Abstract
Broad-range amplification and sequencing of the bacterial 16S rRNA gene directly from clinical specimens are offered as a diagnostic service in many laboratories. One major pitfall is primer cross-reactivity with human DNA which will result in mixed chromatograms. Mixed chromatograms will complicate subsequent sequence analysis and impede identification. In SYBR green real-time PCR assays, it can also affect crossing threshold values and consequently the status of a specimen as positive or negative. We evaluated two conventional primer pairs in common use and a new primer pair based on the dual priming oligonucleotide (DPO) principle. Cross-reactivity was observed when both conventional primer pairs were used, resulting in interpretation difficulties. No cross-reactivity was observed using the DPOs even in specimens with a high ratio of human to bacterial DNA. In addition to reducing cross-reactivity, the DPO principle also offers a high degree of flexibility in the design of primers and should be considered for any PCR assay intended for detection and identification of pathogens directly from human clinical specimens.
INTRODUCTION
Sequencing of the bacterial 16S rRNA gene has demonstrated tremendous value for bacterial taxonomy as well as for routine identification of cultured isolates (5, 17). Primers intended for these purposes have been extensively validated with regard to the target range and discriminatory properties of the generated sequences (1, 3, 7, 9). Today, as a culture-independent supplement to diagnostic bacteriology, broad-range amplification and sequencing of the bacterial 16S rRNA gene directly from clinical specimens are also established in many laboratories. Despite the many articles signifying the usefulness of this approach (8, 11, 14, 18, 19, 21), primer cross-reactivity with human DNA is rarely addressed. Coamplification of human DNA can complicate subsequent sequence analysis by causing mixed chromatograms (10, 12). Crossing threshold (CT) values of real-time PCR assays utilizing SYBR green may also be affected and, consequently, the interpretation of a specimen as positive or negative.
The aim of this study was to identify primer candidates for broad-range amplification of the recommended (5, 6) V1-V3 area of the 16S rRNA gene without significant cross-reactivity with human DNA. Having observed cross-reactivity with well-established conventional primers (2, 6, 7), we proceeded to examine the use of a new primer pair based on the dual priming oligonucleotide (DPO) principle. A DPO consists of two functional segments with distinct annealing properties connected by five consecutive deoxyinosine bases, called a poly(I) linker (4). The 5′ segment is the longest (18 to 25 bp) and crucial for positioning and stable annealing of the primer. The 3′ segment is short (6 to 12 bp) and will only bind if there is already stable annealing of the 5′ end. The short length and low annealing temperature of the 3′ segment result in a low tolerance for mismatches and are reported to ensure target-specific extension.
MATERIALS AND METHODS
Clinical specimens.
The types of specimens selected for this study were assumed to contain high levels of human DNA. In total, residual material from 50 clinical specimens was included, as follows: various abscesses (n = 24), bile (n = 2), blood/hematoma (n = 2), bone (n = 3), peritoneal fluid (n = 1), pleural fluid/empyema (n = 6), heart valve (n = 2), various soft tissues (n = 9), and synovial fluid (n = 1). Fifteen specimens were defined as broad-range PCR negative, and 35 were defined as broad-range PCR positive based on previous investigations. The PCR-negative specimens were sequenced as a toughened validation since a suitable primer pair should be able to amplify low-level background bacterial DNA from the reagents without coamplification of the abundant human DNA in the specimens. Corresponding culture results were available for all specimens.
PCR primers.
Two common primer pairs for sequence-based identification of bacterial isolates were selected. In this paper they will be referred to as Bosshard (forward [Bact-8s-20], 5′-AGA-GTT-TGA-TCM-TGG-CTC-AG-3′; reverse [pD], 5′-GTA-TTA-CCG-CGG-CTG-CTG-3′) (2, 7) and CLSI (forward [4F], 5′-TTG-GAG-AGT-TTG-ATC-MTG-GCT-C-3′; reverse [534R], 5′-TAC-CGC-GGC-TGC-TGG-CAC-3′) (6). Both primer pairs target the first 500 bp of the bacterial 16S rRNA gene. An alternative combination using the forward CLSI primer together with the reverse Bosshard primer was also used. The above primers were ordered from Sigma-Aldrich (St. Louis, MO).
The DPOs are given in Table 1 (DPO16S-forward and DPO16S-reverse). The 5′ segments were based on the Bosshard forward and the CLSI reverse primers, except that the 3′-terminal base was removed for both primers, and the three 5′-terminal bases were removed from the CLSI reverse primer. The DPOs were ordered from Exiqon (Vedbaek, Denmark).
Table 1.
Presentation of the 16SDPO-forward and 16SDPO-reverse primers together with the most common and aberrant sequence variants in their binding areas
| Primer name and target organism | Sequenceb |
|---|---|
| 16SDPO-forward | 5′-AGAGTTTGATCMTGGCTCA-I-I-I-I-I-AACGCT-3′ |
| Most bacteriaa | 5′-AGAGTTTGATCMTGGCTCA-G-R-D-Y-D-AACGCT-3′ |
| Atopobium spp. | 5′------Y--------------I-I-I-I-I--------3′ |
| Chlamydiales | 5′----A-------T---T----I-I-I-I-I--------3′ |
| Borrelia spp. | 5′------------------T--I-I-I-I-I--------3′ |
| Bifidobacteriales | 5′-G----C----T---------I-I-I-I-I--------3′ |
| Coxiella burnetii | 5′-----------T---------I-I-I-I-I--------3′ |
| 16SDPO-reverse | 5′-CGCGGCTGCTGGCA-I-I-I-A-I-TTRGC-3′ |
| Most bacteriaa | 3′-CGCGGCTGCTGGCA-C-R-D-A-D-TTRGC-5′ |
| Chlamydiales | 3′----A-----------I-I-I-A-I-------5′ |
| Mycoplasma pneumoniae | 3′-----A----------I-I-I-A-I-------5′ |
| Ehrlichia spp. | 3′----------------I-I-I-A-I---T---5′ |
| Anaplasma phagocytophilum | 3′----------------I-I-I-A-I---T---5′ |
| Jonquetella anthropi | 3′----------------I-I-I-A-I---T---5′ |
Consensus sequence based on the most commonly observed sequence variants.
Boldface letters indicate a locked nucleic acid. D is A, G, or T; M is A or C; R is A or G; Y is C or T; I is deoxyinosine.
Pre-PCR treatment of clinical specimens.
Total DNA was extracted from the clinical specimens using the following protocol. Between 200 and 800 μl of sample material was added to a bead-containing tube (SeptiFast Lysis Kit; Roche, Mannheim, Germany), together with 400 μl of bacterial lysis buffer (BLB; Roche). For liquid specimens with low viscosity, 800 μl was the maximum capacity of the bead-containing tube. For other specimens 400 μl was used if available. The lowest volume that would still provide 400 μl of supernatant for the subsequent DNA purification was 200 μl, and that was the lowest volume accepted for all specimens. A negative extraction control containing lysis buffer and 400 μl of PCR-grade water was included in every batch of samples. The specimens were run two times for 45 s in a FastPrep machine (Cepheid, Sunnyvale, California) at a speed setting of 6.5. After a short spin, 400 μl of supernatant was transferred to a MagNa Pure Compact automated extractor (Roche), and DNA was extracted and purified using the total nucleic acid program according to the manufacturer's instructions. The resulting 50 μl of eluate was stored at −80°C until used.
PCR conditions.
PCRs were performed in a 25-μl reaction tube on a SmartCycler real-time apparatus (Cepheid, Sunnyvale, CA). The PCR mixture consisted of 12.5 μl of ExTaq SYBR Master Mix (TaKaRa, Otsu, Japan), 0.4 μM each primer, 8.5 μl of PCR-grade water, and 2 μl of extracted DNA. The PCR thermal profile included an initial polymerase activation step of 10 s at 95°C, followed by 45 cycles of 10 s at 95°C (melt), 15 s at 64°C (annealing, conventional) or 62°C (annealing, DPO), and 20 s at 72°C (extension).
To compare CT values directly, an alternative PCR mixture was used consisting of 5 μl of LightCycler FastStart DNA MasterPLUS SYBR green I (Roche), 0.4 μM each primer, 16 μl of PCR-grade water, and 2 μl of extracted DNA. The thermal profile for this PCR included an initial polymerase activation step of 600 s at 95°C, followed by 45 cycles of 10 s at 95°C (melt), 15 s at 64°C (annealing, conventional) or 62°C (annealing, DPO) and 25 s at 72°C (extension).
All PCRs were optimized to determine the most stringent running conditions that did not compromise sensitivity.
Definition of a positive specimen.
A positive specimen was defined as reaching the fluorescence threshold value (CT) ≥3 cycles before the negative control (14, 23). All specimens, both positives and negatives, were run until the amplification curve had reached the plateau level.
Sequencing.
The PCR products were spun out of the Smart Cycler reaction tubes into a 1.5-ml Eppendorf tube and cleaned up using an ExoSAP-IT enzymatic degradation kit (Affymetrix, Santa Clara, CA). Sequencing was performed using an ABI Prism 1.1 BigDye sequencing kit and an ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA).
Interpretation of chromatograms and detection of cross-reactivity.
Mixed DNA chromatograms were analyzed using the RipSeq Mixed web application (Isentio, Bergen, Norway) (13). Nonmixed chromatograms were analyzed with a standard BLAST search against the GenBank database following the interpretation criteria provided by the CLSI (6).
Coamplification of human DNA was detected by carefully examining the chromatograms for unexpected ambiguity or deviant chromatogram lengths.
RESULTS
In silico evaluation of primers.
The DPO primers were aligned against GenBank references for 168 species from 131 genera to discover variations within the primer binding sites that could affect PCR amplification efficacy (see Table S1 in the supplemental material). The alignment revealed no alternative sequence variants in the binding area for the 16SDPO-forward 3′ segment while sequence variants for the 16SDPO-reverse 3′ segment were found within three genera (Anaplasma, Ehrlichia, and Jonquetella). Primer mismatches against the 16SDPO 5′ segments correspond to those seen for the Bosshard forward and CLSI reverse primers, respectively. The aberrant sequence variants in the DPO primer binding areas are listed in Table 1.
Evaluation of conventional primer pairs in clinical specimens.
A preliminary investigation on cross-reactivity for all primer pairs was performed by amplifying bacterial DNA from a subset of 13 clinical specimens (Table 2, samples 1 to 13). Visual examination of the resulting chromatograms revealed coamplification of various human DNA fragments with all the conventional primer pairs. The bacterial 16S rRNA amplicons had lengths around 500 bp as expected. Extra amplicons from human DNA longer than 500 bp were easily detected, while shorter amplicons produced an unusually high chromatogram complexity in the 5′ end followed by a drop back to the expected complexity (Fig. 1). With the Bosshard primers a 260-bp fragment corresponding to a portion of human chromosome 5 was found in 8 out of 13 specimens. In addition, various more prominent fragments with lengths ranging from 120 to 220 bp were observed in some of the chromatograms. With the CLSI primers, cross-reactivity was observed in reverse chromatograms as a 660-bp fragment corresponding to a part of human chromosome 9 in eight specimens. This cross-reactivity was quite strong, and the human fragment dominated in all chromatograms where it was present. The logical next step was to combine the CLSI forward and Bosshard reverse primers. The CLSI forward primer was then found to amplify a strong 120-bp fragment from three specimens representing a part of human chromosome 10.
Table 2.
Evaluation of the new dual priming oligonucleotides
| Sample no.d | Specimen | PCRe | Findings (based on sequence)f | Fg | Rg | Culture |
|---|---|---|---|---|---|---|
| 1 | Abscess, lung | Neg | Bacterial backgroundb,c | ✓ | ✓ | Aspergillus sp. |
| 2 | Tissue, prosthetic joint | Neg | Bacterial backgrounda | ✓ | ✓ | No growth |
| 3 | Abscess, bone | Pos | Kingella kingaea,b,c | ✓ | ✓ | Kingella kingae |
| 4 | Abscess, brain | Pos | Streptococcus intermediusa,b | ✓ | ✓ | Streptococcus intermedius |
| 5 | Abscess, kidney | Pos | Escherichia colia,b,c | ✓ | ✓ | Escherichia coli |
| 6 | Abscess, liver | Pos | Enterococcus faecium, Escherichia coli | ✓ | ✓ | Enterococcus faecium, Escherichia coli |
| 7 | Abscess, subcutaneous | Pos | Enterococcus faecium, Finegoldia magnaa | ✓ | ✓ | Enterococcus faecium, Finegoldia magna |
| 8 | Bile | Pos | Streptococcus anginosus | ✓ | ✓ | Streptococcus anginosus |
| 9 | Pleural fluid | Pos | Streptococcus pyogenesa,b | ✓ | ✓ | Streptococcus pyogenes |
| 10 | Tissue | Pos | Staphylococcus epidermidis | ✓ | ✓ | Staphylococcus epidermidis |
| Pacemaker pouch | Propionibacterium acnesa,b | ✓ | ✓ | |||
| 11 | Tissue prosthetic joint | Pos | Staphylococcus aureusb | ✓ | ✓ | Staphylococcus aureus |
| 12 | Tissue prosthetic joint | Pos | Bacteroides fragilisa,b | ✓ | ✓ | Bacteroides fragilis |
| 13 | Tissue subcutaneous | Pos | Morganella morganii | ✓ | ✓ | Morganella morganii |
| 14 | Abscess, aorta | Neg | Bacterial background | ✓ | ✓ | No growth |
| 15 | Abscess, neck | Neg | Bacterial background | ✓ | ✓ | No growth |
| 16 | Abscess, presacral | Neg | Propionibacterium acnes (high), bacterial background (low) | ✓ | ✓ | No growth |
| 17 | Abscess, subcutaneous | Neg | Bacterial background | ✓ | ✓ | No growth |
| 18 | Heart valve | Neg | Staphylococcus aureus (high), bacterial background (low) | ✓ | ✓ | Staphylococcus aureus |
| 19 | Heart valve | Neg | Bacterial background | ✓ | ✓ | No growth |
| 20 | Peritoneal fluid | Neg | Escherichia coli | ✓ | ✓ | Escherichia coli |
| 21 | Pleural fluid | Neg | Streptococcus intermedius (high), bacterial background (low) | ✓ | ✓ | No growth |
| 22 | Pleural fluid | Neg | Bacterial background | ✓ | ✓ | No growth |
| 23 | Subdural hematoma | Neg | Bacterial background | ✓ | ✓ | No growth |
| 24 | Synovial fluid | Neg | Bacterial background | ✓ | ✓ | No growth |
| 25 | Tissue, prosthetic joint | Neg | Staphylococcus aureus (high), bacterial background (low) | ✓ | ✓ | Staphylococcus aureus |
| 26 | Tissue, surgical wound | Neg | Staphylococcus aureus (high), bacterial background (low) | ✓ | ✓ | Staphylococcus aureus |
| 27 | Abscess, abdominal | Pos | Haemophilus parahaemolyticus | ✓ | ✓ | No growth |
| 28 | Abscess, brain | Pos | Streptococcus intermedius, Fusobacterium nucleatum | ✓ | ✓ | Streptococcus intermedius |
| 29 | Abscess, brain | Pos | Fusobacterium nucleatum | ✓ | ✓ | No growth |
| 30 | Abscess, brain | Pos | Aggregatibacter aphrophilus | ✓ | ✓ | Aggregatibacter aphrophilus |
| 31 | Abscess, hip | Pos | Bacteroides fragilis | ✓ | ✓ | Bacteroides fragilis |
| 32 | Abscess, kidney | Pos | Citrobacter koseri | ✓ | ✓ | Citrobacter koseri |
| 33 | Abscess, liver | Pos | Propionibacterium acnes | ✓ | ✓ | Propionibacterium acnes |
| 34 | Abscess, liver | Pos | Escherichia coli, Clostridium perfringens | ✓ | ✓ | Escherichia coli |
| 35 | Abscess lung | Pos | Lactobacillus gasseri | ✓ | ✓ | Lactobacillus gasseri |
| 36 | Abscess pancreas | Pos | Neisseria sp. | ✓ | ✓ | No growth |
| 37 | Abscess pancreas | Pos | Mycobacterium tuberculosis | ✓ | ✓ | Mycobacterium tuberculosis |
| 38 | Abscess retroperitoneal | Pos | Enterococcus faecium, Klebsiella pneumoniae | ✓ | ✓ | Enterococcus faecium |
| 39 | Abscess shoulder | Pos | Finegoldia magna | ✓ | ✓ | Finegoldia magna |
| 40 | Abscess subcutaneous | Pos | Streptococcus dysgalactiae | ✓ | ✓ | Streptococcus dysgalactiae |
| 41 | Bile | Pos | Escherichia coli | ✓ | ✓ | Escherichia coli |
| 42 | Biopsy bone | Pos | S. aureus | ✓ | ✓ | Staphylococcus aureus |
| 43 | Biopsy bone | Pos | Enterobacter asburiae/hormaecei | ✓ | ✓ | Enterobacter sp. |
| 44 | Biopsy bone | Pos | Staphylococcus aureus, Staphylococcus epidermidis | ✓ | ✓ | Staphylococcus aureus |
| 45 | Heart blood | Pos | Clostridium septicum | ✓ | ✓ | Clostridium septicum |
| 46 | Pleural fluid | Pos | Escherichia coli | ✓ | ✓ | Escherichia coli |
| 47 | Pleural fluid | Pos | Staphylococcus intermedius | ✓ | ✓ | Staphylococcus intermedius |
| 48 | Pleural fluid | Pos | Prevotella pleuritidis | ✓ | ✓ | Eikenella corrodens, Campylobacter gracilis |
| 49 | Tissue, mediastinum | Pos | Enterococcus faecalis | ✓ | ✓ | Enterococcus faecalis |
| 50 | Tissue, prosthetic joint | Pos | Staphylococcus epidermidis | ✓ | ✓ | Staphylococcus epidermidis |
Cross-reactivity with human DNA observed for Bosshard primers.
Cross-reactivity with human DNA observed for CLSI primers.
Cross-reactivity with human DNA observed for the Bosshard forward/CLSI reverse primer combination.
Samples consist of 15 PCR-negative samples (1 to 2 and 14 to 26) and 35 PCR-positive samples (3 to 13 and 27 to 50). Samples 1 to 13 were used for the initial primer evaluation that also included the conventional primer pairs.
Pos, positive; Neg, negative.
Bacterial background indicates for the most part mixed chromatograms dominated by Pseudomonas spp.
F, forward sequence; R, reverse sequence. The check mark indicates that no unexpected fragments/cross-reactivity was observed.
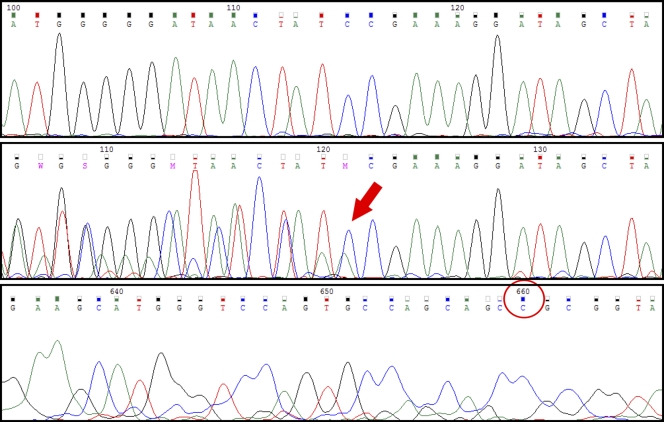
Fig 1.
Examples of various cross-reactivities using different primer sets for sample 3 (Table 2). The top chromatogram was obtained using the DPO primer set. It displays no ambiguity and represents the 16S rRNA gene of Kingella kingae only. The middle chromatogram shows amplification using the CLSI forward/Bosshard reverse primer combination. The area shown is identical to that of the top chromatogram. Coamplification of human DNA gives ambiguity up to bp 121 (red arrow). Thereafter, the 16S rRNA gene of K. kingae continues as a pure sequence. The bottom chromatogram shows amplification using the CLSI primer set. Terminal part of the reverse chromatogram is shown, and the length of the chromatogram is 667 bp (red circle), significantly longer than the expected 16S rRNA amplicon length of 500 bp. The chromatogram represents only human DNA, and the 16S rRNA gene of K. kingae could not be detected.
Evaluation of DPO primers in clinical specimens.
Coamplification of human DNA was not observed in the initial investigation of the new DPOs (Table 2, samples 1 to 13). We therefore proceeded to perform an extended evaluation on the remaining 37 clinical specimens (Table 2, samples 14 to 50). No coamplification of human DNA was observed in any of the resulting chromatograms. For 6 of the 15 specimens defined as negative based on previous and current PCR CT values, the resulting chromatograms still represented potentially relevant findings. These were Escherichia coli in a peritoneal fluid specimen; Propionibacterium acnes in a presacral abscess; Staphylococcus aureus in one periprosthetic tissue specimen, one surgical wound specimen, and one heart valve; and Streptococcus intermedius in a pleural fluid specimen. The detection of E. coli in one specimen and S. aureus in three specimens was confirmed by culture.
Human DNA cross-reactivity and real-time PCR positivity.
A direct comparison between the various primers was performed to explore the consequences of primer cross-reactivity with human DNA and its impact on the specimen CT values by running three specimens and two controls in parallel (Table 3). The DPO16S PCR and the conventional PCRs were found to be equally effective based on CT values for the two controls and specimen 1, a strong positive specimen that had not displayed any cross-reactivity with the conventional primers. CT values were also equal for specimen 2. For this specimen the Bosshard and DPO assays resulted in pure Bacteroides fragilis 16S chromatograms, but the CLSI chromatograms revealed significant coamplification of human DNA. For specimen 3, CT values differed. Based on CT values alone, the specimen was found to be positive when amplified with the CLSI and the Bosshard PCRs and negative when amplified with the DPO PCR. The CLSI chromatograms, however, contained only human DNA sequences with no detectable bacterial 16S rRNA genes. In the Bosshard chromatograms S. aureus could be identified using the RipSeq Mixed software, but multiple other fragments with equal signal strengths were also present. The DPO PCR reached CT two and four cycles after the CLSI and Bosshard PCRs, respectively, but displayed no cross-reactivity. The DPO chromatograms represented only S. aureus (dominant) and Pseudomonas spp. (low secondary peaks representing bacterial background).
Table 3.
A direct comparison between all three primer pairs
| Control or sample type (specimen no.)a | Primer set | CTc | Result forward |
Result reverse |
||
|---|---|---|---|---|---|---|
| Organismd | Cross-reactivitye | Organismd | Cross-reactivitye | |||
| Klebsiella 16S plasmids (strong positive) | CLSI | 15.6 | ||||
| Bosshard | 15.8 | |||||
| DPO | 15.3 | |||||
| Extraction control (weak positive)b | CLSI | 30.9 | ||||
| Bosshard | 31.1 | |||||
| DPO | 31.7 | |||||
| Tissue, subcutaneous (1) | CLSI | 12.5 | M. morganii | − | M. morganii | − |
| Bosshard | 12.3 | M. morganii | − | M. morganii | − | |
| DPO | 12.3 | M. morganii | − | M. morganii | − | |
| Abscess, hip (2) | CLSI | 26.4 | B. fragilis | ++ | BND | +++ |
| Bosshard | 26.3 | B. fragilis | − | B. fragilis | − | |
| DPO | 26.8 | B. fragilis | − | B. fragilis | − | |
| Periprosthetic tissue hip (3) | CLSI | 25.3 | BND | +++ | BND | +++ |
| Bosshard | 27.2 | S. aureus | +++ | S. aureus | +++ | |
| DPO | 29.2 | S. aureus | − | S. aureus | − | |
Parallel amplifications of two controls and three clinical samples were performed. All samples were run in duplex.
The negative extraction control functions as a weak positive because of background bacterial DNA in the reagents.
Values are the average of two runs.
M. morganii, Morganella morganii; S. aureus, Staphylococcus aureus; B. fragilis, Bacteroides fragilis; BND, bacterial 16S gene not detectable.
Cross-reactivity is scored as follows: −, no detectable cross-reactivity; ++, cross-reactivity with chromatogram peak heights equal to bacterial 16S gene peak heights; +++, cross-reactivity with chromatogram peak heights higher than bacterial 16S gene peak heights or cross-reactivity with multiple fragments with equal heights to bacterial DNA.
DISCUSSION
Broad-range amplification and sequencing of the bacterial 16S rRNA gene directly from clinical specimens can be offered as a diagnostic service in laboratories. However, cross-reactivity with human DNA is a major pitfall and may prevent laboratories from embracing broad-range amplification as a viable clinical test. Primers therefore need to be customized for this type of analysis to avoid significant problems in the subsequent interpretation of results.
Primers intended for use directly on patient specimens should be tested against a panel of clinical specimens. Traditional PCR primers may not be very discriminative, and unspecific amplification can occur even with multiple primer mismatches in specimens with a low target-to-nontarget ratio. In addition, major parts of the human genome are represented by only a few references in GenBank, and little is known about how variable these are from one individual to another.
Fifteen of the 50 specimens in this study were defined as negative based on PCR CT values from the present and previous investigations (Table 2, samples 1 to 2 and 14 to 26). The finding of potentially relevant bacterial DNA upon sequencing in six of these PCR-negative samples reflects the reduced sensitivity for a universal PCR in samples containing low levels of pathogen DNA since a positive sample can only be reliably defined if it contains significantly more bacterial DNA than the negative control (i.e., the background/contaminant bacterial DNA from the reagents) (23). The small differences between pathogen DNA levels and background DNA levels in these specimens were demonstrated by background DNA being detectable as low secondary peaks in most of the chromatograms. The development of DNA-free reagents will hopefully contribute to improved sensitivity in the future (16, 22).
Evaluating primer binding close to the 5′ end of the 16S rRNA gene is problematic since the primer binding site is often missing or of poor quality in GenBank references. It is also likely that for some of the references where the 5′ end primer binding site is present, it represents the primer used in the amplification and not necessarily the actual sequence. Sequence variants in the binding area for the Bosshard forward primer have been evaluated by Frank and coworkers (9). Our findings are in concordance with theirs except that we do not find support in GenBank for primer mismatches against Sphingomonas paucimobilis or against any of the medically relevant Campylobacter spp. For the DPOs a single 5′ segment mismatch is unlikely to affect amplification. However, the three 5′ segment mismatches observed for the forward 16SDPO against Chlamydiales and Bifidobacteriales will probably impair stable primer annealing. The variant 3′ segment binding site for 16SDPO-reverse found in Anaplasma phagocytophilum, Ehrlichia spp., and Jonquetella anthropi will also hinder amplification. To include the latter variant, the R (G or A) in position 3 from the primer 3′ end can be replaced with a D (all but C). The increased ambiguity could theoretically introduce cross-reactivity with human DNA and will necessitate a new validation. No broad-range primer pair will match all relevant species (20). It is important that laboratories are aware of primer binding site variants and eventually possess alternative primers for specimens where suspicion of infection remains high despite a negative PCR.
Dual priming oligonucleotides are in many aspects easier to design than standard primers (4). The only problem we encountered was the short lengths of the traditionally used primer binding areas (around 20 bp) since a typical DPO will have a length of 29 to 42 bp. We found that shorter than the recommended 5′ segments could be used in GC-rich areas, as seen in the reverse DPO. In the forward DPO we increased a low 5′-segment annealing temperature by introducing two locked nucleic acids (15). As demonstrated in Table 1, the poly(I) linker offers some flexibility since it can be used to cover a short variable section in between two conserved areas. In the reverse primer we demonstrate a variant where the fourth deoxyinosine base in the poly(I) linker is replaced by a standard base (an A). By doing this, the 3′ segment could be reduced to 5 bp without reducing PCR efficacy. We believe that a match with the interrupted poly(I) will result in some reduction of the discriminatory properties of the subsequent 3′ segment, whereas a mismatch will increase instability and lead to even more efficient blocking of unspecific elongation.
For the parallel run (Table 3) we used a different PCR master mix. This was because of a tendency for primer-dimer formation in the extraction controls with the TaKaRa Master Mix when amplification was performed using the conventional primers. Interestingly, in concordance with Chun et al. (4), this was not found to be a problem with the DPO primers.
In SYBR green assays any increase in double-stranded DNA will increase fluorescence emission. Consequently coamplification of human DNA may also have an impact on specimen CT values. It is likely that this effect will be largest for weak positive specimens. We believe this to be the explanation for the CT inconsistencies for specimen 3 in Table 3. Although in this specific example the lower CT value for the Bosshard primers revealed what was actually a relevant bacterial finding, it nevertheless demonstrates the risk for a true-negative specimen to be erroneously defined as a weak positive. The effect of cross-reactivity was even more pronounced for the CLSI primers, but here the resulting chromatograms represented human DNA alone.
An alternative approach to reduce cross-reactivity with human DNA is so-called targeted isolation of bacterial DNA (10). Through various methods, nonbacterial DNA can be selectively removed in the extraction process. Although this has proven to be efficient, it will not eliminate the need for more specific primers since a complete removal of human DNA cannot be expected. Concern has also appropriately been raised about a concomitant significant reduction of bacterial DNA (12). Importantly, these methods are developed for liquid specimens like blood and synovial fluids and will not necessarily work with tissue or viscous pus specimens.
We have used the dual priming oligonucleotide principle to design a new set of broad-range 16S rRNA gene primers. No coamplification of human DNA was observed when the primers were tested against a panel of 50 human clinical specimens. This is particularly important in SYBR green real-time assays where cross-reactivity not only will result in problems for the subsequent chromatogram analysis but also can affect specimen CT values. The DPO design should make this an attractive primer set for clinical laboratories, where false-negative and false-positive PCRs can be reduced by preventing human DNA cross-reactivity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Cathy Petti for her careful review of the manuscript.
The study was supported by the Western Health Region of Norway and Isentio AS.
The RipSeq algorithm used for interpretation of the mixed DNA chromatograms is owned by Isentio AS. Ø.K. is a coowner and the chief marketing officer of this company. K.S. is a senior application scientist in this company.
Footnotes
Published ahead of print 25 January 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Baker GC, Smith JJ, Cowan DA. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541. [DOI] [PubMed] [Google Scholar]
- 2. Bosshard PP, Zbinden R, Altwegg M. 2002. Turicibacter sanguinis gen. nov., sp. nov., a novel anaerobic, Gram-positive bacterium. Int. J. Syst. Evol. Microbiol. 52:1263–1266 [DOI] [PubMed] [Google Scholar]
- 3. Chakravorty S, Helb D, Burday M, Connell N, Alland D. 2007. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69:330–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chun JY, et al. 2007. Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res. 35:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clarridge JE., III 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases Clin. Microbiol. Rev. 17:840–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clinical and Laboratory Standards Institute . 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing. Approved standard MM18-A. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843–7853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fournier PE, et al. 2010. Comprehensive diagnostic strategy for blood culture-negative endocarditis: a prospective study of 819 new cases. Clin. Infect. Dis. 51:131–140 [DOI] [PubMed] [Google Scholar]
- 9. Frank JA, et al. 2008. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74:2461–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Handschur M, Karlic H, Hertel C, Pfeilstocker M, Haslberger AG. 2009. Preanalytic removal of human DNA eliminates false signals in general 16S rDNA PCR monitoring of bacterial pathogens in blood. Comp. Immunol. Microbiol. Infect. Dis. 32:207–219 [DOI] [PubMed] [Google Scholar]
- 11. Harris KA, Hartley JC. 2003. Development of broad-range 16S rDNA PCR for use in the routine diagnostic clinical microbiology service. J. Med. Microbiol. 52:685–691 [DOI] [PubMed] [Google Scholar]
- 12. Horz HP, Scheer S, Huenger F, Vianna ME, Conrads G. 2008. Selective isolation of bacterial DNA from human clinical specimens. J. Microbiol. Methods 72:98–102 [DOI] [PubMed] [Google Scholar]
- 13. Kommedal O, Karlsen B, Saebo O. 2008. Analysis of mixed sequencing chromatograms and its application in direct 16S rRNA gene sequencing of polymicrobial samples. J. Clin. Microbiol. 46:3766–3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kommedal O, Kvello K, Skjastad R, Langeland N, Wiker HG. 2009. Direct 16S rRNA gene sequencing from clinical specimens, with special focus on polybacterial samples and interpretation of mixed DNA chromatograms. J. Clin. Microbiol. 47:3562–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Latorra D, Arar K, Hurley JM. 2003. Design considerations and effects of LNA in PCR primers. Mol. Cell. Probes 17:253–259 [DOI] [PubMed] [Google Scholar]
- 16. Muhl H, Kochem AJ, Disque C, Sakka SG. 2010. Activity and DNA contamination of commercial polymerase chain reaction reagents for the universal 16S rDNA real-time polymerase chain reaction detection of bacterial pathogens in blood. Diagn. Microbiol. Infect. Dis. 66:41–49 [DOI] [PubMed] [Google Scholar]
- 17. Petti CA. 2007. Detection and identification of microorganisms by gene amplification and sequencing. Clin. Infect. Dis. 44:1108–1114 [DOI] [PubMed] [Google Scholar]
- 18. Rantakokko-Jalava K, et al. 2000. Direct amplification of rRNA genes in diagnosis of bacterial infections. J. Clin. Microbiol. 38:32–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vandercam B, et al. 2008. Amplification-based DNA analysis in the diagnosis of prosthetic joint infection. J. Mol. Diagn. 10:537–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y, Qian PY. 2009. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One 4:e7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Welinder-Olsson C, et al. 2007. Comparison of broad-range bacterial PCR and culture of cerebrospinal fluid for diagnosis of community-acquired bacterial meningitis. Clin. Microbiol. Infect. 13:879–886 [DOI] [PubMed] [Google Scholar]
- 22. Wellinghausen N, et al. 2009. Diagnosis of bacteremia in whole-blood samples by use of a commercial universal 16S rRNA gene-based PCR and sequence analysis. J. Clin. Microbiol. 47:2759–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zucol F, et al. 2006. Real-time quantitative broad-range PCR assay for detection of the 16S rRNA gene followed by sequencing for species identification. J. Clin. Microbiol. 44:2750–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.