Abstract
Here we report the biological and molecular characterization of a virulent genotype VII Newcastle disease virus (NDV) circulating in Venezuela and the assessment of the vaccination efficacy under field conditions compared to controlled rearing conditions. Biological pathotyping showed a mean embryo dead time of 50 h and an intracerebral pathogenicity index of 1.86. Sequence-based phylogenetic analysis demonstrated that the virus belongs to genotype VII in class II (a genotype often found in Asia and Africa), representing the first report of the presence of this genotype in the continent of South America. A vaccine-challenge trial in commercial broilers reared in fields or in a experimental setting included dual (live/killed) priming of 1-day-old chicks plus two live NDV and infectious bursal disease virus (IBDV) field vaccinations at days 7 and 17, followed by a very stringent genotype VII NDV challenge at day 28. Serology for NDV and IBDV, bursal integrity, and protection against NDV lethal challenge were assessed. At 28 days, field vaccinates showed significantly lower NDV (1,356 versus 2,384) and higher IBD (7,295 versus 1,489) enzyme-linked immunosorbent assay (ELISA) antibody titers than the experimentally reared birds. A lower bursal size and bursa-body weight ratio (P < 0.05) and higher bursa lesion score were also detected in the field set. Only 57.1% of field vaccinates survived the lethal challenge, differing (P < 0.05) from 90.5% survival in the experimental farm. Overall, results confirmed the presence of the genotype VII viruses in South America and suggest that field-associated factors such as immunosuppression compromise the efficacy of the vaccination protocols implemented.
INTRODUCTION
Newcastle disease virus (NDV) is one of the most important infectious agents in veterinary medicine and the causative agent of Newcastle disease (ND), which affects commercial poultry and causes important economical losses (2). The virus belongs to the family Paramyxoviridae, subfamily Paramyxovirinae, in the genus Avulavirus (15), which encompasses a diverse group of single-stranded, negative-sense, nonsegmented RNA viruses. Due to variations in virulence and host susceptibility, the symptoms of NDV infection in domestic species (chicken, turkey, goose, duck, and pigeon) range from unapparent to severe; infection causes respiratory, enteric, and nervous system disease, leading to high mortality rates (2).
Antigenic and genetic diversity within the NDV isolates is recognized (1). Different genotypes of NDV circulate throughout the world, albeit they are all members of a unique avian paramyxovirus group 1 (APMV-1) serotype. Since 1926 and based on nucleotide sequence, 9 class I NDV and 10 distinct lineages of class II NDV (I to IX and XI) strains have been identified (6, 11, 17). Molecular characterization is of paramount importance for the epidemiology studies required in the development and adaptation of control strategies (10). Genotypes V, VI, and VII of virulent viruses are the predominant genotypes circulating worldwide (16, 17); of these, genotype VII is particularly important given that it has been associated with many of the most recent outbreaks in Asia, Africa, and the Middle East (10, 11, 12, 28, 29).
Vaccination of commercially reared birds is the best way to reduce losses resulting from NDV infection (2, 21). NDV vaccine strains of genotypes I and II are used to control detrimental effects of subclinical forms of the disease and severe clinical disease during outbreaks (2, 23, 27). Additionally, control of risk factors, including immunosuppressive agents, biosecurity breaks, inadequate management practices, and harsh environments, together with common sense, is required to diminish the economic impact of this and other poultry diseases (5). The aim of this work was to fully characterize a novel genotype VII virulent NDV from Venezuela and to evaluate the efficacy of a currently used commercial vaccination scheme under field and controlled rearing conditions.
MATERIALS AND METHODS
Field virus.
A NDV isolate that was negative in hemagglutination inhibition tests for avian influenza virus and avian adenoviruses (VEN-611) was obtained from a field outbreak (May 2008) in commercial pullet flocks experiencing high mortality rates and showing ND clinical signs. Virus isolation was performed using standard virus isolation procedures and specific-pathogen-free (SPF) embryonated eggs (3).
Biological pathogenicity assessment.
The pathogenic evaluation of the isolate was carried out using standard assay methods to determine the intracerebral pathogenicity index (ICPI) in 1-day-old chicks (3). Briefly, 1-day-old chicks were inoculated intracerebrally with 0.1 ml of a 1:10 dilution of infective allantoic fluid. Chicks were monitored during an 8-day observation period and scored as normal (0), sick or paralyzed (1), and dead (2). Total scores were determined, and the mean daily score was calculated to obtain the ICPI. Mean death time (MDT) determinations were performed as previously reported using SPF embryonated chicken eggs (3).
Vaccine viruses.
The NDV vaccine strains used included a Villegas-Glisson/University of Georgia (VG/GA) strain (AVINEW) for initial live vaccination, an Ulster strain (GALLIMUNE ND) inactivated in oil adjuvant for day-old subcutaneous (SC) application, and strain LaSota and an intermediate live infectious bursal disease virus (IBDV) strain (BURSABLEN) for field vaccination. All commercial vaccines were from Merial Select, Inc., Gainesville, GA, and were applied following the manufacturer's instructions.
Viral RNA extraction and amplification.
NDV-positive allantoic fluid was inactivated and transported to the Southeast Poultry Research Laboratory (Athens, GA) for molecular studies, using Flinders technology filter paper (FTA cards; Whatman International Ltd., Springfield Mill, United Kingdom). Fifty microliters of each NDV-positive allantoic fluid sample was added to the matrix in the card, and RNA extraction was performed as previously explained (22). Reverse transcriptase PCR (RT-PCR) amplification of the complete coding region for the F gene was performed using a Qiagen OneStep RT-PCR kit (Qiagen, Valencia, CA) as previously described (24).
Sequence analysis.
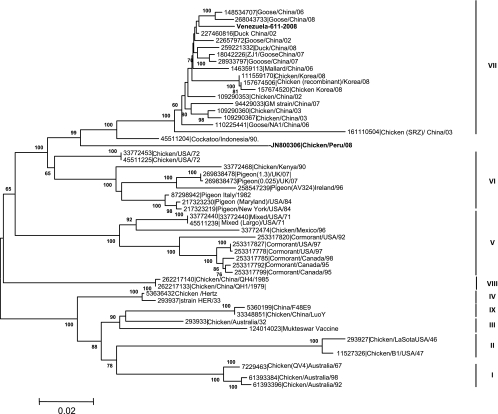
Amplified products were separated on a 1% agarose gel; the bands were excised and eluted using a QIAquick gel extraction kit (Qiagen Valencia, CA) and then sequenced at the DNA sequencing facility at Southeast Poultry Research Laboratory in Athens, GA. An ABI BigDye Terminator version 1.1 sequencing kit (Applied Biosystems, Foster City, CA) was used, and products were run on an ABI 3730XL DNA analyzer (Applied Biosystems). Sequences were edited and aligned with the DNAStar Lasergene 8.0 program (Madison, WI) using the Clustal W algorithm. Determinations of percentages of amino acid identity for the Fusion protein were performed by obtaining a comparison of the full fusion protein of the Venezuelan isolate to those of vaccine and reference strains. The optimal tree with the sum of branch length = 1.10958014 is shown (Fig. 1). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (150 replicates) are shown next to the branches (8). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method and the Kimura 2-parameter method (data not shown), and the units represent the number of base substitutions per site. The rate of variation among sites was modeled with a gamma distribution (shape parameter = 1). Codon positions included were the 1st, 2nd, 3rd, and noncoding positions. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were a total of 1,653 positions in the final data set. Phylogenetic analyses were conducted using MEGA5 software (26).
Fig 1.
Complete F gene nucleotide comparison and evolutionary relationship of the Venezuela virus to reference isolates. The evolutionary history was inferred using the neighbor-joining method as described in Materials and Methods. The numbers used in the phylogenetic trees represent the GenInfo (GI) sequence identification numbers in GenBank and are followed by a brief description of the viruses. The tree is drawn to scale, with branch lengths presented in the same units as those used for the evolutionary distances used to infer the phylogenetic tree.
Vaccine-challenge trial.
To evaluate the level of protection conferred by the NDV vaccination protocol recommended to producers in Venezuela, a vaccine-challenge trial using three groups of commercial broilers was designed and implemented as follows. At day 1, two (2) groups were vaccinated by spray with live strain VG/GA followed by subcutaneous (SC) application of 0.1 ml of the Ulster strain inactivated in oil adjuvant (G01 and G02). The third group was the unvaccinated control (G03). The G01 and G03 groups were transported to a controlled environment (experimental farm) with water and feed provided at libitum. The group G02 birds were wing banded and transferred to a 20,000-bird commercial chicken house. Birds in groups G01 and G02 were vaccinated against NDV and IBDV at days 7 and 17 by the use of drinking water following the manufacturer's instructions. At 1, 14, 21, and 28 days of age, 10 birds per group were bled and assessed for ND and IBDV response to vaccination by the use of an enzyme-linked immunosorbent assay (ELISA), with FlockCheck Newcastle disease and IBDV antibody tests performed following the instructions of the manufacturer (IDEXX, Westbrook, ME). A total of 63 commercial boilers (three groups of seven birds per treatment with three replicates) were used in the challenge. Birds were challenged at 28 days by the intraocular route with 0.1 ml of the local genotype VII virus-infected allantoic fluid diluted 104 in phosphate-buffered saline (PBS) to a 50% tissue culture infective dose (CID50)/ml in 0.1 ml. Birds were observed up to 42 days of age for survival and clinical manifestations.
Bursal integrity assessment.
At 14, 21, and 28 days of age, three birds per group were weighed, euthanatized by cervical dislocation, and subjected to necropsy. The bursae were collected, weighed, and evaluated for the presence of macroscopic lesions, and a portion of each was fixed immediately by immersion in 10% neutral buffered formalin for 24 h. Tissues were then processed and embedded in paraffin, using routine histological techniques. The relative bursa/body weight ratio was obtained using the following formula: relative bursa weight = (bursa weight/body weight) × 1,000. The extent of bursal histological damage was graded on a scale from 1 to 4 as previously described (24): briefly, 1 = normal to 10% follicular atrophy; 2 = focal, mild scattered cell depletion up to 10% to 30% follicular atrophy; 3 = multifocal follicular atrophy at 30% to 70%; 4 = diffuse atrophy of >70% of the follicles or any evidence of acute necrosis.
Statistical analysis.
All statistical analysis was performed using Sigma Stat 3.0 software. Serology data are presented as mean titers. Group means were analyzed by analysis of variance (ANOVA) with Tukey's multiple-comparison test. Significance is reported at the level of P ≤ 0.05.
Nucleotide sequence accession number.
The full F protein gene sequence has been reported to GenBank (accession number JQ319052).
RESULTS AND DISCUSSION
In this trial, biological pathotyping confirmed that the Venezuelan virus is velogenic; 50-h MDT and 1.86 ICPI results indicate high virulence and are similar to previously reported values for Venezuelan NDV from domestic and waterfowl origins (19). Additionally, we previously reported comparable results for virulent NDV isolates obtained between 1996 and 2006 in Mexico, where the MDT test results ranged between 39.7 and 61.5 h and the lowest and highest intracerebral pathogenicity index (ICPI) values were 1.59 and 1.94, respectively (24).
The molecular approach for NDV identification and pathotyping using reverse transcriptase PCR (RT-PCR) followed by direct sequencing and analysis of the fusion protein gene cleavage site is currently used for NDV research and surveillance (1, 7, 10, 16, 17, 18, 25). In this trial, the full F protein gene was sequenced and reported to GenBank (see above). The full gene sequence was compared to those of reference strains belonging to genotypes I to VII of class II. Most of the North and Central American NDV virulent isolates reported during the last decade belonged to class II in genotype V (United States, Mexico [cormorants]) or VI ([pigeons]), supporting the idea of a local evolutionary trend with regional viral dissemination (16, 17, 25). The phylogenetic analysis for the Venezuelan strain shown in Fig. 1 demonstrated that the isolate belongs to genotype VII in class II, representing the first report of the presence of this genotype in the continent of South America. Additional work, including a higher number of archived and current Venezuelan NDV isolates, is required to assess whether VII is the sole genotype present in the country or whether there are other genotypes circulating.
A comparison of the percentages of amino acid identity between the Venezuelan full F protein and those of reference strains resulted in similarities of up to 98.6% (1.4% amino acid differences) with genotype VII viruses, mainly several goose isolates from China, suggesting a close relationship between the Venezuelan isolate and Asian genotype VII isolates. Amino acid differences of up to 5.8% from a Peru 2008 isolate suggest that those isolates are more distantly related and not likely part of the same introduction. As shown in Table 1, the Venezuelan strain F protein was up to 12.4% divergent from that of the LaSota vaccine strain that belongs to genotype II in class II and is commonly used in vaccination programs.
Table 1.
Estimates of evolutionary divergence between amino acid sequences of the full fusion protein of the Venezuelan isolate and those of commercial vaccines and reference strains of different genotypesa
| Strain | % amino acid substitutions0 compared to: |
|
|---|---|---|
| 293927 LaSota vaccine | Venezuela 2008 | |
| JN800306 Venezuela 2008 | 12.4 | 0.0 |
| 148534707 Goose/China/2006 | 12.7 | 1.4 |
| 146359113 Mallard/China/2006 | 11.8 | 1.8 |
| 268043733 Goose/China/2008 | 12.8 | 1.8 |
| 28933797 Goose/China/2007 | 13.3 | 1.8 |
| 110225441 Goose/NA1/China/2006 | 12.0 | 1.9 |
| 18042226 ZJ1/Goose/China/2007 | 13.5 | 1.9 |
| 109290367 Chicken/China/2003 | 12.1 | 2.3 |
| 109290353 Chicken/China/2002 | 12.8 | 2.3 |
| 45511225 Chicken/USA/1972 | 10.1 | 4.3 |
| 217323230 Pigeon (MD)/USA/1984 | 10.9 | 5.2 |
| 269838473 Pigeon/UK/2007 | 10.9 | 5.2 |
| Chicken/Peru/2008 | 12.6 | 5.8 |
| 258547239 Pigeon (AV324)/Ireland/1996 | 12.1 | 6.6 |
| 262217133 Chicken/China/QH1/1979 | 10.4 | 6.6 |
| 53636432 Chicken/Hertz | 8.6 | 6.6 |
| 161110504 Chicken (SRZ)/China/2003 | 7.3 | 7.0 |
| 45511239 Mixed (Largo, MD)/USA/1971 | 12.4 | 8.0 |
| 253317778 Cormorant/USA/1997 | 12.9 | 8.4 |
| 61393396 Chicken/Australia/1992 | 8.5 | 8.7 |
| 33348851 Chicken/China | 8.2 | 8.8 |
| 293933 Chicken/Australia/1932 | 9.2 | 8.8 |
| 7229463 Chicken (QV4)/Australia/1967 | 7.4 | 9.0 |
| 253317820 Cormorant/USA/1992 | 12.3 | 9.3 |
| 11527326 B1 vaccine | 0.9 | 12.2 |
| 293927 LaSota vaccine | 0.0 | 12.4 |
The percentages of amino acid substitutions between sequences are shown. The numbers used to identify the viruses represent the GenBank GenInfo (GI) sequence identification numbers. Analyses were conducted using the JTT matrix-based model as implemented in MEGA5 (26). The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). All positions containing gaps and missing data were eliminated. There were a total of 551 positions in the final data set.
Regardless of the genotype differences between NDV strains circulating worldwide, all NDV isolates belong to the same serotype; if given correctly, ND vaccines prepared with any NDV should protect poultry from clinical disease and mortality in the event of a virulent challenge (21, 23, 27). Current vaccination strategies do protect against mortality; however, existing vaccines have proven to be incapable of completely stopping viral excretion after infection, leading to high recirculation of the virus in the environment and to a viral population suitable to mutations and adaptive changes (17, 18).
In Venezuela, a rigorous vaccination program is applied, including several live and killed boosts during a breeder's life to provide high maternal antibodies (ELISA titers above 13,000 at 1 day of age) and protect against egg production losses and clinical disease. Additionally, the offspring receives dual (live/killed) priming at 1 day of age, followed by two (days 7 and 17) field revaccinations. Nonetheless, outbreaks in vaccinated broilers reared under field conditions occur and are responsible for high mortalities and production losses in the broiler and layer industry.
The serological response and protection against a genotype VII lethal challenge conferred by the current vaccine program implemented in Venezuela were tested under experimental and commercial conditions; the results are summarized in Table 2. At day 28, the average NDV serological responses measured as ELISA antibody titers were 2,384 and 1,356 for experimental and commercial farms, respectively, indicating a significant (P < 0.05) decrease in the level of humoral immunity provided by the NDV vaccination program in the birds raised in the commercial set. The antibodies measured in the commercial farm suggest an early IBDV field infection, with antibody levels of 10,506 and 7,295 at 21 and 28 days, respectively. In the group reared under controlled conditions, the IBDV antibodies were at a level of 1,489 at 28 days, which is within the range of previously reported levels for challenge-free birds after administration of two live vaccines (4). The vaccine-challenge trial results showed that all the naïve controls died during the observation period, validating the challenge. Furthermore, only 57% of the birds from the commercial farm survived, differing (P < 0.05) from the 90.5% survival rate observed in the experimental farm (Table 2).
Table 2.
Serological response and protection conferred by the vaccine program in place against a genotype VII lethal challenge
| Group | Vaccination protocol (genotype VII Venezuelan isolate challenge virus; n = 21 birds) | No. of birds with positive results/total no. of birds tested (% protection) | Mean ELISA titera |
|||||
|---|---|---|---|---|---|---|---|---|
| NDV |
IBDV |
|||||||
| Day 14 | Day 21 | Day 28 | Day 14 | Day 21 | Day 28 | |||
| G01 | Exptl farm (live/killed day 1 + live day 7 and 17) | 2/21 (90.5) | 856a | 1,713a | 2,384a | 573a | 10,506a | 7,295a |
| G02 | Commercial farm (live/killed day 1 + live day 7 and 17) | 9/21 (57) | 908a | 89b | 1,356b | 160b | 141b | 1,489b |
| G03 | Exptl farm (challenged, nonvaccinated) | 21/21 (0) | 140b | 53b | 22c | 134b | 77b | 52c |
Serology data are presented as mean titers. Group means were analyzed by ANOVA with Tukey's multiple-comparison test. Different letters within columns indicate significant differences (P < 0.05) between antibody levels. ELISA antibody titers were 2,343 and 1,043 for experimental and commercial farms, respectively, indicating a significant (P < 0.05) decrease in the level of humoral immunity provided by the NDV vaccination program in the birds raised in the commercial set. For IBDV, the antibodies measured in the commercial farm suggest an early IBDV field infection with antibody levels of 10,506 and 7,295 at 21 and 28 days, respectively. In the group reared under controlled conditions, the IBDV antibody level was 1,489.
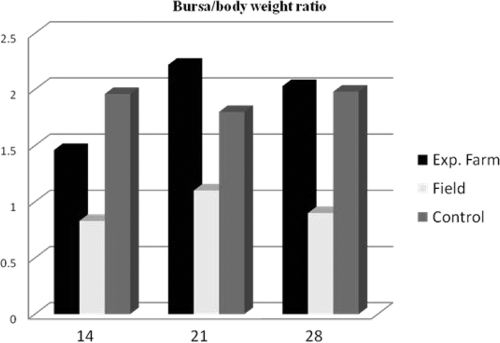
The differences in humoral immune response and protection observed are noteworthy (Fig. 2); additional results show that at the challenge age (28 days), the bursa/body weight ratios were 2.03 and 0.9 in the experimental set and the field, respectively, suggesting the presence of better-sized and healthier bursae in the experimental birds. The bursa of Fabricius is the main target organ for the IBDV. The virus replicates in actively dividing IgM-positive B cells (20). Infection results in lymphoid depletion and severe atrophy of the bursa as the predominant features of the pathogenesis of this disease, eliciting a suboptimal immune response to live attenuated vaccines (13).
Fig 2.
Bursa/body weight ratio. The bursal index was significantly (P < 0.05) higher for the experimental farm vaccinates than for the field vaccinates.
In this trial, the histopathology evaluation of the bursa in the field group demonstrated severe lymphoid depletion of the follicles, an increased amount of stroma between follicles, and severe follicular atrophy consistent with grade 4 in the bursal lesion scores at 28 days; the experimental farm birds showed a bursal score of 1.5 at the same age. The grading system is a diagnostic tool for IBDV infection and provides valuable information on the predicted immune status of the flock (4, 13, 20). All together, these results suggest that, despite vaccination, the field birds developed IBD, compromising the ability of the birds to mount an adequate immune response to the NDV vaccination and challenge.
Therefore, and based on the differences between the experimental and the field sets, the extent of the vaccine failure in the commercial farm group may be explained by field environmental and/or immunosuppressive factors affecting the efficacy of the vaccine (9). An additional plausible explanation for the significantly higher protection observed in the group subjected to experimental conditions is that the NDV and IBDV field vaccination procedures (implemented by field personnel in a population of 20,000 birds) could have been impaired, affecting vaccine efficacy and emphasizing the well-known fact that proper vaccination procedures are required for vaccination success (14, 27).
Additional experiments increasing the number of birds challenged are required to assess the cause of the lack of 100% protection in chickens reared under experimental conditions after the intensive vaccination program.
In summary, the presence of a velogenic NDV belonging to genotype VII has been confirmed in Venezuela. The differences in protection observed in the field-vaccinated birds suggest that management, environmental, and/or immunosuppressive factors are affecting ND control and vaccine efficacy in the country.
ACKNOWLEDGMENTS
This work was supported by USDA-ARS CRIS (6612-32000-049-00D).
We acknowledge Dawn Williams-Coplin for excellent technical assistance and the South Atlantic Area Sequencing Facility for nucleotide sequencing.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print 11 January 2012
REFERENCES
- 1. Aldous EW, Mynn JK, Banks J, Alexander DJ. 2003. A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol. 32:239–256 [DOI] [PubMed] [Google Scholar]
- 2. Alexander DJ, Senne DA. 2008. Newcastle disease, other avian paramyxoviruses, and pneumovirus infections, p 75–98 In Saif YM, Fadly AM, Glisson JR, McDougald LR, Nolan LK, Swayne DE. (ed), Diseases of poultry, 12th ed Blackwell Publishing, Ames, IA [Google Scholar]
- 3. Alexander DJ, Swayne DE. 1998. Newcastle disease virus and other avian paramyxoviruses, p 156–163 In Swayne DE, Glisson JR, Jackwood MW, Pearson JE, Reed WM. (ed), A laboratory manual for the isolation and identification of avian pathogens, vol 4 The American Association of Avian Pathologists, Kennett Square, PA [Google Scholar]
- 4. Banda, Alejandro Villegas P, Purvis L, Perozo F. 2008. Protection conferred by coarse spray vaccination against the challenge with infectious bursal disease virus in commercial broilers. Avian Dis. 52:297–301 [DOI] [PubMed] [Google Scholar]
- 5. Bermudez A. Principles of disease prevention: diagnosis and control, p 75–98 In Saif YM, Fadly AM, Glisson JR, McDougald LR, Nolan LK, Swayne DE. (ed), Diseases of poultry, 12th ed Blackwell Publishing, Ames, IA [Google Scholar]
- 6. Czeglédi A, et al. 2006. Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res. 120:36–48 [DOI] [PubMed] [Google Scholar]
- 7. Ding Z, et al. 2010. Genetic analysis of avian paramyxovirus-1 (Newcastle disease virus) isolates obtained from swine populations in China related to commonly utilized commercial vaccine strains. Virus Genes 41:369–376 [DOI] [PubMed] [Google Scholar]
- 8. Felsenstein J. 1992. Estimating effective population size from samples of sequences: a bootstrap Monte Carlo integration method. Genet. Res. 60:209–220 [DOI] [PubMed] [Google Scholar]
- 9. Hoerr F. 2010. Clinical aspects of immunosuppression in poultry. Avian Dis. 54:2–15 [DOI] [PubMed] [Google Scholar]
- 10. Khan ST, Rehmani, Rue C, Miller P, Afonso CL. 2010. Phylogenetic and pathological characterization of Newcastle disease virus isolates from Pakistan. J. Clin. Microbiol. 48:1892–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim LM, King DJ, Suarez DL, Wong CW, Afonso CL. 2007. Characterization of class I Newcastle disease virus isolates from Hong Kong live bird markets and detection using real-time reverse transcription-PCR. J. Clin. Microbiol. 45:1310–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu XF, Wan HQ, Ni XX, Wu YT, Liu WB. 2003. Pathotypical and genotypical characterization of strains of Newcastle disease virus isolated from outbreaks in chicken and goose flocks in some regions of China during 1985–2001. Arch. Virol. 148:1387–1403 [DOI] [PubMed] [Google Scholar]
- 13. Lukert PD, Saif YM. 2003. Infectious bursal disease, p 161–179 In Saif YM, Barnes HJ, Fadly AM, Glisson JR, McDougald LR, Swayne DE. (ed), Diseases of poultry, 11th ed Iowa State University Press, Ames, IA [Google Scholar]
- 14. Marangon S, Busani L. 2007. The use of vaccination in poultry production. Rev. Sci. Tech. 26:265–274 [PubMed] [Google Scholar]
- 15. Mayo MA. 2002. Virus taxonomy—Houston 2002. Arch. Virol. 147:1071–1076 [DOI] [PubMed] [Google Scholar]
- 16. Miller PJ, Decanini EL, Afonso CL. 2010. Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infect. Genet. Evol. 10:26–35 [DOI] [PubMed] [Google Scholar]
- 17. Miller PJ, Kim LM, Ip HS, Afonso CL. 2009. Evolutionary dynamics of Newcastle disease virus. Virology 391:64–72 [DOI] [PubMed] [Google Scholar]
- 18. Miller PJ, King DJ, Afonso CL, Suarez DL. 2007. Antigenic differences among Newcastle disease virus strains of different genotypes used in vaccine formulation affect viral shedding after a virulent challenge. Vaccine 25:7238–7246 [DOI] [PubMed] [Google Scholar]
- 19. Moscoso H, Brett M, Fernandez R, Hofacre CL. 2007. Molecular analysis Newcastle disease virus in avian species from Venezuela, p 117 Proceedings of the 144th American Veterinary Medical Association National Convention, Washington, DC, 14 to 18 July 2007 American Veterinary Medical Association, Washington, DC [Google Scholar]
- 20. Müller H, Islam MR, Raue R. 2003. Research on infectious bursal disease—the past, the present and the future. Vet. Microbiol. 97:153–165 [DOI] [PubMed] [Google Scholar]
- 21. OIE 2009. Manual of diagnostic tests and vaccines for terrestrial animals: mammals, birds and bees, 5th ed, vol 1, part 2, chapter 2.3.14, p 576–589 Office international des eìpizooties, Paris, France: [PubMed] [Google Scholar]
- 22. Perozo F, Villegas P, Estevez C, Alvarado I, Purvis LB. 2006. Use of FTA filter paper for the molecular detection of Newcastle disease virus. Avian Pathol. 35:93–98 [DOI] [PubMed] [Google Scholar]
- 23. Perozo F, Villegas P, Dolz R, Afonso CL, Purvis L. 2008. The VG/GA strain of Newcastle disease virus: mucosal immunity, protection against lethal challenge and molecular analysis. Avian Pathol. 37:237–245 [DOI] [PubMed] [Google Scholar]
- 24. Perozo F, Merino R, Afonso CL, Villegas P, Calderon N. 2008. Biological and phylogenetic characterization of virulent Newcastle disease virus circulating in Mexico. Avian Dis. 52:472–479 [DOI] [PubMed] [Google Scholar]
- 25. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 26. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Villegas P. 1998. Viral diseases of the respiratory system. Poult. Sci. 77:1143–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Z, et al. 2006. Genotyping of Newcastle disease viruses isolated from 2002 to 2004 in China. Ann. N. Y. Acad. Sci. 1081:228–239 [DOI] [PubMed] [Google Scholar]
- 29. Yu L, Wang Z, Jiang Y, Chang L, Kwang J. 2001. Characterization of newly emerging Newcastle disease virus isolates from the People's Republic of China and Taiwan. J. Clin. Microbiol. 39:3512–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]