Abstract
Streptomyces cacaoi subsp. cacaoi, a Gram-positive, branching filamentous bacteria, was isolated from a scalp infection in a patient from Pondicherry, India. Phenotypic tests identified the isolate as a Streptomyces species, but 16S rRNA sequence analysis provided the species identification required for tracking of this emerging pathogen.
CASE REPORT
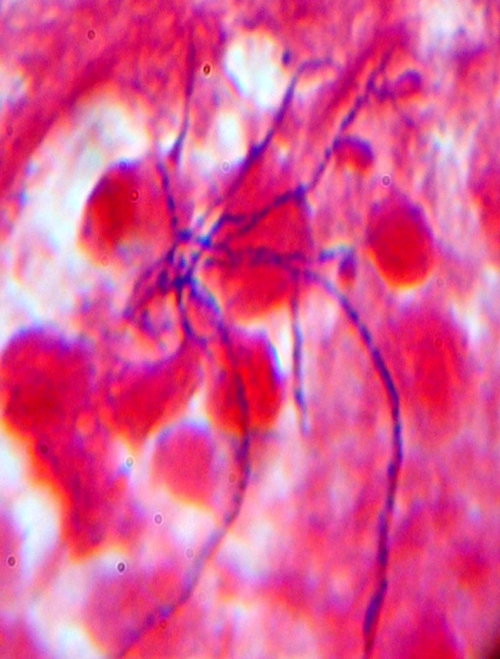
A31-year-old female patient presented to medical care at Aarupadai Veedu Medical College Hospital, Pondicherry, India, in December 2007 with a history of swelling on the left parietal region of the scalp 2 to 3 months following trauma experienced while carrying a log on her head. Symptoms reported were pain in the region, fever, weight loss, and anorexia for 3 months. Examination confirmed a soft, tender swelling on the left parietal region of the scalp. The swelling was aspirated and yielded dense pus positive for leukocytes and Gram-positive branched filamentous bacilli (Fig. 1). The aspirate was cultured to 5% sheep blood agar and MacConkey agar media, and the culture was incubated aerobically at 37°C for 48 h. The sheep blood agar grew a pure culture of small, chalky-white, hard, betahemolytic colonies. No growth was observed on the MacConkey media in 48 h. The Gram stain of the colonies demonstrated Gram-positive branched filamentous bacilli. The organism was presumptively identified as an actinomycetes based on its Gram stain and gross morphology on blood agar.
Fig 1.
Gram stain of pus aspirate from scalp swelling, demonstrating Gram-positive, filamentous, non-spore-forming bacilli (Gram stain magnification, ×1,000).
A computed tomographic scan of the patient's head showed a 6-by-4.5-cm soft-tissue swelling of 1.5-cm maximal thickness on the subgaleal plane of the left parietal scalp region. No intracranial lesion was visible. The patient tested positive for HIV antibodies by enzyme-linked immunosorbent assay (ELISA) in two rapid HIV test kits (HIV Tri-dot [Biomed Industries, Himachal Pradesh, India] and HIV Comb [J. Mitra & Co. Pvt. Ltd., New Delhi, India]). The abscess was aseptically drained, and the patient was treated with trimethoprim-sulfamethoxazole (160 to 800 mg orally twice daily) and referred to the government hospital in Pondicherry, India, for standard antiretroviral therapy (ART) and further evaluation. The patient was later lost for follow-up; therefore, the response to therapy was unknown. The bacterial isolate was grown on brain heart infusion agar (Hi-Media Laboratories, Mumbai, India) and sent to the Actinomycetes Reference Laboratory at the Centers for Disease Control and Prevention (CDC), Atlanta, GA, for identification and further testing.
At the CDC, the patient's isolate was cultured aerobically at 35°C with heart infusion agar plates with 5% rabbit blood (Becton, Dickinson and Company, Sparks, MD) and with Trypticase soy broth. The isolate exhibited hard, volcano-shaped colonies that developed distinctive white aerial and substrate hyphae after 72 h of aerobic growth. The organism grew at between 25°C and 45°C with optimum growth at 35°C and presented Gram-positive, branched, filamentous bacillus forms without fragmentation. The organism stained acid fast negative by the modified Kinyoun method. The following phenotypic characteristics were determined using methods described previously (2). The strain utilized (with acid production) adonitol, l-arabinose, cellobiose, i-erythritol, d-fructose, d-galactose, d-glucose, glycerol, lactose, maltose, d-mannitol, mannose, melibiose, raffinose, l-rhamnose, salicin, trehalose, and d-xylose but did not utilize dulcitol, d-sorbitol, or sucrose. Although the type strain of Streptomyces cacaoi subsp. cacaoi (DSM 40057) is l-rhamnose negative, we determined our isolate to be l-rhamnose positive. The organism decomposed casein, esculin, hypoxanthine, xanthine, and urea but did not decompose tyrosine. The organism did not grow in lysozyme broth, produce arylsulfatase, or reduce nitrate. In summary, based on morphological, physiological, and biochemical results, the organism resembled a Streptomyces species. To further characterize this isolate, the CDC laboratory performed susceptibility and molecular studies.
The susceptibility of the isolate to different antimicrobials was determined by using a commercial broth microdilution panel (PML Microbiologicals Inc., Wilsonville, OR) with cation-supplemented Mueller-Hinton broth inocula. Testing was performed and read according to CLSI guidelines (1). MIC testing indicated susceptibility to amikacin (1 μg/ml), imipenem (1 μg/ml), and linezolid (2 μg/ml) and resistance to ceftriaxone (>128 μg/ml), ciprofloxacin (4 μg/ml), clarithromycin (16 μg/ml), tigecycline (8 μg/ml), and trimethoprim-sulfamethoxazole (4/76 μg/ml). Intermediate resistance was observed to amoxicillin-clavulanate (16/8 μg/ml) and minocycline (4 μg/ml).
Molecular identification was performed by PCR amplification of a 1,450-bp sequence of the 16S rRNA gene as described by Morey et al. (7) except for the modification of primers FL1 (5′-CCGAATTCGTCGACAACAGAGTTTGATCCTGGCTCAG-3′) and RL1 (5′-CCCGGGATCCAAGCTTACGGCTACCTTGTTACGACTT-3′). BLAST software (nucleotide-nucleotide BLAST [http://www.blast.ncbi.nlm.nih.gov/Blast.cgi]) was used to identify related sequences in GenBank. Sequence analysis of the 1,450-bp nucleotide was performed using a GenBank BLAST search. The 16S rRNA gene sequence for our isolate had 100% similarity to the 1,479-bp sequence of the type strain, Streptomyces cacaoi subsp. cacaoi (GenBank accession number AB184115), within the GenBank database. The next closest match in GenBank was to the type strain of Streptomyces violaceoruber (GenBank accession number AY999815), with 99.37% similarity.
The species S. cacaoi subsp. cacaoi, isolated from cacao beans, was originally described by Waksman in 1932, emended by Waksman and Henrici in 1948 (9), and further emended by Lanoot et al. in 2002 as S. cacaoi subsp. cacaoi (5).
Historically, the phenotypic characterization of Streptomyces species was based on macroscopic and microscopic morphology and pigmentation. Although these phenotypic characteristics are useful in assigning a bacterium to the genus Streptomyces, identification to the species level has evolved to a more polyphasic approach that relies heavily on molecular evaluation (5).
Because of difficulties in determining the pathogenic role of the ubiquitous Streptomyces, Kapadia et al. (4) have given guidelines to assist in the diagnosis of true infection: (i) isolation of the agent from sterile site, (ii) direct microscopic identification, and (iii) exclusion of other causes. While Streptomyces species are not a common cause of deeply invasive human infections, other than mycetoma attributed to Streptomyces somaliensis, there are increasing reports from nonmycetomic invasive infections (6). Recent reports include the following: Kapadia et al. (4) reported on 47 patient isolates and found six invasive infections in the study group. Five Streptomyces isolates were presumptively identified to the species level, and one isolate was identified only to the genus level by 16S rRNA gene sequence analysis in that study (4). In a recent case report (8), a traumatic inoculation caused a superficial infection, but even with 16S rRNA gene sequence analysis, the streptomycete could be identified only to the genus level, suggesting a new species of Streptomyces (8). To our knowledge, no human clinical case of S. cacaoi subsp. cacaoi has been published before in medical literature. We speculate that in the past, similar Streptomyces infections were underdiagnosed because of technical and methodological limitations, including the lack of molecular techniques and the use of short (500-bp) 16S rRNA sequences for BLAST analysis. In the Aerobic Actinomycetes Laboratory of the CDC, we can identify most clinical Streptomyces isolates to the species level when using 1,450-bp 16S rRNA gene sequences for our GenBank searches. Although Streptomyces systematics is gradually evolving, the large number of Streptomyces species remains a major obstacle in establishing phenotypic identification schemes. An average of 11% of the human clinical isolates submitted to the CDC each year are identified as belonging to the genus Streptomyces, which is composed of more than 500 validated species (http://www.bacterio.cict.fr/s/streptomycesa.html).
There have been no prospective controlled studies of antibiotic regimens for treatment of Streptomyces infections. The currently recommended empirical therapy for Streptomyces actinomycetoma infection is trimethoprim-sulfamethoxazole and is commonly used worldwide. There have been two recent reports of in vitro susceptibility studies on Streptomyces isolates not identified to the species level. In an extensive review by Rose et al. (8) of 92 isolates, the susceptibility testing results indicated a significant degree of resistance to many agents: 79% of isolates were resistant to ceftriaxone, 75% of isolates to sulfamethoxazole alone and to erythromycin, 65% to trimethoprim-sulfamethoxazole, and 54% to ciprofloxacin; all isolates tested were susceptible to amikacin (92 of 92 isolates) and linezolid (41 of 41 isolates).
In another recent study conducted in the Sudan, the author studied 18 strains of Streptomyces species isolated from human and animal actinomycetoma cases by the use of a few phenotypic tests and antimicrobial susceptibility profiles (3). The results of this study showed a broad range of phenotypic patterns (13 out of 18 isolates) and 15 different antimicrobial susceptibility profiles. More than 90% of the isolates were susceptible to novobiocin, gentamicin, doxycycline, and fusidic acid; all isolates were resistant to sulfamethoxazole.
The diversity of phenotypic and antimicrobial susceptibility profiles suggests different etiologic agents, and, importantly, the resistance to sulfamethoxazole and/or trimethoprim-sulfamethoxazole in both of those studies (3, 8) suggests that the empirical regimens for Streptomyces infections should be reconsidered. These data may aid in formulating new recommendations to include amikacin, linezolid, novobiocin, gentamicin, doxycycline, or fusidic acid for treatment in combination with trimethoprim-sulfamethoxazole until a strain-specific antimicrobial susceptibility study is performed. Although our patient's susceptibility data showed resistance to trimethoprim-sulfamethoxazole after initiation of the use of these antimicrobial agents as therapy, we were unable to review the effectiveness, as the patient was lost to follow-up.
In summary, this organism was convincingly visualized in the direct specimen, isolated in pure culture, and identified as S. cacaoi subsp. cacaoi, the first isolate from an invasive human infection. The patient's immunocompromised state may have contributed to her infection. The introduction of molecular techniques applied to a bacterial isolate dramatically improves the ability to identify a rare species that may occasionally cause disease, especially in a taxon such as Streptomyces, which is comprised of over 500 species.
Nucleotide sequence accession number.
The GenBank accession number for the 16S rRNA gene sequence of our isolate is GU144523.
ACKNOWLEDGMENTS
Use of trade names and commercial sources is for identification only and does not imply endorsement from the Centers for Disease Control and Prevention (CDC).
Footnotes
Published ahead of print 25 January 2012
REFERENCES
- 1. CLSI/NCCLS 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. Approved standard M24-A. National Committee for Clinical Laboratory Standards, Wayne, PA: [PubMed] [Google Scholar]
- 2. Conville PS, Witebsky FG. 2007. Nocardia, Rhodococcus, Gordonia, Actinomadura, Streptomyces, and other aerobic actinomycetes, p 515–542 In Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA. (ed), Manual of clinical microbiology, 9th ed ASM Press, Washington, DC [Google Scholar]
- 3. Hamid ME. 2011. Variable antibiotic susceptibility patterns among Streptomyces species causing actinomycetoma in man and animals. Ann. Clin. Microbiol. Antimicrob. 10:24 doi:1186/1476-0711-10-24 (formerly doi:10.1073/pnas.041593698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kapadia M, Rolston KVI, Han XY. 2007. Invasive Streptomyces infections. Am. J. Clin. Pathol. 127:619–624 [DOI] [PubMed] [Google Scholar]
- 5. Lanoot B, et al. 2002. The search for synonyms among streptomycetes by using SDS-PAGE of whole cell proteins. Emendation of the species Streptomyces aurantiacus, Streptomyces cacaoi subsp. cacaoi, Streptomyces caeruleus and Streptomyces violaceus. Int. J. Syst. Evol. Microbiol. 52(Pt. 3):823–829 [DOI] [PubMed] [Google Scholar]
- 6. McNeil MM, Brown JM. 1994. The medically important aerobic actinomycetes: epidemiology and microbiology. Clin. Microbiol. Rev. 7:357–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morey RE, et al. 2006. Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J. Clin. Microbiol. 44:3510–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rose CE, III, Brown JM, Fisher JF. 2008. Brain abscess caused by Streptomyces infection following penetration trauma: case report and results of susceptibility analysis of 92 isolates of Streptomyces species submitted to the CDC from 2000 to 2004. J. Clin. Microbiol. 46:821–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Waksman S, Henrici A. 1943. The nomenclature and classification of the actinomycetes. J. Bacteriol. 46:337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]