Abstract
We compared bacillary loads after splitting sputum specimens by chemical (N-acetyl-l-cysteine [NALC]) and mechanical homogenization by vortexing with sterile glass beads. NALC and vortexing with glass beads were equally effective at homogenizing sputum specimens, resulting in an equal distribution of tubercle bacilli in the aliquots.
TEXT
Interest in and development of new diagnostic tools for tuberculosis (TB) have increased dramatically over the past decade. Surprisingly little attention has been paid to the optimal method to compare diagnostic tests using sputum specimens, the specimens most commonly collected for TB diagnosis globally. As part of a study to compare two methods of microscopy to detect acid-fast bacilli (AFB), we aimed to determine an optimal method to split sputum specimens evenly. We preferred this approach to that of comparing specimens obtained on different days or at different times due to the known variability in bacillary load, most often reflected in the AFB semiquantification, among specimens obtained at different points in time. The macroscopic appearance of sputum samples may vary considerably, and many laboratories use a time-honored descriptive classification of mucoid, mucopurulent, purulent, and bloody (5). Purulent sputum specimens have long been recognized as having a higher likelihood of being positive for AFB on microscopy (5). Accordingly, laboratory personnel have been instructed to sample the most purulent portion of the specimens to maximize recovery of tubercle bacilli (4). Mycobacteria may also clump within specimens (2), which could in turn result in an unequal distribution of bacilli upon splitting. Given these recognized sources of variability both between and within sputum specimens, we aimed to use a method that would most evenly split the specimen based on quantitative culture. We compared splitting after homogenization by chemical mucolysis versus that by mechanical homogenization by vortexing with sterile glass beads.
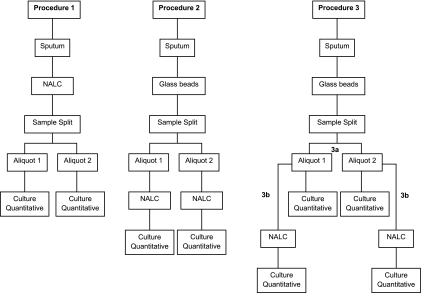
The protocol was approved by the institutional review board. The study was conducted from August 2007 to May 2009. A total of 103 adults (18 to 65 years) with 2+ (42%) or 3+ (58%) acid-fast bacillus smear-positive pulmonary TB were enrolled. These diagnoses were confirmed by the cultivation of Mycobacterium tuberculosis. In order to evaluate the efficacy of chemical and mechanical procedures to digest sputum, native sputum samples were subjected to one of the following procedures (Fig. 1). In procedure 1, 43 samples were treated with N-acetyl-l-cysteine (NALC) (50 mg/ml) in a volume equivalent to 10% of the total sample volume for 15 min at room temperature, homogenized by vortexing, split into two equal aliquots and cultured on selective Middlebrook 7H11 agar plates as described previously (3). In procedure 2, 42 samples were vigorously vortexed with sterile glass beads (4 mm in diameter) three times, 1 min each, split into two equal aliquots, treated with NALC, and cultured on selective Middlebrook 7H11 agar plates. In procedure 3, 18 samples were homogenized with sterile glass beads for 15 min in a vortex and split into two equal aliquots; 1 ml of each aliquot was serially diluted and inoculated into selective Middlebrook 7H11 agar plates (procedure 3a); the remaining volume was treated with NALC, serially diluted, and inoculated into selective Middlebrook 7H11 medium (procedure 3b). To avoid contamination, digested sputum samples were plated on selective Middlebrook 7H11 agar plates containing amphotericin, polymyxin B, carbenicillin, and trimethoprim. Using a micropipettor and changing pipette tips between dilutions, plates were inoculated in duplicate with 30-μl droplets of each dilution (101 to 105). Mycobacterial colonies were counted in each of the two 30-μl drops at the dilution that yields a readable count. The number of M. tuberculosis organisms per ml in the processed sputum sediment was calculated using the following formula: CFU/ml sediment = no. of colonies per 60 μl (both drops) × 16.7 (1,000 μl/60 μl) × 1/dilution. Plates inoculated on procedures 1, 2, and 3 were examined weekly, and colonies were counted on plates with dilutions yielding 10 to 50 visible colonies. Data were expressed as log10 CFU per ml of undiluted sputum. All isolates were identified as Mycobacterium tuberculosis according to the standard methods (4). The analysis of the results was made through the GraphPad Prism software program, version 5.0-2007, with a 95% confidence interval.
Fig 1.
Chemical and mechanical procedures used for digestion, splitting, and mycobacterial culture of sputum samples.
The mean sputum volumes collected for procedures 1, 2, and 3 were 10.1 ml (standard deviation [SD] of 3.6), 9.9 ml (SD of 3.4), and 10.6 ml (SD of 2.5), respectively. Most samples were mucopurulent (68.6%), while a minority were classified as mucoid (31.4%) according to macroscopic judgment. Overall, the results of the bacillary loads after splitting sputum in aliquots were comparable. As shown in Table 1, no significant statistical difference was observed among bacillary loads of the three different splitting procedures. Based on the Spearman's rank correlation coefficient, the correlation was highly significant between the aliquot pairs for all the procedures (P < 0.0001).
Table 1.
Comparison of sputum bacillary loads after splitting sputum samples
| Procedure | Mean bacillary load (log10 CFU/ml) ± SD |
P valuei | rsj | |
|---|---|---|---|---|
| Aliquot 1 | Aliquot 2 | |||
| 1 | 5.66 ± 0.92a | 5.65 ± 0.94b | 0.76 | 0.99 |
| 2 | 5.54 ± 0.91c | 5.5 ± 0.94d | 0.22 | 0.98 |
| 3a | 6.09 ± 0.76e | 6.1 ± 0.71f | 0.61 | 0.98 |
| 3b | 6.09 ± 0.67g | 6.1 ± 0.78h | 0.43 | 0.97 |
Load range, 3.69897 to 7.63682 log10 CFU/ml.
Load range, 3.22185 to 7.42597 log10 CFU/ml.
Load range, 3.69897 to 7.12494 log10 CFU/ml.
Load range, 3.22185 to 7.42597 log10 CFU/ml.
Load range, 3.69897 to 7.06695 log10 CFU/ml.
Load range, 3.92082 to 7.06695 log10 CFU/ml.
Load range, 3,92082 to 6,82391 log10 CFU/ml.
Load range, 3.52288 to 7.12494 log10 CFU/ml.
Wilcoxon test.
Spearman's coefficient.
To assess the performance of new diagnostic tools, clinical trials are generally conducted in the target study population (1). However, in some circumstances, the results, in particular their accuracy and reproducibility, do not reflect the true performance of a new test because of either study design or sample collection (1). The propensity of mycobacteria to clump together may explain their irregular distribution in a clinical sample (6). In our study addressed to this question, the CFU counts after splitting sputum in aliquots were similar and consistent using different digestion procedures. These results indicate that NALC and vortexing with glass beads were equally effective at homogenizing sputum specimens, resulting in an equal distribution of tubercle bacilli in the aliquots. Since there are no publications available in this context, it was not possible to draw direct comparisons with previous studies. Neither paucibacillary specimens nor large amounts of sputum (>10 ml) were included in our study. However, we considered that intrapatient and interpatient variability of bacillary load would be more likely to be detected in AFB smear-positive sputum specimens under routine conditions.
These data support the hypothesis that chemical and physical procedures for splitting sputum samples are equally effective and do not alter the concentrations of viable mycobacteria.
ACKNOWLEDGMENTS
This study was supported by contract UNICEF/UNDP/World Bank/WHO, TDR ID number A 30499, CNPq/INCT number 573548/2008-0, and Fundação de Apoio à Ciência e Tecnologia do Espírito Santo (FAPES), process number 37552686/2007.
Footnotes
Published ahead of print 25 January 2012
REFERENCES
- 1. Dinnes J, et al. 2007. A systematic review of rapid diagnostic tests for diagnosis of tuberculosis infection. Health Technol. Assess. 11:1–311 [DOI] [PubMed] [Google Scholar]
- 2. Ieven M, Goosens H. 1997. Relevance of nucleic acid amplification techniques for diagnosis of respiratory tract infections in the clinical laboratory. Clin. Microbiol. Rev. 10:242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnson JL, et al. 2006. Early and extended early bactericidal activity of levofloxacin, gatifloxacin and moxifloxacin in pulmonary tuberculosis. Int. J. Tuberc. Lung Dis. 10:605–612 [PubMed] [Google Scholar]
- 4. Kent PT, Kubica GP. 1985. Public health mycobacteriology: a guide for the level III laboratory. Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 5. Laird AT. 1909. A method for increasing the diagnostic value of sputum reports. JAMA LII:294–295 [Google Scholar]
- 6. Thwaites GE, et al. 2004. Comparison of conventional bacteriology with nucleic acid amplification (Amplified Mycobacterium Direct Test) for diagnosis of tuberculous meningitis before and after inception of antituberculosis chemotherapy. J. Clin. Microbiol. 42:996–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]