Abstract
Using a newly developed Plasmodium vivax merozoite surface protein 1 gene (Pvmsp1) heteroduplex tracking assay, we genotyped 107 P. vivax infections in individuals from Cambodia, 45 of whom developed recurrent parasitemia within 42 days. The majority of isolates were polyclonal, but recurrent parasitemias displayed fewer variants compared to initial parasitemias. Two Pvmsp1 gene variants occurred more frequently in the initial genotypes of those who developed recurrent parasitemia, representing the first time P. vivax variants associated with a higher risk of relapse have been described.
TEXT
Plasmodium vivax is the most prevalent malaria species outside Africa and causes significant morbidity in the developing world. It uniquely maintains its transmission in a wide range of latitudes by causing periodic relapses through reactivation of liver stage parasites called hypnozoites. Being able to identify those at risk for relapse has the potential to guide clinical management and drug policy (4, 9). However, studying P. vivax relapse in areas of endemicity has been difficult because genotyping methods have not been able to precisely differentiate relapse from new infection or recrudescence due to treatment failure.
It is known that P. vivax infections are frequently polyclonal, even in relatively low-transmission settings (3, 7). We sought to exploit this complexity of infection to better describe genotypic patterns of relapsing parasites and search for genotypic variants associated with relapse. To do this, we developed a P. vivax heteroduplex tracking assay (HTA) to evaluate pretreatment and recurrent parasitemias from 107 patients treated with chloroquine monotherapy in Chumkiri, Cambodia, 45 of whom developed recurrent P. vivax infection within 42 days of therapy.
HTAs have previously been shown to be more sensitive than nested PCR for determining the multiplicity of infection (MOI) in malaria infections due to their ability to detect both sequence and size polymorphisms (11). We developed an HTA targeting the P. vivax merozoite surface protein 1 gene (Pvmsp1), a highly polymorphic gene that is commonly used for genotyping P. vivax malaria. The probe was constructed as previously described, using primers based on a previously published assay and genomic DNA obtained from MR4 (Nicaragua strain, catalog no. MRA-340; MR4, Manassas, VA) (10, 11) (GenBank accession number JN674534). Pvmsp1 PCR products were amplified using the same primers from patient DNA that was extracted from filter paper blood spots. HTAs were performed as previously described. All gels were run with probe alone and nontemplate control lanes.
Patient samples were acquired from a clinical trial that enrolled persons ≥1 year old presenting with uncomplicated P. vivax malaria to the Chumkiri health center in Kampot Province, Cambodia, between August 2006 and December 2007 (14). In this study, subjects were given a total dose of 25 mg/kg of body weight of chloroquine base over 3 days with directly observed therapy, followed with weekly blood smears until day 42. Antirelapse therapy with primaquine was not given, as this is not part of Cambodian national guidelines. The 45 subjects who developed recurrent P. vivax infection during the 42-day follow-up were retreated with chloroquine. Chumkiri is a low-transmission area, with most of the malaria acquired in the surrounding mountains and forest. Thus, these patients had a low, but not negligible, chance of reinfection.
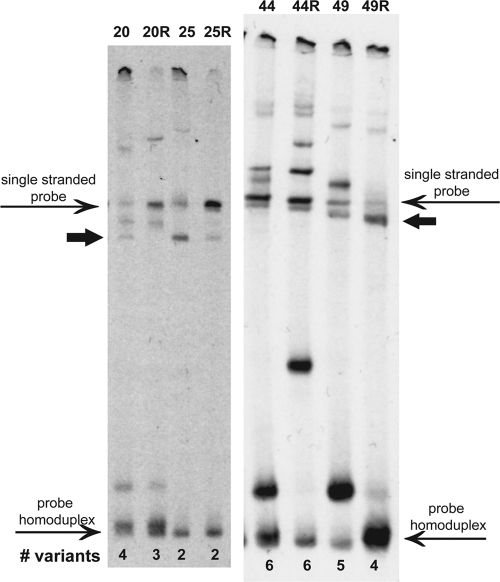
Molecular analysis was done on 107 enrollment (day 0) samples and 44 available recurrent P. vivax samples collected on the day of recurrence. HTA bands were considered unique Pvmsp1 variants if they were not in the single-stranded probe or probe homoduplex lanes (Fig. 1). Multiplicity of infection was determined by counting the number of unique variants in each isolate. To assign identities to individual variants, the relative migration distance (Rf) was calculated for each variant by dividing the distance migrated by the band by the distance migrated by the probe homoduplex. Rf units were divided among 20 bins of 0.05 Rf units (8).
Fig 1.
Pvmsp1 HTAs for P. vivax isolates from selected Cambodian subjects with relapsing infections. Bands that were not in the single-stranded probe or probe homoduplex lanes were considered unique Pvmsp1 variants. Paired initial and recurrent infections are shown, with “R” indicating the recurrent sample. The variant with an Rf of 0.35 is indicated by a thick arrow in both gels.
The Pvmsp1 HTA detected a high proportion of polyclonal infections among the clinical samples. Among the day 0 isolates, 89/107 (83%) displayed multiple heteroduplex bands, with a range of 1 to 7 msp1 variants detected within a single isolate. The median MOI was 3, with a mean MOI of 2.82. A total of 16 distinct variants were identified, with six common variants accounting for 88% of the total (n = 302 bands). The different combinations of these variants revealed 64 unique genotypes among the 107 isolates.
Among the 44 available paired samples from recurrences, there was a high degree of genetic relatedness between the initial and recurrent samples. In 86% (38/44) of the pairs, at least half of the variants detected in the recurrent parasitemia were also detected in the initial parasitemia (examples in Fig. 1). Since none of the recurrences occurred prior to day 28, when failure due to chloroquine resistance is expected (15), the genetic relatedness of these 38 pairs suggests that they represent relapses or less likely, recrudescence. Analysis of these 38 pairs showed a significant reduction in MOIs between initial and recurrent infections (mean MOIs of 3.13 and 2.26, respectively; P < 0.001 by a paired t test). This suggests that relapsing genotypes arising from hypnozoites are still often polyclonal but more restricted in their multiclonality.
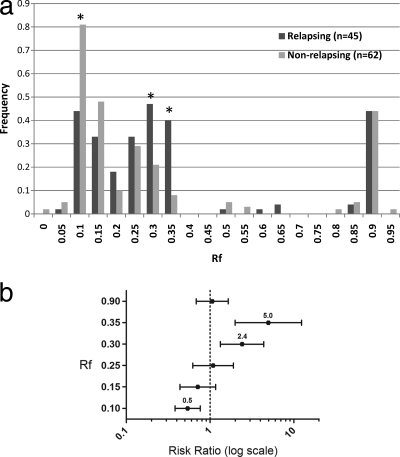
When the genotypes from initial infections of 45 “relapsers” and 62 “nonrelapsers” (those who did and did not develop recurrent parasitemia within 42 days, respectively) were compared, certain variants appeared to be associated with subsequent relapse (Fig. 2a). Specifically, variants with Rfs of 0.30 and 0.35 were 2.4 and 5.0 times more likely, respectively, to appear in the initial genotype of a relapsing infection (95% confidence intervals [CI] of 1.3 to 4.4 and 2.0 to 12.4, respectively; P values of 0.003 and <0.001, respectively, by Fisher's exact test) (Fig. 2b). The presence of either of these two variants in an initial genotype “predicted” relapse with 78% sensitivity and 73% specificity. The presence of an Rf of 0.35 alone was 92% specific for predicting relapse within 42 days in our cohort. In contrast, a variant with an Rf of 0.10 was less likely to be associated with a relapsing infection (risk ratio [RR], 0.5; 95% CI, 0.4 to 0.8; P < 0.001).
Fig 2.
(a) Frequency of Pvmsp1 variants in relapsing (n = 45) and nonrelapsing (n = 62) P. vivax malaria infections in 107 Cambodian patients. The frequencies of variants with Rfs of 0.10, 0.30, and 0.35 were significantly different between relapsing and nonrelapsing strains. (b) Risk ratios for the association of Pvmsp1 variants with relapsing infection. Out of the six common variants identified by the Pvmsp1 HTA, variants with Rfs of 0.35 and 0.30 were positively associated with relapsing infections, while an Rf of 0.10 was associated with nonrelapsing infections. Error bars represent the 95% confidence intervals of the relative risk ratio.
On average, subjects who developed recurrent parasitemia within 42 days were younger than those who did not (means of 19 years of age in relapsers and 22 years of age in nonrelapsers; P = 0.02 by a t test) but did not otherwise differ from their nonrelapsing counterparts in terms of gender, baseline parasitemia, or parasite clearance times. They likewise demonstrated no difference in MOI at enrollment (mean MOIs of 2.89 in relapsers and 2.76 in nonrelapsers).
These data illustrate the novel finding that different parasite subpopulations may display different propensities for relapse. Relapse patterns of P. vivax parasites are known to differ between broad geographic regions, with tropical strains relapsing more frequently and at shorter 3- to 4-week intervals compared to the longer intervals (up to 8 to 9 months later) seen in strains from subtropical/temperate zones (1). This is thought to reflect environmental adaptations based on the availability of mosquito vectors. Our findings support a genetic basis to relapse, with certain parasite variants possibly being more susceptible to the triggers that reactivate hypnozoites or more likely to remain in the hypnozoite form after sporozoite inoculation, thus devoting a greater fraction of their numbers to future relapse (5). Alternatively, these variants may have evolved a better mechanism for immune evasion in the primary infection, allowing them to once again become patent in a relapsed infection.
The main limitation to our study is our reliance on samples from an area of P. vivax endemicity in which it is not possible to prove definitively that a recurrent infection is a relapse, recrudescence, or reinfection. Relapses are expected to comprise a significant portion of the recurrences in this study, as primaquine was not given at enrollment, and the majority of recurrences (43/45) occurred between day 35 and day 42, a pattern which is consistent with previously reported relapse intervals in this region (12). There were no recurrent parasitemias prior to day 28 in the cohort, suggesting a limited role for recrudescence among the recurrences. Indeed, chloroquine resistance has not been reported from this study area to date. Finally, the incidence of malaria in Chumkiri based on passive case detection at health centers was approximately 15 cases/1,000 person-years in 2008, suggesting that reinfection should be relatively uncommon (Cambodian National Malaria Center, unpublished data). Recurring genotypes with fewer than two shared variants and multiple novel variants, suggestive of reinfection, made up just 14% (6/44) of recurrent infections, a rate that is similar to the estimated Plasmodium falciparum reinfection rate of 13% (4/31) reported in the same trial that was based on standard PCR genotyping methods for P. falciparum (14).
Previous studies on relapsing P. vivax infections that were able to compare initial and recurrent genotypes from subjects who had left areas of P. vivax endemicity have concluded that the genotypes of relapsing parasites differ from those of the initial infection in over half of cases (6, 13). However, the nested PCR and microsatellite genotyping methods employed in these studies are limited in their ability to detect minority variants and fully assess polyclonal infections. Our strategy of uncovering multiple variants at a single gene locus and comparing these complex mixtures of variants across time affords a greater appreciation of the relatedness of initial and relapsing parasite populations. Data obtained using this method suggest that P. vivax relapses arise from the activation of multiple and diverse hypnozoites, not single hypnozoite clones as has been postulated (2).
Using an HTA, we have shown that clinical P. vivax infections are often composed of complex mixtures of multiple variants, and some variants are more prone to relapse than others. Having a greater appreciation of the polyclonality of P. vivax infections may help us recognize genotypic patterns of relapsing parasite populations and perhaps even identify individuals at greater risk for relapsing malaria who would benefit from terminal prophylaxis with primaquine.
ACKNOWLEDGMENTS
We thank the participants in the clinical trial from which samples were collected, as well as Preab Ratha and Preab Saroth of Kampot Provincial Health Department and Socheat Duong of the Cambodian National Malaria Center.
This work was supported by the U.S. Department of Defense Global Emerging Infections Surveillance and Response System (DoD-GEIS) Program (for funding of the clinical trial) and grants from the National Institutes of Health (AI076785 to S.R.M. and AI089819 to J.J.J.). J.T.L. was supported by a National Institutes of Health Infectious Disease Pathogenesis Research Training Grant (5T32AI0715132).
The views expressed in this paper are those of the authors and do not represent the official position of the U.S. Department of Defense.
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Bray RS, Garnham PCC. 1982. The life-cycle of primate malaria parasites. Br. Med. Bull. 38:117–122 [DOI] [PubMed] [Google Scholar]
- 2. Chen N, Auliff A, Rieckmann K, Gatton M, Cheng Q. 2007. Relapses of Plasmodium vivax infection result from clonal hypnozoites activated at predetermined intervals. J. Infect. Dis. 195:934–941 [DOI] [PubMed] [Google Scholar]
- 3. Cui L, et al. 2003. Genetic diversity and multiple infections of Plasmodium vivax malaria in Western Thailand. Am. J. Trop. Med. Hyg. 68:613–619 [DOI] [PubMed] [Google Scholar]
- 4. Douglas NM, et al. 2011. Plasmodium vivax recurrence following falciparum and mixed species malaria: risk factors and effect of antimalarial kinetics. Clin. Infect. Dis. 52:612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hollingdale MR, Collins WE, Campbell CC. 1986. In vitro culture of exoerythrocytic parasites of the North Korean strain of Plasmodium vivax in hepatoma cells. Am. J. Trop. Med. Hyg. 35:275–276 [DOI] [PubMed] [Google Scholar]
- 6. Imwong M, et al. 2007. Relapses of Plasmodium vivax infection usually result from activation of heterologous hypnozoites. J. Infect. Dis. 195:927–933 [DOI] [PubMed] [Google Scholar]
- 7. Karunaweera ND, et al. 2008. Extensive microsatellite diversity in the human malaria parasite Plasmodium vivax. Gene 410:105–112 [DOI] [PubMed] [Google Scholar]
- 8. Kwiek JJ, et al. 2007. Estimating true antimalarial efficacy by heteroduplex tracking assay in patients with complex Plasmodium falciparum infections. Antimicrob. Agents Chemother. 51:521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin JT, et al. 2011. Plasmodium falciparum gametocyte carriage is associated with subsequent Plasmodium vivax relapse after treatment. PLoS One 6:e18716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maestre A, et al. 2004. Inter-allelic recombination in the Plasmodium vivax merozoite surface protein 1 gene among Indian and Colombian isolates. Malar. J. 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ngrenngarmlert W, et al. 2005. Measuring allelic heterogeneity in Plasmodium falciparum by a heteroduplex tracking assay. Am. J. Trop. Med. Hyg. 72:694–701 [PubMed] [Google Scholar]
- 12. Pukrittayakamee S, Imwong M, Looareesuwan S, White NJ. 2004. Therapeutic responses to antimalarial and antibacterial drugs in vivax malaria. Acta Trop. 89:351–356 [DOI] [PubMed] [Google Scholar]
- 13. Restrepo E, Imwong M, Rojas W, Carmona-Fonseca J, Maestre A. 2011. High genetic polymorphism of relapsing P. vivax isolates in northwest Colombia. Acta Trop. 119:23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rogers WO, et al. 2009. Failure of artesunate-mefloquine combination therapy for uncomplicated Plasmodium falciparum malaria in southern Cambodia. Malar. J. 8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. World Health Organization 2010. Guidelines for the treatment of malaria, 2nd ed WHO Press, Geneva, Switzerland [Google Scholar]