Abstract
Leptotrichia spp. are anaerobic, pencil-shaped, Gram-negative rods that are part of the normal oral and intestinal human flora. Although not typically considered pathogenic, invasive Leptotrichia infections have been reported in immunosuppressed patients. A perceived rise in the identification of Leptotrichia spp. at our institution prompted a retrospective evaluation of these infections. Laboratory and clinical records were reviewed to identify Leptotrichia culture-positive patients. Over a 5-year period, 68 Leptotrichia-positive specimens were identified. Of these, 21% (14/68) were identified in original samples submitted from 13 different patients at our institution, and the remainder (79% [54/68]) were unknown isolates referred from outside hospitals for molecular identification. All in-house Leptotrichia were identified from blood cultures. Only 64% (9/14) of these grew on solid media, and 5 were a part of polymicrobial bacteremias containing other enteric pathogens. All local patients were receiving chemotherapy and a majority received hematopoietic stem cell transplant (HSCT) (11/13). All had neutropenic fever with symptoms of mucositis and/or enteritis. Most of the HSCT patients (73% [8/11]) were autologous recipients hospitalized after recent high-dose chemotherapy for multiple myeloma. L. hongkongensis, a novel species, was found in the majority of myeloma cases (63% [5/8]). In conclusion, we suggest that Leptotrichia spp. may be an underappreciated cause of bacteremia, particularly in multiple myeloma patients receiving cytotoxic chemotherapy for autologous HSCT. In our cohort, these infections were associated with neutropenic fever from an enteric source, and most isolates remained sensitive to standard antibiotics.
INTRODUCTION
High-dose chemotherapy followed by hematopoietic stem-cell transplantation (HSCT), is a well-established treatment modality for a variety of hematologic malignancies; however, infectious complications remain a major barrier to the overall success of this procedure. HSCT recipients are at particularly high risk for the development of invasive bacterial infections as a result of regimen-related neutropenia and cytotoxic damage to the oral and gastrointestinal mucosa. Mucositis severity, specifically, is an independent predictor of anaerobic bloodstream infection (BSI) following HSCT (11).
Leptotrichia spp. are fastidious anaerobic, pencil-shaped, Gram-negative rods that reside in the mouths, intestines, and female genital tracts of humans (20). Traditionally considered to be nonpathogenic, Leptotrichia species have occasionally been reported to cause invasive disease in HSCT patients and other immunocompromised hosts (6, 14, 18). The overall incidence of Leptotrichia infections in at-risk patient populations, however, may be underestimated. These organisms are notoriously difficult to recover from blood culture and may retain crystal violet, which can lead to their misidentification as Gram-positive rods. Commercially available phenotypic identification systems also have problems classifying these organisms (12). We routinely use 16S rRNA partial gene sequencing for the identification of anaerobic bacteria (16), which allowed us to recognize an increase in the number of Leptotrichia isolates identified by our laboratory. The purpose of the present study was to evaluate the incidence of Leptotrichia infections identified by ARUP Laboratories over a 5-year period and to review the occurrence of this infection at our institution.
(A portion of these data were presented at the 2010 American Society for Microbiology meeting in San Diego, CA, and at the 2010 Infectious Disease Society of America meeting in Vancouver, British Columbia, Canada.)
MATERIALS AND METHODS
Clinical isolates.
The Associated Regional and University Pathologists (ARUP) laboratories database was searched for all isolates identified as Leptotrichia spp. between January 2005 and December 2010. Specimen source, Gram stain result, the presence or absence of growth on solid media, and antimicrobial susceptibility patterns (when available) were recorded. ARUP is a national reference laboratory that also operates as the primary microbiology laboratory for the University of Utah Health Care (UUHC) system. Isolated organisms were submitted from outside institutions within the United States for molecular identification, whereas UUHC organisms were identified from primary clinical specimens. Medical records were reviewed for UUHC patients, under a protocol approved by the University of Utah Institutional Review Board. The following clinical information was assessed for adult patients: past medical history, presenting signs and/or symptoms of infection, diagnostic imaging, microbiology results, and antimicrobial therapies.
Blood culture.
UUHC patient blood samples were inoculated into one Bactec Plus aerobic bottle (resin media) and a Bactec standard anaerobic bottle (BD, Franklin Lakes, NJ). Cultures were incubated for 5 days in a Bactec 9240 blood culture system (BD) and were monitored automatically every 10 min for the presence of microbial growth. All positive anaerobic blood cultures were analyzed by Gram stain and subcultured on two sheep blood agar (SBA) plates (Hardy Diagnostics, Santa Maria, CA). One SBA plate was incubated at 37°C in an anaerobic chamber, and the second SBA plate was incubated in 5% CO2 atmosphere at 37°C. For samples that did not yield growth on the SBA plate, an additional chocolate and brucella blood agar plate, supplemented with hemin and vitamin K (Hardy Diagnostics), was inoculated and incubated for 5 days. Growth from the anaerobic SBA plate was directly referred for DNA sequencing.
Molecular identification.
DNA was extracted from isolated colonies using PrepMan Ultra sample preparation reagent (Applied Biosystems, Carlsbad, CA) according to the kit recommendations. DNA was analyzed using partial sequencing of a 500-bp 5′ region of the 16S rRNA gene as previously described (15). Blood cultures broths confirmed to be positive by Gram stain for bacteria morphologically consistent with Leptotrichia spp. but negative for growth on solid media were directly extracted for sequencing. Briefly, a 1.0-ml volume of positive blood was pelleted at 5,200 × g for 2 min, resuspended in 1.0 ml of sterile phosphate-buffered saline, and pelleted again (repeat twice). The final suspension was then extracted for sequencing as described above.
Antimicrobial susceptibility.
Antibiotic susceptibility testing was performed in real-time by physician request using the Clinical and Laboratory Standards Institute (CLSI) broth microdilution reference method for anaerobic organisms (4). In addition, 18 viable isolates were subcultured from frozen storage and analyzed retrospectively. Isolates were tested against a standard drug panel that included ampicillin-sulbactam, cefoxitin, meropenem, metronidazole, penicillin, and clindamycin (Trek Diagnostic Systems, Cleveland, OH). Leptotrichia isolates were also screened for β-lactamase activity using a Cefinase disk (Becton Dickinson Microbiology Systems, Cockeysville, MD). There are currently no established interpretive breakpoints for Leptotrichia spp.; however, antimicrobial susceptibility was inferred based on CLSI breakpoints for anaerobic organisms other than Bacteroides fragilis group (5).
Sequence and phylogenetic analysis.
Sequence-based identifications were performed in accordance with the CLSI guidelines (3). For the present study, Leptotrichia sequences were reanalyzed using the most up-to-date version of our 16S reference database (SmartGene IDNS version 3.6.1; Zug, Switzerland). A multiple sequence alignment of our isolates and Leptotrichia type strains was performed using CLUSTAL W in MEGA 5 (17). A phylogenetic analysis was then conducted using the neighbor-joining method with 500 bootstrap replications with MEGA5 software.
Statistical analysis.
Statistical analysis was performed using Analyze-It Software for Microsoft Excel (version 2.26). The proportions of patients with Leptotrichia bacteremia were compared using the Fisher exact test for small cell sizes. Summary statistics were used to describe the distribution of Leptotrichia infection at our institution.
RESULTS
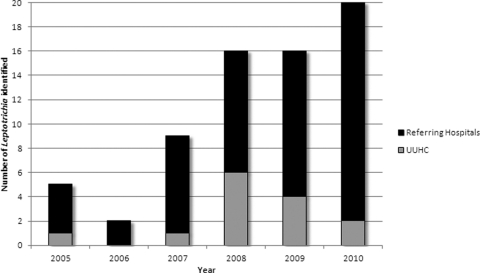
Over the 5-year study period, 68 Leptotrichia-positive cultures were obtained from 67 different patients. Most of these (79% [54/68]) were organisms referred to ARUP from outside institutions for molecular identification (Fig. 1). The majority of organisms (91% [62/68]) came from blood cultures. The remaining six isolates came from a variety of sources including: wounds (n = 3), respiratory (n = 2), and amniotic fluid (n = 1).
Fig 1.
Distribution of Leptotrichia isolates identified at ARUP between January 2005 and December 2010.
Leptotrichia microbiology.
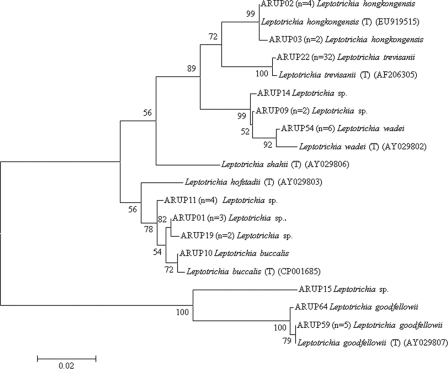
Seventy-five percent (51/68) of all Leptotrichia were identified to the species level, with L. trevisanii being the most frequently encountered species (47% [32/68]). The remaining species-level identifications included six isolates each of L. wadei, L. goodfellowii, and L. hongkongensis and a single isolate of L. buccalis. Overall, 17 Leptotrichia isolates could only be identified to genus level by partial sequencing of the 16S rRNA gene. Several of these isolates grouped in distinct clades, bearing similarity to L. buccalis, L. goodfellowii, or L. wadei (Fig. 2).
Fig 2.
Neighbor-joining dendrogram of unique partial 16S rRNA gene sequences (first 500 bp) for 64 Leptotrichia clinical isolates and 7 reference type strains. Branch support is recorded at nodes as a percentage of 500 bootstrap iterations. The scale bar indicates nucleotide substitutions per site. Type strains are indicated by the species name, a “(T)”, and the corresponding GenBank accession number. Clinical isolates are indicated by a specimen number, followed by the number of clinical isolates sharing the sequence and the reported identification.
Of the 14 organisms identified from primary specimens at UUHC, 36% (5/14) failed to grow on solid media. All of the nonculturable UUHC Leptotrichia were retrospectively identified as L. hongkongensis. One isolate that failed to grow on initial plating was repeatedly subcultured to several broth (Bactec standard anaerobic ± human blood) and solid (Columbia sheep blood, chocolate, and Brucella blood + hemin + vitamin K) media but was found to grow only in broth containing human blood. Only one multiple myeloma patient in the UUHC cohort had L. honkongensis successfully subcultured from their blood culture bottle to chocolate agar.
Antimicrobial susceptibility testing was performed for 39 isolates (Table 1). The majority appeared highly susceptible to the antibiotics tested, and there was no obvious difference in susceptibility patterns for a particular species. However, two L. trevisanii isolates displayed metronidazole MICs greater than 32 μg/ml, and one of these isolates also had a cefoxitin MIC of 32 μg/ml. A single isolate of L. goodfellowii also had an MIC of 16 for metronidazole. None of the tested organisms possessed detectable β-lactamase activity.
Table 1.
Leptotrichia antimicrobial susceptibility test resultsa
| Antibiotic | Modal MIC (μg/ml) | MIC range (μg/ml)b | No. of isolates tested |
|---|---|---|---|
| Ampicillin/sulbactam | <0.5/0.25 | ≤0.5/0.2–1/0.5 | 39 |
| Cefoxitin | <1 | <1–32 | 39 |
| Clindamycinc | <0.5 | <0.5–2 | 38 |
| Meropenem | <0.25 | <0.25 | 39 |
| Metronidazole | 1 | ≤0.5–≥32 | 39 |
| Penicillin | <0.06 | ≤0.06–0.5 | 39 |
| β-Lactamase | Negative | Negative | 38 |
The Leptotrichia species distribution included 21 L. trevisanii, 4 L. wadei, and 4 L. goodfellowii isolates and 10 isolates that could not be identified to the species level.
There are currently no established interpretive breakpoints for Leptotrichia spp.
Clindamycin susceptibility was performed upon specific request.
Clinical case histories.
In all, 14 cases of Leptotrichia BSI were identified from 13 unique UUHC patients. Approximately a third of cases (36% [5/14]) were part of polymicrobial bacteremias involving other pathogens (i.e., Enterococcus faecium, viridans group Streptococcus, Streptococcus infantis, Bacteroides urealyticus, or Fusobacterium nucleatum). All UUHC patients had received high-dose chemotherapy immediately prior to the onset of bacteremia; twelve had hematological cancers, and one was being treated for a solid tumor malignancy.
More than half of the UUHC hematology patients (67% [8/12]) were undergoing autologous HSCT (auto-HSCT) specifically for the treatment of multiple myeloma. Patients in the myeloma cohort had all been treated with a melphalan-based induction regimen that also included the novel chemotherapeutic agents thalidomide and bortezomib. L. hongkongensis was the most common single species identified from multiple myeloma patients (63%, 5/8). However, the test for association between L. hongkongensis and myeloma chemotherapy for auto-HSCT did not reach statistical significance (P = 0.4).
The multiple myeloma subgroup presented with similar clinical signs and symptoms of infection. All were neutropenic and had fever that was associated with nausea, vomiting, diarrhea, and abdominal pain despite prophylactic oral levofloxacin and fluconazole. In addition, two patients were hemodynamically unstable at the time of hospital admission: one grew L. hongkongensis and viridans group Streptococcus, and the other had L. hongkongensis identified by sequencing directly from the blood culture broth. Of note, one of the hypotensive patients experienced two separate episodes of L. hongkongensis bacteremia following tandem auto-HSCT separated by 3 months. Several myeloma patients (3/8) had computerized axial tomography confirmed necrotizing enterocolitis with negative Clostridium difficile enzyme immunoassay testing from stool specimens. L. hongkongensis was identified in the blood of two of these patients, and L. wadei was isolated from the third. All of the patients in this cohort survived their respective episodes of Leptotrichia BSI, with negative repeat blood cultures following the initiation of empirical meropenem as per our institution's protocol for the management of neutropenic fever.
The remaining five UUHC patients from were receiving therapy for acute myelogenous leukemia (AML) (n = 2), acute lymphoblastic leukemia (n = 1), T-cell-rich large B-cell lymphoma (n = 1), and esophageal cancer (n = 1). Both of the AML patients received allogeneic HSCT, while the lymphoma patient received auto-HSCT. All five patients presented initially for the evaluation of neutropenic fever; however, only the hematology patients had received prophylactic antimicrobials. At the time of hospital admission, all five had severe oral mucositis and were treated empirically with meropenem with or without vancomycin. Unlike the multiple myeloma cohort, only one patient exhibited diarrhea, which was negative by both enteric stool culture and PCR for C. difficile. The Leptotrichia species identified in these patients were variable, with two unspeciated isolates and one of each L. hongkongensis, L. trevisanii, and L. goodfellowii. Like the multiple myeloma cohort, these patients all survived these episodes of bacteremia.
DISCUSSION
Anaerobic BSIs have increasingly been recognized as an important sequela of high-dose chemotherapy, with severe alimentary tract mucositis identified as a predisposing risk-factor (11). While members of the genus Leptotrichia have been isolated from neutropenic patients (1, 6, 12–14, 19), they likely remain an underappreciated cause of BSI due to inherent difficulties with conventional laboratory identification methods (12).
We observed an increase in invasive Leptotrichia infections at ARUP in 2007, followed by a subsequent rise in our own hospital in 2008. The reasons for this are probably multifactorial. First, regular use of 16S rRNA partial gene sequencing significantly expands the number of anaerobic organisms that can be accurately identified (10, 16). It is also well recognized that Leptotrichia are not reliably identified by a commonly used phenotypic identification system (RapID ANA II; Remel, Lenexa, KS) (12, 14). Furthermore, the number of at-risk patients has also expanded in recent years with the routine use of high-dose cytotoxic chemotherapy for HSCT and the treatment of other malignancies. In recognition of the increasing numbers of auto-HSCT patients admitted with severe mucositis, enteric bacteremia, and sepsis, antimicrobial prophylactic strategies were revised at our institution in 2009. These revisions may account for the subsequent decline in Leptotrichia infections from our transplant center in 2010 (Fig. 1).
Although not generally regarded as drug-resistant organisms, Leptotrichia antimicrobial susceptibility patterns are largely undefined. Our data suggest that these organisms remain highly susceptible to standard agents. Some of the drugs used routinely for prophylaxis or empirical treatment of neutropenic fever, however, lack robust anaerobic activity (e.g., levofloxacin, cefepime, or ceftazidime). Patients with neutropenic oral mucositis and/or enterocolitis are at increased risk for anaerobic infection and should therefore be treated with a broad-spectrum antimicrobial regimen that has anaerobic coverage (9).
Seven Leptotrichia species have been validly described. Of these, L. buccalis and L. goodfellowii were implicated in severe infections, including endocarditis, L. trevisanii, L. wadei, and L. hongkongensis were associated with bacteremia, and L. hofstadii and L. shahii have been isolated from oral wounds (2, 7, 8, 21). It is interesting that, although previously only described as a cause of BSI in two cases (6, 18), L. trevisanii accounted for half of our isolates (32/68 [48%]). It is unclear whether this organism has been regularly identified at other institutions but not reported or whether it is simply not as pathogenic as other species (e.g., L. goodfellowii and L. buccalis). In addition, we identified multiple isolates only to the genus level despite high-quality sequencing results. The phylogenetic analysis does not cluster these isolates within any of the currently described species (Fig. 2), suggesting that some of these organisms may in fact represent previously undescribed species.
Of particular interest was the identification of six cases of L. hongkongensis bacteremia in our UUHC cohort, five of which were isolated from multiple myeloma patients. Although the apparent association between L. hongkongensis and auto-HSCT for multiple myeloma did not reach statistical significance, likely due to our small sample size, the observation is clinically important. L. hongkongensis was only recently described and to date, limited information exists on the clinical spectrum of associated illness (21). Four of the L. hongkongensis sequences in the present study were identical to the type strain (GenBank accession no. EU919515, listed as Leptotrichia sp. strain HKU24) (21), and two were identical to a Leptotrichia sp. isolated from the blood of a patient undergoing treatment for AML (GenBank accession no. AF189244) (12) (Fig. 2). These two clusters of L. hongkongensis sequences were 99.8% identical (differing by only 2 bp) to each other, which suggests that the organism in Patel et al. (12) was most likely L. hongkongensis. Consistent with this notion is the fact that their isolate also failed to grow on initial subculture to multiple solid media. The Patel et al. report (12), in addition to our six cases of bacteremia with L. hongkongensis in a single BMT unit, suggests that this species may be of particular concern in patients undergoing high-dose chemotherapy for HSCT, and its presence may be suggested by the characteristic Gram stain and failure to grow on subculture to solid media. Widespread recognition of these clinical and laboratory observations may lead to better understanding of the incidence of this organism.
In conclusion, Leptotrichia spp. are emerging pathogens in neutropenic patients receiving high-dose chemotherapy. At our institution, there was an apparent association between Leptotrichia BSI and the chemotherapeutic agents used for multiple myeloma patients undergoing auto-HSCT. In particular, L. hongkongensis was shown to be the predominant species in this population, further defining the clinical relevance of this recently described species. Molecular methods have greatly improved the clinical laboratories' ability to identify these potential pathogens, which have fortunately remained susceptible to most antimicrobial agents.
ACKNOWLEDGMENTS
We thank Mary Lampas and Stephanie Sanders for assistance with the medical record review. We thank Keith E. Simmon for his review of these data. We also thank the clinical microbiology laboratory at ARUP for providing antimicrobial susceptibility testing.
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Blairon L, et al. 2006. A 62-month retrospective epidemiological survey of anaerobic bacteraemia in a university hospital. Clin. Microbiol. Infect. 12:527–532 [DOI] [PubMed] [Google Scholar]
- 2. Caram LB, et al. 2008. Leptotrichia endocarditis: report of two cases from the International Collaboration on Endocarditis (ICE) database and review of previous cases. Eur. J. Clin. Microbiol. Infect. Dis. 27:139–143 [DOI] [PubMed] [Google Scholar]
- 3. CLSI 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline MM18-A. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 4. CLSI 2007. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard, 7th ed Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. CLSI 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Cooreman S, et al. 2011. Bacteraemia caused by Leptotrichia trevisanii in a neutropenic patient. Anaerobe 17:1–3 [DOI] [PubMed] [Google Scholar]
- 7. Duperval R, Beland S, Marcoux JA. 1984. Infective endocarditis due to Leptotrichia buccalis: a case report. Can. Med. Assoc. J. 130:422–424 [PMC free article] [PubMed] [Google Scholar]
- 8. Eribe ER, Olsen I. 2008. Leptotrichia species in human infections. Anaerobe 14:131–137 [DOI] [PubMed] [Google Scholar]
- 9. Freifeld AG, et al. 2011. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 52:e56–e93 [DOI] [PubMed] [Google Scholar]
- 10. Justesen US, et al. 2010. 16S rRNA gene sequencing in routine identification of anaerobic bacteria isolated from blood cultures. J. Clin. Microbiol. 48:946–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lark RL, et al. 2001. Risk factors for anaerobic bloodstream infections in bone marrow transplant recipients. Clin. Infect. Dis. 33:338–343 [DOI] [PubMed] [Google Scholar]
- 12. Patel JB, et al. 1999. Bacteremia caused by a novel isolate resembling Leptotrichia species in a neutropenic patient. J. Clin. Microbiol. 37:2064–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reig M, Baquero F, Garcia-Campello M, Loza E. 1985. Leptotrichia buccalis bacteremia in neutropenic children. J. Clin. Microbiol. 22:320–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwartz DN, Schable B, Tenover FC, Miller RA. 1995. Leptotrichia buccalis bacteremia in patients treated in a single bone marrow transplant unit. Clin. Infect. Dis. 20:762–767 [DOI] [PubMed] [Google Scholar]
- 15. Simmon KE, Croft AC, Petti CA. 2006. Application of SmartGene IDNS software to partial 16S rRNA gene sequences for a diverse group of bacteria in a clinical laboratory. J. Clin. Microbiol. 44:4400–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Simmon KE, Mirrett S, Reller LB, Petti CA. 2008. Genotypic diversity of anaerobic isolates from bloodstream infections. J. Clin. Microbiol. 46:1596–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tee W, Midolo P, Janssen PH, Kerr T, Dyall-Smith ML. 2001. Bacteremia due to Leptotrichia trevisanii sp. nov. Eur. J. Clin. Microbiol. Infect. Dis. 20:765–769 [DOI] [PubMed] [Google Scholar]
- 19. Weinberger M, Wu T, Rubin M, Gill VJ, Pizzo PA. 1991. Leptotrichia buccalis bacteremia in patients with cancer: report of four cases and review. Rev. Infect. Dis. 13:201–206 [DOI] [PubMed] [Google Scholar]
- 20. Winn WC, Koneman EW. 2006. Koneman's color atlas and textbook of diagnostic microbiology, 6th ed Lippincott/The Williams & Wilkins Co., Philadelphia, PA [Google Scholar]
- 21. Woo PC, et al. 2010. Leptotrichia hongkongensis sp. nov., a novel Leptotrichia species with the oral cavity as its natural reservoir. J. Zhejiang Univ. Sci. B 11:391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]