Abstract
Here, we describe a transmission event of hepatitis C virus (HCV) among injection drug users. Next-generation sequencing (NGS) was used to assess the intrahost viral genetic variation. Deep amplicon sequencing of HCV hypervariable region 1 allowed for a detailed analysis of the structure of the viral population. Establishment of the genetic relatedness between cases was accomplished by phylogenetic analysis. NGS is a powerful tool with applications in molecular epidemiology studies and outbreak investigations.
TEXT
It has been estimated that ∼130 million individuals are infected with hepatitis C virus (HCV) worldwide. Approximately three million new infections occur annually (2), many of which will develop into severe liver disease (8, 11). The most common risk factors associated with HCV infection are intravenous drug use, blood transfusions (before the advent of screening for HCV) and, to a lesser extent, nosocomial transmission (9, 19). In Mexico, the prevalence of HCV ranges between 0.74 and 1.5% among blood donors and ∼2% in asymptomatic individuals (12, 17). Transmission associated with blood transfusion is considered the most common source of HCV infection in the country (15, 16). Use of illegal intravenous drugs has been widely recognized as a major risk associated with HCV infection (13); however, this mode of transmission is not considered among the most important risk factors in Mexico (16). Thus, the true scope of the problem of HCV infection in Mexico is, most likely, not fully recognized.
Identification of HCV transmission is of critical importance in implementing measures aimed at preventing virus spread. HCV transmission networks are difficult to identify for several reasons. First, acute HCV infections are usually asymptomatic; therefore, establishing when a transmission event has occurred is rather challenging (4). Second, the lack of laboratory tests capable of distinguishing acute from chronic infections further complicates the identification of related cases. Finally, the characteristically long incubation periods among symptomatic cases make difficult to link related cases to a common source of infection. In addition, molecular approaches required to assess the intrahost viral genetic variation are quite sophisticated, time-consuming, and expensive. Thus, straightforward, inexpensive platforms suitable for molecular epidemiology studies are much needed. Next-generation sequencing (NGS) allows for cost-effective probing of virus populations at an unprecedented level of detail. The massively parallel sequencing approach can detect low-frequency mutations, providing a snapshot of the entire virus population, which makes it a viable alternative to other conventional methodologies (2).
In this work, we describe the identification, by using an NGS approach, of an HCV transmission event between individuals reporting the use of illegal injection drugs. Initially, two HCV cases (patients A and B), identified at the same medical center but at different times, were enrolled as part of a larger study addressing resistance to anti-HCV interferon treatment. As part of the protocol, baseline serum samples were obtained from both individuals before the start of treatment. Simultaneously, six different HCV patients (patients 1 through 6), with no history of drug use, enrolled at the same medical center, and cases epidemiologically unrelated to patients A and B were included as controls. None of the patients had previously received antiviral therapy. There was no coinfection with HBV or HIV among these individuals. Viral genotyping was performed by sequencing of the NS5B region as reported elsewhere (5). Table 1 summarizes the patient characteristics.
Table 1.
Patient characteristics
| Patient identifier | Gender | Age (yr) | HCV infecting genotype | Viral titer (IU/ml) | No. of sequence reads |
|---|---|---|---|---|---|
| A | Male | 33 | 1b | 3,680,000 | 10,644 |
| B | Male | 31 | 1b | 2,090,000 | 10,839 |
| 1 | Male | 56 | 1b | 2,090,000 | 10,593 |
| 2 | Male | 61 | 1a | 7,900,000 | 11,156 |
| 3 | Male | 58 | 1a | 1,400,000 | 10,254 |
| 4 | Male | 53 | 1a | 3,056,540 | 11,406 |
| 5 | Male | 63 | 1a | 7,190,000 | 8,263 |
| 6 | Male | 56 | 1a | 16,300,000 | 10,569 |
The intrahost viral genetic variation in each patient was assessed by deep amplicon sequencing of the HCV hypervariable region 1 (HVR-1) using the 454 GS FLX platform. Briefly, total RNA was extracted from all serum samples and cDNA was generated using a SuperScript Vilo cDNA synthesis kit (Invitrogen, Carlsbad, CA). Amplification of the HVR-1 region was accomplished by nested PCR as reported elsewhere (14). The first-round PCR products were further amplified with “fusion primers” composed of the 454 primer keys, with different multiple identifiers for each sample, and the HCV HVR-1-specific primers. The resulting PCR products were purified by agarose gel electrophoresis on SizeSelect e-gels (Invitrogen). Purified amplicons were quantified and mixed at equimolar concentrations. Deep sequencing was performed with a 454/Roche GS FLX instrument using the titanium chemistry (Roche Applied Science, Indianapolis, IN). The sequence reads were processed and denoised as described elsewhere (6). Only long reads covering the entire length of the amplicon were included in the sequence analysis. Additionally, a comprehensive nucleotide alignment, containing 358 HCV HVR-1 representative sequences from both subgenotypes, 1a and 1b (140 and 218, respectively), was obtained from the Los Alamos National Laboratory HCV database (http://hcv.lanl.gov/content/sequence/NEWALIGN/align.html). Multiple alignments were performed using MAFFT (8). Phylogenetic analysis was conducted using the neighbor joining method (Kimura 2-parameter) as implemented in MEGA5 (18).
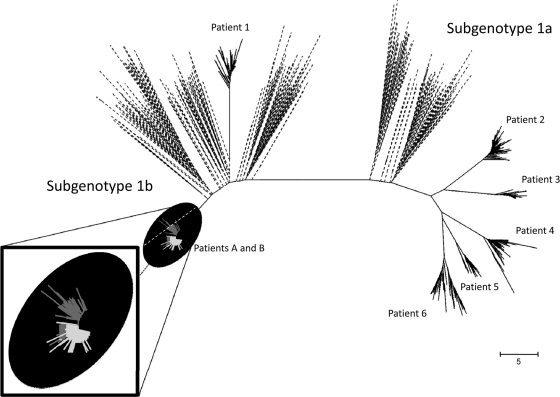
The resulting phylogenetic analysis is presented in Fig. 1. Patients A, B, and 1 belonged to subgenotype 1b, while the remaining five patients were infected with subgenotype 1a. Sequences belonging to patients A and B clearly shared the same sequence space, revealing significant intermixing between viral variants from the two patients. Analysis of the structure of the viral population allowed the establishment of genetic relatedness between the corresponding viral variants linking the two individuals. The genetic distances between the two related cases ranged between 0.0 and 0.041691 (average genetic distance, 0.014845). Sequences from all other unrelated cases occupied a completely different sequence space. The average genetic distance between unrelated cases was significantly higher (P < 0.05). The closest average genetic distance between sequences belonging to patient A and any other sequence, excepting those belonging to patient B, was 0.2057. Similarly, the closest distance between sequences from patient B and any other sequence not belonging to patient A was 0.2162. The closeness between both viral populations found among patients A and B might be the reflection of a rather recent transmission. A later interview with the two patients showed that the two individuals were acquainted. Both reported the use of intravenous drugs; however, no syringe sharing, sexual contact, or blood transfusion was acknowledged by either individual. Thus, while the use of drugs remained the main associated risk factor, the exact mode of transmission could not be established.
Fig 1.
Phylogenetic analysis. Genetic distances (Kimura 2-parameter) were used to conduct the corresponding phylogenetic analysis (neighbor-joining method) of HCV HVR-1 sequences from all HCV cases studied. Sequences from related cases are color coded (patient A, light gray; patient B, dark gray) and magnified in the window in the lower left corner. Unrelated local HCV cases are indicated by solid black lines. HVR-1 sequences from the Los Alamos National Laboratory HCV database are shown in black discontinuous lines.
While injection drug use has not yet been recognized as a major risk for HCV infection in Mexico, the increasing accessibility of affordable illegal drugs remains a growing problem in the country (3). Thus, in the near future, HCV infection associated with networks of injection drug users (IDUs) can potentially become one of the main modes of viral transmission in Mexico. Identification of such networks is of importance since it can help the implementation of preventive and control measures (health education, opiate substitution therapy, etc.) aimed at preventing transmission by reducing unsafe injection (1, 11). The current treatment guidelines do not exclude IDUs due to the fact that IDUs seem to exhibit viral responses comparable to those seen among ex- or non-IDUs, despite the possibility of subsequent reinfection (7). Recently, modeling studies have suggested that even modest rates of anti-HCV treatment among active IDUs could effectively reduce virus transmission (10). Thus, identifying HCV cases among IDU networks and extending therapy coverage to such populations is likely to have a positive impact on HCV control, especially with the improved sustained viral responses rates seen with the new direct antiviral agents.
NGS approaches are powerful methods that allow a rather comprehensive analysis of the intrahost viral genetic variation (6). Additionally, the simplicity and increasing accessibility to such technologies is likely to revolutionize the field of molecular epidemiology. The advantages over conventional methods, such as consensus sequencing, bacterial cloning, and endpoint limiting dilution, are noteworthy (6). Moreover, the parallel development of quite a large number of software and algorithms capable of handling the massive amount of data generated by NGS platforms is likely to expedite the implementation of such approaches in a variety of settings in the near future. Thus, NGS is expected to significantly improve molecular epidemiology studies and HCV outbreak investigations.
Footnotes
Published ahead of print 1 February 2012
REFERENCES
- 1. Advisory Council on the Misuse of Drugs 2009. The primary prevention of hepatitis C among injecting drug users. Advisory Council on the Misuse of Drugs, London, United Kingdom [Google Scholar]
- 2. Beerenwinkel N, Zagordi O. 2011. Ultra-deep sequencing for the analysis of viral populations. Curr. Opin. Virol. 1:413–418 [DOI] [PubMed] [Google Scholar]
- 3. Boddiger D. 2010. Mexico eager to reduce demand for illicit drugs. Lancet 375:15–16 [DOI] [PubMed] [Google Scholar]
- 4. Centers for Disease Control and Prevention 1998. Recommendations for prevention and control of hepatitis C virus (HCV) infection and HCV-related chronic disease. MMWR Recomm. Rep. 47(RR-19):1–39 [PubMed] [Google Scholar]
- 5. Fischer GE, et al. 2010. Hepatitis C virus infections from unsafe injection practices at an endoscopy clinic in Las Vegas, Nevada, 2007–2008. Clin. Infect. Dis. 51:267–273 [DOI] [PubMed] [Google Scholar]
- 6. Fonseca-Coronado S, et al. 2012. Specific detection of naturally occurring hepatitis C virus mutants with resistance to telaprevir and boceprevir (protease inhibitors) among treatment-naive infected individuals. J. Clin. Microbiol. 50:281–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hellard M, Sacks-Davis R, Gold J. 2009. Hepatitis C treatment for injection drug users: a review of the available evidence. Clin. Infect. Dis. 49:561–573 [DOI] [PubMed] [Google Scholar]
- 8. Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9:286–298 [DOI] [PubMed] [Google Scholar]
- 9. Lavanchy D. 2011. Evolving epidemiology of hepatitis C virus. Clin. Microbiol. Infect. 17:107–115 [DOI] [PubMed] [Google Scholar]
- 10. Martin NK, et al. 2011. Can antiviral therapy for hepatitis C reduce the prevalence of HCV among injecting drug user populations? A modeling analysis of its prevention utility. J. Hepatol. 54:1137–1144 [DOI] [PubMed] [Google Scholar]
- 11. Martin NK, Vickerman P, Hickman M. 2011. Mathematical modelling of hepatitis C treatment for injecting drug users. J. Theor. Biol. 274:58–66 [DOI] [PubMed] [Google Scholar]
- 12. Mendez-Sanchez N, et al. 2005. Prevalence of hepatitis C infection in a population of asymptomatic people in a checkup unit in Mexico City. Dig. Dis. Sci. 50:733–737 [DOI] [PubMed] [Google Scholar]
- 13. Nelson PK, et al. 2011. Global epidemiology of hepatitis B and hepatitis C in people who inject drugs: results of systematic reviews. Lancet 378:571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ramachandran S, Xia GL, Ganova-Raeva LM, Nainan OV, Khudyakov Y. 2008. End-point limiting-dilution real-time PCR assay for evaluation of hepatitis C virus quasispecies in serum: performance under optimal and suboptimal conditions. J. Virol. Methods 151:217–224 [DOI] [PubMed] [Google Scholar]
- 15. Rivera-Lopez MR, Zavala-Mendez C, Arenas-Esqueda A. 2004. Prevalence for seropositivity for HIV, hepatitis B and hepatitis C in blood donors. Gac. Med. Mex. 140:657–660 (In Spanish.) [PubMed] [Google Scholar]
- 16. Romero-Figueroa S, et al. 2012. Risk factors associated with hepatitis C virus infection in an urban population of the State of Mexico. Arch. Virol. 157:329–332 [DOI] [PubMed] [Google Scholar]
- 17. Sosa-Jurado F, et al. 2010. Hepatitis C virus infection in blood donors from the state of Puebla, Mexico. Virol. J. 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thompson ND, Perz JF, Moorman AC, Holmberg SD. 2009. Nonhospital health care-associated hepatitis B and C virus transmission: United States, 1998–2008. Ann. Intern. Med. 150:33–39 [DOI] [PubMed] [Google Scholar]