomics. 1995;27:124–130. doi: 10.1006/geno.1995.1014. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y B, Zhu J S, Zierath J R, Shen H Q, Baron A D, Kahn B B. Diabetes. 1999;48:310–320. doi: 10.2337/diabetes.48.2.310. [DOI] [PubMed] [Google Scholar]
- 31.Klaman L D, Boss O, Peroni O D, Kim J K, Martino J L, Zabolotny J M, Moghal N, Lubkin M, Kim Y B, Sharpe A H, et al. Mol Cell Biol. 2000;20:5479–5489. doi: 10.1128/mcb.20.15.5479-5489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Withers D J, Gutierrez J S, Towery H, Burks D J, Ren J M, Previs S, Zhang Y, Bernal D, Pons S, Shulman G I, et al. Nature (London) 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 33.Kim J K, Gavrilova O, Chen Y, Reitman M L, Shulman G I. J Biol Chem. 2000;275:8456–8460. doi: 10.1074/jbc.275.12.8456. [DOI] [PubMed] [Google Scholar]
- 34.Salmon D M, Flatt J P. Int J Obes. 1985;9:443–449. [PubMed] [Google Scholar]
- 35.Johnson J E, Wold B J, Hauschka S D. Mol Cell Biol. 1989;9:3393–3399. doi: 10.1128/mcb.9.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruning J C, Michael M D, Winnay J N, Hayashi T, Horsch D, Accili D, Goodyear L J, Kahn C R. Mol Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 37.Zisman A, Peroni O D, Abel E D, Michael M D, Mauvais-Jarvis F, Lowell B B, Wojtaszewski J F, Hirshman M F, Virkamaki A, Goodyear L J, et al. Nat Med. 2000;6:924–928. doi: 10.1038/78693. [DOI] [PubMed] [Google Scholar]
- 38.Michael M D, Kulkarni R N, Postic C, Previs S F, Shulman G I, Magnuson M A, Kahn C R. Mol Cell. 2000;6:87–97. [PubMed] [Google Scholar]
- 39.Chang P Y, Benecke H, Le Marchand-Brustel Y, Lawitts J, Moller D E. J Biol Chem. 1994;269:16034–16040. [PubMed] [Google Scholar]
- 40.Lauro D, Kido Y, Castle A L, Zarnowski M J, Hayashi H, Ebina Y, Accili D. Nat Genet. 1998;20:294–298. doi: 10.1038/3112. [DOI] [PubMed] [Google Scholar]
- 41.Kim J K, Michael M D, Previs S F, Peroni O D, Mauvais-Jarvis F, Neschen S, Kahn B B, Kahn C R, Shulman G I. J Clin Invest. 2000;105:1791–1797. doi: 10.1172/JCI8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neel B G, Tonks N K. Curr Opin Cell Biol. 1997;9:193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.