Abstract
Rapid diagnostic tests (RDTs) represent important tools to diagnose malaria infection. To improve understanding of the variable performance of RDTs that detect the major target in Plasmodium falciparum, namely, histidine-rich protein 2 (HRP2), and to inform the design of better tests, we undertook detailed mapping of the epitopes recognized by eight HRP-specific monoclonal antibodies (MAbs). To investigate the geographic skewing of this polymorphic protein, we analyzed the distribution of these epitopes in parasites from geographically diverse areas. To identify an ideal amino acid motif for a MAb to target in HRP2 and in the related protein HRP3, we used a purpose-designed script to perform bioinformatic analysis of 448 distinct gene sequences from pfhrp2 and from 99 sequences from the closely related gene pfhrp3. The frequency and distribution of these motifs were also compared to the MAb epitopes. Heat stability testing of MAbs immobilized on nitrocellulose membranes was also performed. Results of these experiments enabled the identification of MAbs with the most desirable characteristics for inclusion in RDTs, including copy number and coverage of target epitopes, geographic skewing, heat stability, and match with the most abundant amino acid motifs identified. This study therefore informs the selection of MAbs to include in malaria RDTs as well as in the generation of improved MAbs that should improve the performance of HRP-detecting malaria RDTs.
INTRODUCTION
Malaria rapid diagnostic tests (RDTs) are lateral flow immunochromatographic tests that detect parasitic antigens circulating in the blood of malaria-infected patients. Currently, the tests most widely used for diagnosis of Plasmodium falciparum infection target the parasite antigen histidine-rich protein 2 (HRP2).
HRP2 is produced in abundance during the asexual cycle and in early gametocyte stages of P. falciparum parasites (5, 6, 9, 10, 12). It has an unusual structure in that it contains multiple contiguous repeats that are rich in alanine, histidine, and aspartic acid, which comprise 37%, 34%, and 10% of the protein, respectively (9, 12). These features make HRP2 an ideal biomarker for immunodetection. A number of monoclonal antibodies (MAbs) against HRP2 have been developed for use in HRP2-detecting RDTs. A major determinant of the performance of these HRP2-detecting malaria RDTs is the binding characteristics of the MAbs. It is plausible that the number of potential target epitopes in the HRP protein of a parasite isolate could be an important determinant of the binding affinity of a MAb, where a higher epitope frequency may result in greater test sensitivity. The frequency or distribution of the target epitopes present in a particular parasite population may have an impact on the efficiency of detection of parasitemia in this population and, hence, the sensitivity of RDTs that use this antibody. An ideal epitope to be targeted by the signal and capture antibodies in an RDT would be one that is present in all isolates (high prevalence) and has a high copy number in each isolate (high abundance). It is also plausible that due to the extensive polymorphism of HRP2 (2, 3), certain target epitopes could be present in higher or lower frequencies among isolates in certain geographic regions, thereby influencing RDT performance.
In previous studies (2, 3, 8), we have investigated various factors that may influence the performance of HRP2-detecting RDTs, including sequence variation of the pfhrp2 gene (2, 3). To explore the potential influence of MAb specificity on the performance of RDTs, we have defined the epitopes recognized by four HRP2-specific MAbs (3A4, 4A5, 2G12-1C12, and 1E1-A9 [8]). We now report a comprehensive survey of HRP2 epitopes, including study of a further eight available P. falciparum HRP (PfHRP)-specific MAbs. We first identify the epitope recognized by each of these MAbs. Then, using derived sequences of the pfhrp2 genes from 448 P. falciparum isolates from 9 different geographic regions, the frequency of the 12 epitopes (eight identified in this work plus those from the four previously studied MAbs [8]) was determined, and geographic variation in the frequency of these epitopes was investigated. Furthermore, a bioinformatic approach was undertaken to identify the most abundant short amino acid motifs which are encoded by the population of pfhrp2 genes and to investigate the relationship of these motifs to the epitopes recognized by the mapped MAbs. As available data indicate that most antibodies specific for HRP2 also recognize the smaller histidine-rich protein HRP3 (8), we undertook analysis of epitope and motif frequency in the derived amino acid sequences from 99 pfhrp3 genes. In addition, the heat stability of the MAbs when blotted onto nitrocellulose membranes was investigated. It was expected that these analyses would provide further insights into the suitability of the current MAbs and the need to produce new MAbs that recognize epitopes which are present at a higher frequency than those recognized by currently available MAbs.
MATERIALS AND METHODS
P. falciparum parasite cultivation.
Eight P. falciparum lines, including 4 originating from Papua New Guinea (PNG) (AN143, FCQ41, D10, and FCQ64), 2 from Solomon Islands (N70 and SJ15), and 1 each from Thailand (INDO21) and the Philippines (PH4), were cultured in vitro using standard methods (11). Parasites were synchronized using a standard sorbitol treatment method (7), harvested at ring stage (100% rings in 1,000 erythrocytes), and used for protein extraction.
MAbs.
Seven MAbs (PTL-3, A6-4, N7, C1-13, C2-3, TC10, S2-5, and K2-1) generated against HRP2 were supplied by National Bioproducts Institute, South Africa. One MAb was purchased from Genway Biotech, Inc., USA (catalogue no. 20-783-74344). All MAbs were resuspended in phosphate-buffered saline (PBS) at a concentration of 1 mg/ml.
Protein extraction.
An aliquot of 1 × 107 infected erythrocytes was collected from each culture, and protein extracts were prepared by subjecting cells to three freeze-thaw cycles, followed by a 30-min incubation on ice in 50 μl of PBS containing 1% Triton X-100 and a protease inhibitor cocktail (proteinase inhibitor mini cocktail tablets; Roche). Samples were centrifuged at 15,000 × g, and the supernatant was collected as a water-soluble protein extract and stored at −20°C.
Dot blot analysis.
To investigate the relative recognition of native parasite proteins by the MAbs, dot blot analysis was undertaken as previously described (8). Briefly, a total parasite protein extract was prepared in PBS in doubling dilutions; 15 μl of each dilution was transferred onto a nitrocellulose membrane by vacuum aspiration in a dot blot apparatus. Membranes were blocked with 2% casein–Tris-buffered saline (TBS) and incubated with the MAbs diluted 1:2,000 in TBS–2% casein. Following washing in TBS-Tween (TBS, 0.5% Tween 20), alkaline phosphatase-conjugated goat anti-mouse immunoglobulin secondary antibody (Sigma) was added in a dilution of 1:5,000 in TBS–2% casein. Incubations with both primary and secondary antibodies were carried out for 1 h at room temperature. Detection of positive signals was undertaken by visualizing chemiluminescence with the CDP-STAR substrate (Amersham Bioscience) and captured on autoradiography film.
Epitope mapping.
Epitope mapping was undertaken as previously described (8) except that an additional 7 peptides were included in the screening process. These were added following bioinformatic analysis of a more comprehensive set of 448 pfhrp2 gene sequences that have since become available (2), instead of the 46 sequences used in our previous analysis of four MAbs (8). In brief, a peptide library was synthesized encompassing a total of 178 peptides, representing combinations of all 448 HRP2 sequences, each of 15 residues and overlapping sequentially by 3 amino acids (aa) (Mimotopes Pty. Ltd., Clayton, Victoria, Australia). Peptides were synthesized with a biotin residue at the N terminus followed by an SGSG spacer. The peptides were coated onto streptavidin (5 μg/ml)-coated 96-well polycarbonate plates (Nunc Maxisorb). Plates were washed with TBS-Tween, blocked with PBS–1% casein, and incubated at room temperature for 1 h with 100 μl of each MAb (diluted 1:1,000 in PBS). Plates were then washed with PBS–0.1% Tween and incubated with a secondary antibody (horseradish peroxidase-conjugated anti-mouse immunoglobulin; Sigma) diluted 1:2,000 in PBS. Plates were developed using freshly prepared ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] substrate solution, and the optical density (OD) at 405 nm was measured after 35 min. The epitope recognized by each MAb was determined by arranging in hierarchical order the OD values obtained with each peptide such that peptides with the highest OD were determined to contain the optimal or complete epitope. Those peptides with a positive signal but a lower OD were judged to contain only partial or suboptimal epitopes.
Once the peptides with the greatest reactivity to each MAb were identified, they were subjected to further mapping by a glycine scan to define the minimum epitope, i.e., the shortest peptide with the highest reactivity to the MAb. Briefly, for each reactive peptide, a set of 9- to 12-mer peptides was synthesized, each of which had 1 amino acid replaced by a glycine residue. In total, 47 peptides were synthesized. All peptides carried biotin plus an SGSG spacer at the N terminus. The reactivity of each of these peptides with each MAb was tested as outlined above. From the peptide reactivity results, the best-recognized epitope for each antibody was determined visually by comparing OD readings of different peptides, with the peptide with the highest OD reading considered to contain the potential epitope. Truncation scans were also performed on MAbs where the glycine scan was unsuccessful in determining the minimum epitopes of the MAb. These entailed synthesis and testing of sets of 3- to 14-mer peptides with each peptide having one less amino acid than previously. A total of 31 truncation peptides were synthesized. Once the potential epitope was determined, the number of repeats of each epitope in the collection of P. falciparum HRP2 sequences (2) was enumerated visually.
Identification and enumeration of repeat sequences.
The amino acid sequences encoded by a total of 448 pfhrp2 genes originating from P. falciparum isolates collected from a range of geographical regions (2) were subjected to analysis for discrete linear motifs ranging in length from 6 to 30 amino acid residues. These motifs were identified using a purpose-designed script (see the supplemental material) provided by Allan McRae and Peter Visscher from the Queensland Statistical Genetics Laboratory, Queensland Institute of Medical Research. This script generated a list of the frequencies of each motif in each parasite isolate, as well as the total frequency of each motif in the 448 sequences. From the computer-generated script, the epitopes recognized by the MAbs were also located. The percentage of isolates containing a given epitope among the 448 strains sequenced was determined. In addition, the total count of the epitope in the isolates (occurrence) and the average count per isolate (frequency) were determined. The amino acid sequences encoded by 99 pfhrp3 genes from P. falciparum isolates were also subjected to analysis for discrete linear motifs ranging in length from 6 to 30 amino acid residues. These motifs were identified using the same script used to analyze pfhrp2. Analysis generated a list of the frequency count of each motif in each individual isolate as well as the total frequency in the 99 HRP3 sequences.
Analysis for regional variation.
To compare the frequency of the epitopes and motifs in parasites from different geographic regions, P. falciparum isolates were grouped into 5 major geographic regions and 19 subregional groups (see Tables 2 to 4 below). Countries with more than 10 isolates represented their own subregional group, while countries that had 10 or fewer isolates were grouped together. A one-way analysis of variance (ANOVA) (with Tukey's test as post hoc analysis) was used to test for differences between the mean frequencies of the motifs between regions. Statistical analysis was performed using the SPSS software package (PASW Statistics version 17.0).
Table 2.
Most frequent HRP2 motifs and regional variability in 448 sequences
| Motif length | Motif | Total frequency (mean) | Geographic variability |
|||
|---|---|---|---|---|---|---|
| Highest frequency |
Lowest frequency |
|||||
| Region (n) | Mean (range) | Region (n) | Mean (range) | |||
| 7 | AHHAADA | 8,231 (18.4) | Caribbean (11) | 20.7 (18–25) | Asian Pacific (84) | 17.9 (13–23) |
| 8 | AHHAADAH | 8,231 (18.4) | Caribbean (11) | 20.7 (18–25) | Asian Pacific (84) | 17.9 (13–23) |
| 9 | AHHAADAHH | 8,226 (18.4) | Caribbean (11) | 20.7 (18–25) | Asian Pacific (84) | 17.8 (13–23) |
| 10 | AHHAADAHHA | 8,226 (18.4) | Caribbean (11) | 20.7 (18–25) | Asian Pacific (84) | 17.8 (13–23) |
| 11 | AHHAHHAADAH | 5,523 (12.3) | Caribbean (11) | 13.2 (8–17) | South America (41) | 11.4 (8–14) |
| 12 | AHHAHHAADAHH | 5,518 (12.3) | Caribbean (11) | 13.2 (8–17) | South America (41) | 11.4 (8–14) |
| 7 | HHAADAH | 8,231 (18.4) | Caribbean (11) | 20.6 (18–25) | Asian Pacific (84) | 17.9 (13–23) |
| 8 | HHAADAHH | 8,226 (18.4) | Caribbean (11) | 20.6 (18–25) | Asian Pacific (84) | 17.8 (13–23) |
| 9 | HHAADAHHA | 8,226 (18.4) | Caribbean (11) | 20.6 (18–25) | Asian Pacific (84) | 17.8 (13–23) |
| 10 | HHAHHAADAH | 5,523 (12.4) | Caribbean (11) | 13.2 (8–17) | South America (41) | 11.4 (8–14) |
| 11 | HHAHHAADAHH | 5,518 (12.4) | Caribbean (11) | 13.2 (8-17) | South America (41) | 11.4 (8–14) |
| 12 | HHAHHAADAHHA | 5,518 (12.4) | Caribbean (11) | 13.2 (8–17) | South America (41) | 11.4 (8–14) |
Table 4.
Length of HRP2 in different geographic regions and subregionsa
| Region and subregion | No. of isolates | Avg aa length (± SE) |
|---|---|---|
| Asian Pacific | 84 | 256 |
| E Timor | 24 | 263 ± 14 |
| Other Asian Pacific | 8 | 242 ± 22 |
| PNG | 17 | 252 ± 20 |
| Solomon Islands | 35 | 256 ± 19 |
| SE Asia | 109 | 254 |
| Cambodia | 31 | 257 ± 16 |
| Other SE Asia | 23 | 256 ± 19 |
| Philippines | 45 | 253 ± 24 |
| China | 10 | 244 ± 15 |
| S America | 41 | 253 |
| Other S America | 13 | 258 ± 13 |
| Peru | 16 | 256 ± 13 |
| Colombia | 12 | 243 ± 22 |
| Africa | 203 | 252 |
| Congo | 13 | 260 ± 16 |
| Nigeria | 80 | 255 ± 18 |
| Madagascar | 17 | 251 ± 24 |
| Kenya | 30 | 249 ± 15 |
| Tanzania | 34 | 248 ± 21 |
| Other Africa | 29 | 243 ± 25 |
| Caribbean | 11 | 246 |
| Other Caribbean | 1 | 275 |
| Haiti | 10 | 243 ± 22 |
Data for regions in boldface; data for subregions in lightface. E, East; S, South; SE, Southeast.
Heat stability profiles of the MAbs.
MAbs were blotted onto nitrocellulose membrane in 5 dilutions using a dot blotter. The concentration of MAbs blotted onto the membrane varied depending on the strength of signal that the MAbs gave in preliminary experiments. All MAb concentrations were quantified to give the same strength of signal on an X-ray film at baseline. The membranes were blocked with 2% casein-PBS and washed 3 times with PBS followed by rinsing in deionized water. The membranes were kept at 30°C, 45°C, and 60°C for 5, 30, and 90 days in a sealed bag.
Reaction with test antibodies was carried out by incubating the various membranes for 1 h at room temperature with 15 ml of biotinylated recombinant PfHRP2 (rPfHRP2; 3 μg/ml) (our unpublished data). Biotinylation of rPfHRP2 was performed by incubating equal moles of biotin (B4501; Sigma-Aldrich) to the same amount of rPfHRP2. The membrane was washed 3 times with PBS-Tween, followed by addition of Neutr-Avidine conjugated to horseradish peroxidase (HRP) (catalogue #31001; Pierce) diluted 1:20,000 in PBS and incubated for 1 h at room temperature. The membrane was washed 3 times in PBS-Tween. Detection was carried out using ECL (GE Healthcare) detection reagent, and the membrane was exposed to X-ray film for 2 min.
RESULTS
All MAbs recognize parasite proteins.
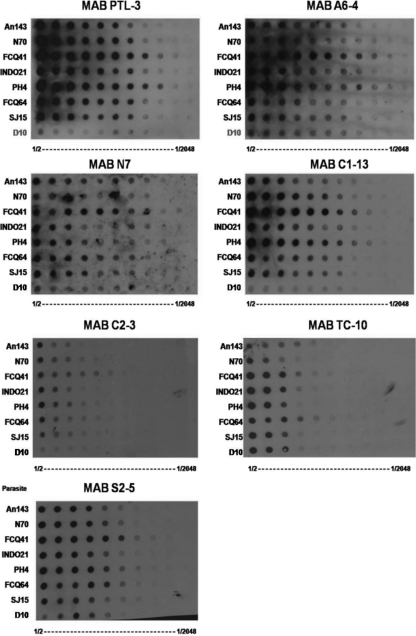
The eight MAbs tested all recognized P. falciparum parasite proteins with varying intensities. Differences in reactivity to different strains were also observed (Fig. 1). MAbs A6-4 and C1-13 had similar patterns of reactivity, binding strongly to parasite isolates FCQ41 and PH4 and moderately to the other isolates. MAbs S2-5 and N7 reacted moderately with all strains, with a slightly higher reactivity to FCQ41. This pattern was also observed for MAb TC-10, except the overall reactivity was lower. MAb C2-3 and MAb Genway reacted weakly to all strains (data not shown). As expected, none of the MAbs tested reacted strongly with FCQ27-D10, a strain that lacks the pfhrp2 gene but contains the pfhrp3 gene.
Fig 1.
Dot blot analysis for seven HRP2-specific MAbs against crude parasite extract from eight different P. falciparum isolates tested in serial dilutions.
Epitope mapping of the MAbs.
In the initial epitope mapping analysis, all 8 MAbs tested positive, with each MAb recognizing a different epitope (Table 1). MAb A6-4 recognized HATDAHHAAA as its major epitope, with possible substitution of D for the last amino acid. MAbs C1-13, N7, S2-5, and C2-3 all recognized similar epitopes with AHHAADAHH as a part of their major epitope. C1-13 recognized the major epitope AHHAADAHHA, while MAb N7 recognized the same epitope except with an additional D at the first residue. MAb S2-5 recognized AHHAADAHHA, with possible substitution of the third A with an S. MAb TC-10 recognized the longest major epitope of TDAHHAADAHHAADA. MAb PTL-3 recognized the major epitope DAHHAHHA with possible substitution of Y for the D. MAb Geneway recognized a different major epitope of AAYAHHAHHAAY.
Table 1.
Major and minimal epitopes of HRP-specific MAbs, occurrence in P. falciparum field isolates, and heat stability
| MAb | Major epitope(s) | Minimum epitopeb |
Reactivity before and after incubation at 37°Cc |
||||
|---|---|---|---|---|---|---|---|
| Epitope | Prevalence in isolates tested (%) | Avg frequency | Day 0 | 30 days | 90 days | ||
| 3A4a | DAHHAHHA | AHHAHHA | 448/448 (100) | 15 | +++++ | +++ | − |
| 2G12-1C12a | DAHHAADAHH | DAHHAADAHH | 448/448 (100) | 10 | +++++ | +++++ | − |
| DAHHVADAHH | |||||||
| 1E1-A9a | YAHHAHHA | AHHAHHV | 445/448 (99.3) | 3 | +++++ | +++ | − |
| DAHHAHHV | |||||||
| 4A5a | Conformational | Conformational | NA | NA | +++++ | ++++ | − |
| A6-4 | HATDAHHAAD | HATDAHH | 448/448 (100) | 5 | +++++ | ++ | − |
| HATDAHHAAA | |||||||
| C1-13 | AHHAADAHHA | AHHAADAHH | 448/448 (100) | 18 | +++++ | +++++ | +++ |
| N7 | DAHHAADAHHA | DAHHAADAHHA | 448/448 (100) | 10 | +++++ | +++ | − |
| PTL-3 | DAHHAHHA | YAHHAHHA | 439/448 (97.9) | 3 | +++++ | +++++ | +++ |
| YAHHAHHA | |||||||
| S2-5 | AHHAADAHH | AHHASDAHHA | 367/448 (82) | 1 | +++++ | − | − |
| AHHASDAHH | |||||||
| TC-10 | TDAHHAADAHHAADA | TDAHHAADAHHAADA | 327/448 (72.9) | 1 | ND | ND | ND |
| C2-3 | AHHAADAHH | HAHHAHHAADAHH | 123/448 (27.4) | 0 | +++++ | ++ | − |
| Genway | AAYAHHAHHAAY | AYAHHAHHAAY | 3/448 (0.01) | 0 | ND | ND | ND |
Epitope mapping previously published (8).
NA, not applicable.
Signal strength was graded on an arbitrary scale from +++++ (maximum) to + (just visible) to − (absent). ND, not determined.
To further define the composition and the minimum size of the epitopes recognized by the MAbs, a glycine scan of the predicted epitopes was performed (Table 1). The minimum epitope recognized by MAb A6-4 was HATDAHH. The removal of the AAD or AAA from the major epitope had minimal effects on the enzyme-linked immunosorbent assay (ELISA) result. Minimum epitopes of MAbs N7 and TC-10 remained the same as the major epitopes, with substitution of the first or the last amino acid significantly decreasing the ELISA result. The minimum epitope recognized by C1-13 was AHHAADAHH. The removal of the last amino acid A from the initial epitope (AHHAADAHHA) had no effect on the reactivity to the MAb. MAb Genway recognized AHHAHHAAY as its minimum epitope. MAb PTL-3 recognized the minimum epitope YAHHAHHA with a greater reactivity than DAHHAHHA. MAb S2-5 recognized AHHASDAHHA better than AHHAADAHHA; both were 1 amino acid longer than the major epitope. A glycine scan of the MAb C2-3 revealed that the minimum epitope recognized by this antibody was also longer than the major epitope. By adding single amino acids one by one to the start and end of the major epitope, we were able to increase the reactivity, hence identifying the minimum epitope. Therefore, the minimum epitope for MAb C2-3 was identified as HAHHAHHAADAHH.
Epitope frequency in P. falciparum isolates.
The distribution of the minimum epitopes recognized by each of the MAbs among the 448 available pfhrp2 sequences (2) was determined, as were the total and average frequencies. The frequencies of each epitope varied among parasite isolates (Table 1). Epitopes for five of the MAbs were present in all 448 isolates (3A4, 2G12-1C12, A6-4, C1-13, and N7) but varied in frequency from an average of 5 (A6-4) to 18 (C1-13). Epitopes for three other MAbs were present in the majority of isolates (1E1-A9, 99.3%; PTL-3, 97.9%; and TC-10, 72.9%) but occurred at a lower frequency with an average of one to three epitopes per isolate. Epitopes of C2-4 and Geneway were less common, being present in 27.4% and 0.01% of isolates, respectively. Where present, they were there at low frequency.
Identification of the most abundant motifs in HRP2.
An alternate approach to defining which strings of repetitive sequence in the HRP2 protein are best matched by available antibodies and if a more abundant linear epitope can be defined for generating novel MAbs is to identify the most abundant amino acid sequence strings, termed here motifs. For this purpose, a computer script was written and used to scan all 448 pfhrp2 gene sequences. As a first step, motifs with sequence lengths ranging from 7 to 12 amino acids that were present in all 448 isolates were identified. Then motifs were ranked by the number of occurrences in 448 isolates (Table 2).
The most abundant HRP2 motif identified had a core motif of AHHAADA. When this motif was progressively lengthened by up to 3 residues to 10 amino acids (AHHAHHDAHHA), its abundance remained unchanged in the 448 isolates, with a total of 8,226 occurrences and an average frequency of 18.4 per isolate. However, the addition of a further histidine residue resulted in a significant drop in the abundance of the motif to 5,523 (Table 2).
The second most abundant core motif was the 7-amino-acid sequence HHAADAH. Compared to the most abundant motif, it is shifted to the right by 1 amino acid residue. It was possible to extend the motif by only 2 amino acid residues before a significant reduction in abundance was observed (Table 2). Both motifs were most frequent in isolates from the Caribbean region and were least frequent in isolates from the Asian Pacific and South American regions (Table 2).
Identification of the most prevalent motifs in HRP3.
Because the repeat structure in the HRP3 protein is very similar to that in HRP2, and HRP2-specific MAbs cross-react with the HRP3 protein (8), the motif-finding analysis was repeated for the available 99 pfhrp3 gene sequences (2). Two highly prevalent HRP3 motif groups were identified (Table 3); each occurred on 1,178 occasions in the 99 isolates. The first HRP3 motif was AHHAANA. It differed from the most abundant HRP2 motif by the substitution of an aspartic acid for an asparagine. It was present at significantly low frequency compared to the HRP2 motif (11.7 versus 18.4 per isolate; P < 0.0001). Increasing the motif length to 11 (AHHAANAHHAN) resulted in only a small reduction (from 1,178 to 1,175) in occurrence in the 99 isolates (Table 3).
Table 3.
Most frequent HRP3 motifs and regional variability in 99 sequences
| Motif length | Motif | Total frequency (mean) | Geographic variability |
|||
|---|---|---|---|---|---|---|
| Highest frequency |
Lowest frequency |
|||||
| Region (n) | Mean (range) | Region (n) | Mean (range) | |||
| 7 | AHHAANA | 1,162 (11.7) | South America (13) | 14.3(10–17) | Asian Pacific (20) | 9.1 (3–13) |
| 8 | AHHAANAH | 1,162 (11.7) | South America (13) | 14.3 (10–17) | Asian Pacific (20) | 9.1 (3–13) |
| 9 | AHHAANAHH | 1,162 (11.7) | South America (13) | 14.3 (10–17) | Asian Pacific (20) | 9.1 (3–13) |
| 10 | AHHAANAHHA | 1,162 (11.7) | South America (13) | 14.3 (10–17) | Asian Pacific (20) | 9.1 (3–13) |
| 11 | AHHAANAHHAA | 1,159 (11.7) | South America (13) | 14.3 (10–17) | Asian Pacific (20) | 9.1 (3–13) |
| 12 | AHHAANAHHAAN | 1,052 (10.6) | South America (13) | 13.3 (9–16) | Asian Pacific (20) | 7.9 (1–12) |
| 7 | HHAANAH | 1,162 (11.7) | South America (13) | 14.3 (10–17) | Asian Pacific (20) | 9.1 (3–13) |
| 8 | HHAANAHH | 1,162 (11.7) | South America (13) | 14.3 (10–17) | Asian Pacific (20) | 9.1 (3–13) |
| 9 | HHAANAHHA | 1,162 (11.7) | South America (13) | 14.3 (10–17) | Asian Pacific (20) | 9.1 (3–13) |
| 10 | HHAANAHHAA | 1,159 (11.7) | South America (13) | 14.3 (10–17) | Asian Pacific (20) | 9.1 (3–13) |
| 11 | HHAANAHHAAN | 1,052 (10.6) | South America (13) | 13.3 (9–16) | Asian Pacific (20) | 7.9 (1–12) |
| 12 | HHAANAHHAANA | 1,052 (10.6) | South America (13) | 13.3 (9–16) | Asian Pacific (20) | 7.9 (1–12) |
The second HRP3 motif (HHAANAH), again a single amino acid shift to the right from the first motif, could be lengthened by up to 3 amino acid residues without significant reduction in occurrence. Both motifs were most prevalent in isolates from South America and were least prevalent in isolates from Asian Pacific regions (Table 3).
Investigation of geographic distribution of HRP motifs.
To investigate the geographic variability in motif abundance, the mean frequencies of selected motifs were compared between geographic regions. For the HRP2 7-amino-acid motif of AHHAADA, significant differences in frequency per parasite were detected (P < 0.001), with the Caribbean isolates having a frequency significantly higher than those observed for isolates from Southeast (SE) Asia, the Asian Pacific, and Africa (P < 0.05) (Table 2; Fig. 2). These same differences occurred for the motifs of AHHAADAH, AHHAADAHH, and AHHAADAHHA. Geographic differences were also detected in the 11- and 12-aa motifs of AHHAHHAADAH and AHHAHHAADAHH (P < 0.001). For both motifs, isolates from SE Asia had significantly more copies than isolates from either the Asian Pacific or South America (P < 0.05) (Fig. 2). Considering the 7-aa HRP3 motif of AHHAANA, the geographic distribution fell into two significantly different groups; South America and Africa had significantly higher frequencies per isolate than SE Asia and the Asian Pacific (P < 0.05) (Fig. 3). These differences were maintained as the motif was extended to 12 aa (Table 3).
Fig 2.
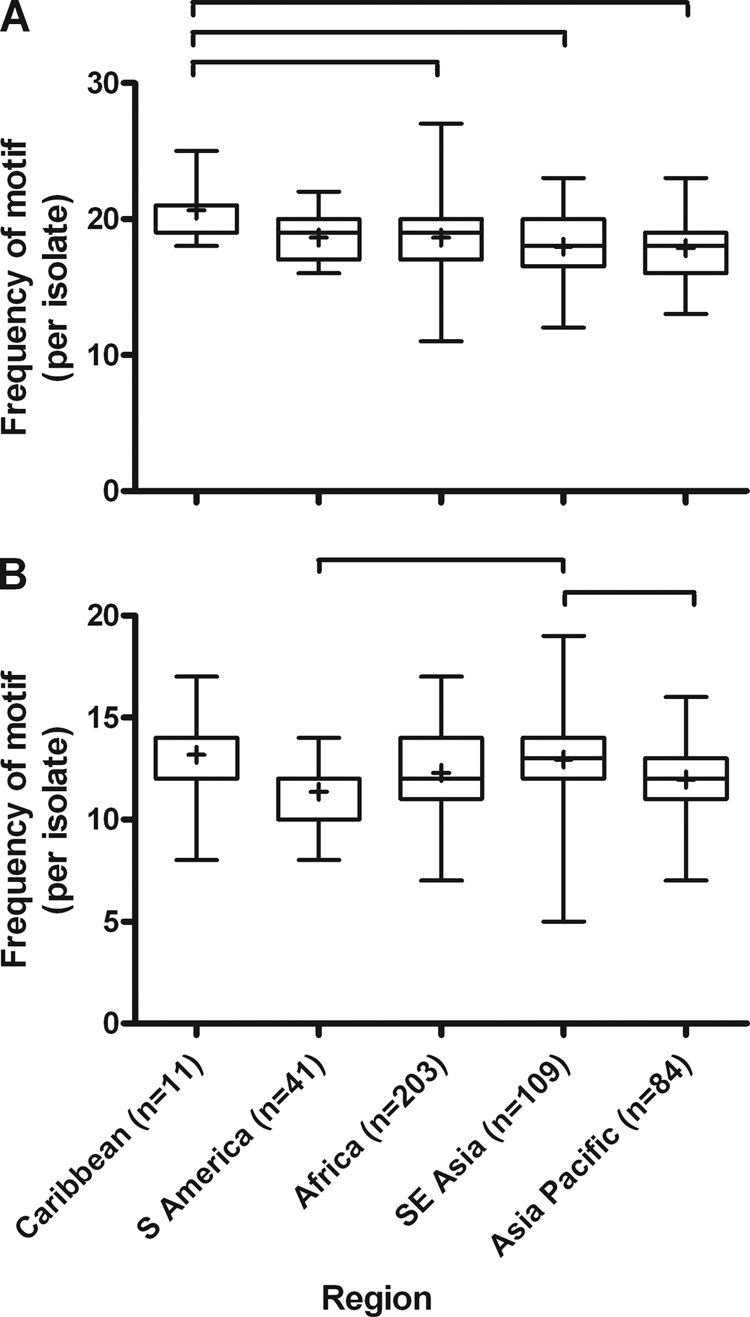
Geographic variation in frequency of HRP2 motifs AHHAADA (A) and AHHAHHAADAH (B). Whiskers on box plot indicate minimum and maximum values. Bracketing illustrates significant differences between regions (P < 0.05).
Fig 3.
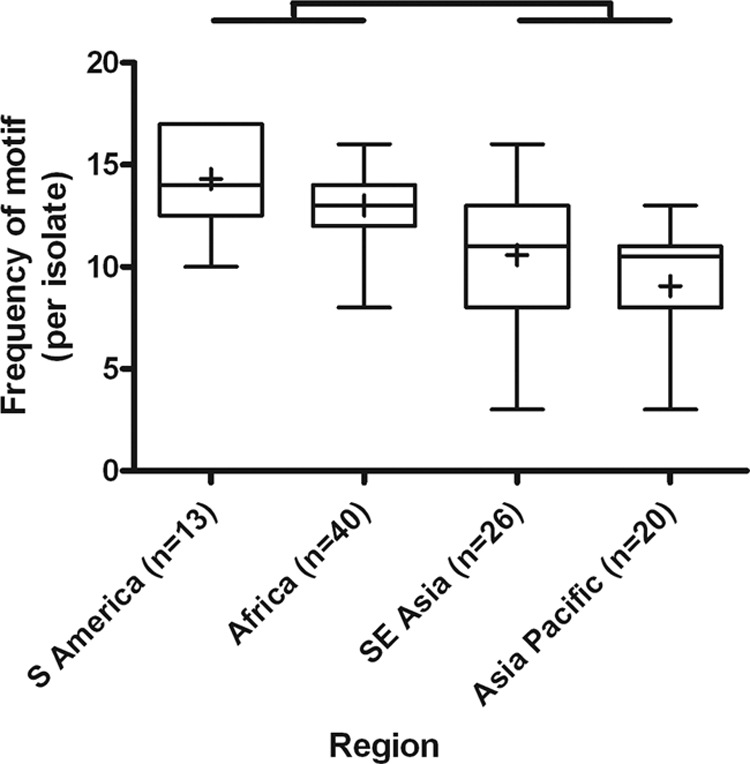
Geographic variation in frequency of HRP3 motif AHHAANA. Whiskers on box plot indicate minimum and maximum values. Bracketing illustrates significant differences between regions (P < 0.05).
Motif length 7.
Four 7-aa motifs (HHAADAH, AHHAAD, AADAHHA, HAADAHH) were found to be predominant in all regions and subregional groups, with each having a total occurrence of over 8,000 in the 448 sequences. These 4 motifs had the highest average frequency in the analysis. In isolates from the Pacific, Cambodia, Colombia, PNG, and Southeast Asia, an additional motif (AHHAHHA) was also predominant. Isolates from China had 2 more predominant motifs (DAHHAHH and HHAHHAA).
Motif length 10.
Only one 10-aa motif (AHHAADAHHA) was present in the predominant subset for all region groups. This motif had a total occurrence of more than 8,000.
Motif length 12.
Two predominant 12-aa motifs were observed in all regional and subregional groups (AHHAHHAADAHH, HHAHHAADAHHA). In Colombia and PNG, an additional three motifs (HHAADAHHAHHA, AHHAADAHHAHH, DAHHAHHAADAH) occurred in their group.
Geographic variation in HRP2 protein length.
The average length of the HRP2 is 253 amino acids (Table 4). Isolates from East Timor had the longest average length of 263 amino acids. Colombian isolates had the shortest average HRP2 at 243 amino acids. Out of the 5 major regional groups, the Asian Pacific group had the longest average HRP2 length at 256 amino acids, while the Caribbean group, with 246 amino acids, had the shortest.
Heat stability of MAbs.
Ten of the 12 MAbs (Genway and TC-10 were unavailable) were blotted onto the nitrocellulose membrane and tested for stability at varying temperatures. The control blot at day 0 (Fig. 4) shows that all 10 MAbs were reactive to labeled HRP2. Following incubation at 60°C for 5 days, all MAbs lost their reactivity. After incubation at 37°C for 30 days, MAbs 2G12-1C12, 4A5, C1-13, and PTL-3 maintained the same level of activity. MAbs 3A4, 1E1, and N7 had lost about 50% of their activity, with only around 3 of the 5 dots still visible. MAbs A6-4 and C2-3 had lost more than 50% of their activity with only faint dots still visible, while MAb S2-5 had lost all reactivity (Fig. 4; Table 1). A similar result was also seen when the membranes were stored at 45°C (Fig. 4). At day 90 for 37°C, MAbs PTL-3 and C1-13 still showed reactivity and ∼60% of the dilution dots were still clearly visible (data not shown). All other MAbs had lost reactivity. For 45°C at day 90, all MAbs had lost their reactivity (data not shown).
Fig 4.
Heat stability of MAbs. MAbs were blotted onto nitrocellulose membrane in 5 dilutions using a dot blotter at concentrations selected to give the same strength of signal on an X-ray film at baseline; membranes were then stored at 30°C, 45°C, and 60°C in a sealed bag for the designated times. Chemiluminescent detection was carried out by incubating membranes with biotinylated recombinant PfHRP2 and then with peroxidase-conjugated avidin.
DISCUSSION
The detection sensitivity, specificity, and useful life of P. falciparum-detecting RDTs depend on the binding affinity and avidity of MAbs to their target epitopes and their durability to heat. While a number of MAbs against HRP2 are commercially available, their epitopes and thermostability have not been systematically compared. In this study, we characterized the epitopes and thermostability of 8 MAbs.
All MAbs recognized native proteins extracted from seven isolates of different geographic origin. Four different dot blot patterns observed among the eight MAbs tested in this paper and three different patterns observed for four MAbs tested previously (8) suggest that the 12 available MAbs recognize different epitopes and that different isolates have different epitopes. This result was supported by epitope mapping that revealed that the major and minimum epitopes recognized by 11 of the 12 MAbs had different amino acid compositions and lengths. The prevalence of these epitopes in 448 isolates was highly variable. Epitopes for 5 of the 12 MAbs (3A4, 2G12, A6-4, C1-13, and PTL-3) were present in all 448 isolates analyzed. Epitopes of another five MAbs were present in 73 to 99% of isolates examined, while epitopes of MAb C2-3 and Genway were present only in less than 25% of the isolates. This suggests that the diagnostic sensitivity of an RDT using MAbs C2-3 and Genway may be lower. If an RDT uses the five MAbs that do not have epitopes in 100% of the isolates, it may also miss a small range of parasites. Among the 5 MAbs that had epitopes in 100% of the 448 isolates, the average frequency of the epitope varied between 5 and 18 per isolate.
It is hypothesized that a specific parasite antigen with more target epitopes would have a higher binding avidity to an antibody than an antigen with fewer epitopes. In such a case, the reactivity of each of the MAbs to a given isolate should correlate with the number of epitopes present in that isolate. This was true for some MAbs. For example, MAb C1-13, which targets an epitope with an average frequency of 18 per isolate, displayed a very high reactivity on dot blot analysis. Likewise, MAbs C2-3 and TC-10 had a very low frequency in all eight parasite isolates tested and had relatively weak dot blot results. This correlated well with our epitope mapping data and analysis. In contrast, however, were the results observed with other MAbs. S2-5 and PTL-3 displayed high reactivity despite having average epitope frequencies of 1 and 3, respectively. Likewise, MAbs A6-4 and N7, which had dot blot results similar to those of C1-13, had very low average epitope frequencies (5 and 10, respectively) compared to C1-13 (frequency of 18 per parasite). It is apparent in these cases that epitope frequency did not correlate with the strength of antibody binding to antigens as judged by dot blot analysis.
Several factors other than epitope frequency could influence the binding of antibodies. First, a consequence of the highly repetitive nature of the HRP2 sequences is that they contain many partial epitopes that are one or several amino acids shorter than the major or minimum epitope. Although we observed that peptides containing these partial epitopes reacted less strongly with the MAbs than peptides containing the optimal and complete epitopes, reactivity was still significant and therefore would likely contribute to the binding of the MAbs to HRP2.
Second, many epitopes allow single amino acid substitutions that have relatively little effect on the binding affinity of the MAbs but may increase the binding avidity of the MAbs because of an increasing number of epitopes. For instance, a single amino acid substitution in the optimal epitope of MAb 1E1-A9 (V to A) results in an increase in the average frequency of target epitopes from 2 to 14 per isolate. A single amino acid substitution in MAb PTL-3 from YAHHAHHA to DAHHAHHA results in the average number of target epitopes increasing from 3 to 12 per isolate. The same applies to MAb S2-5, where substituting the amino acid S with an A results in the average frequency increasing from 1 to 18 per isolate.
Finally, the conformation of the epitopes could also affect the antibody binding. In this epitope frequency study, we analyzed the frequency of epitopes in the protein sequences based on its linear form. However, the dot blot analysis used the native form of HRP2, which may contain both linear and conformational epitopes. Some linear epitopes may be hidden inside the molecule or blocked by adjacent overlapping epitopes and not available as antibody binding sites.
While the mapping and analysis of epitopes for currently available MAbs enable ranking of MAbs for use in RDTs, the study also revealed that most MAbs do not recognize “the ideal motif,” and there may be ways to improve the MAbs by generating new MAbs targeting this. Using a purpose-designed computer script to analyze 448 HRP2 and 99 HRP3 sequences, two ideal motifs were identified in HRP2: a 10-amino-acid motif of AHHAADAHHA and a 9-amino-acid motif of HHAADAHHA. Although the shorter versions of these motifs (7, 8, and 9 amino acids) have a motif frequency count higher than or identical to those of the longer motifs, these were not selected because it is possible that antibodies generated against shorter peptides may have reduced specificity, causing false-positive results. It is important to recognize that these motifs could partially overlap with each another during the calculation. However, in biological conditions, each antibody can bind only to a single target. Thus, the number of times each motif is present in a sequence does not necessarily represent the number of antibodies that could bind to the cognate epitope in the HRP2 protein but rather is an indication of the binding opportunities.
HRP3 has a sequence homology of more than 75% in the tandem repeat region to HRP2 and is recognized by most anti-HRP2 MAbs. Normally, this cross-reactivity may not have a major impact on the sensitivity of the RDTs because of the lower abundance of HRP3 (1). However, in situations of low-level parasitemia or when parasites have pfhrp2 deletions (4), it could enhance the sensitivity of the RDTs. A MAb specifically targeting HRP3 may also be used in combination with MAbs against HRP2 to enhance test sensitivity. We identified an 11-amino-acid motif of AHHAANAHHAA and a 10-amino-acid motif of HHAANAHHAA as optimal targets for HRP3-targetting MAbs. It is of note that the most prevalent motif of HRP3 differs by only a single amino acid residue from the most prevalent HRP2 motif (D to N).
How well epitopes of available MAbs match the ideal motifs can be used as a criterion to assess the suitability of currently available MAbs and whether there is a need to generate new MAbs. MAbs N7 and TC-10 recognize epitopes of 11 and 15 amino acids which contain the ideal 10-aa motif. MAb C2-3 recognizes an epitope of 13 aa, containing the second ideal 11-aa motif as well as the most predominant 9-aa motif. MAb 2G12-1C12 also recognizes an epitope that contains the most prevalent 9-aa motif. MAb C1-13 and S2-5 recognized epitopes very similar to one of the ideal motifs. MAb S2-5 recognizes the predominant 10-aa predicted motif with a single amino acid substitution of a Y, while MAb C1-13 recognizes the predominant 9-aa predicted motif. Both MAbs displayed a strong sensitivity in dot blots against crude parasite extracts. Unfortunately, S2-5 has an unsatisfactory durability to heat and is unlikely to be suitable for use in RDTs. Therefore, as for epitope analysis, the comparison of epitope with ideal motifs also identified C1-13 as the most suitable MAb for use in RDTs. However, although MAb C1-13 recognizes an epitope similar to the ideal motif, none of the MAbs recognized the full length of the most predominant 10-aa motif identified. It would be interesting to explore whether a MAb generated against the full-length ideal motif has better overall sensitivity and specificity.
It is reassuring to note that statistical analysis for geographic distribution in epitope and motif abundance does not indicate that this consideration is a significant factor in test performance. Therefore, it would be expected that MAbs recognizing optimal epitopes would recognize parasites equally well across all regions.
Durability to heat is critical to the usefulness of RDTs in the tropical field setting. In principle, heat stability is determined by the thermostability of the MAbs used in the RDT. When we compared 10 of the 12 MAbs for their heat durability at 37°C for 90 days, 45°C for 30 days, and 60°C for 5 days, three MAbs retained reactivity at 37°C for 30 days, but only two (C1-13 and PTL-3) retained some reactivity at day 90.
When the prevalence and frequency of epitopes were considered together with the heat durability profile, one currently available MAb showed the best potential for use in an RDT: C1-13. This was followed by 2G12 and PTL-3. Although the epitope of PTL-3 appeared in only 97.9% of isolates tested, its partial epitopes may be present in the remaining isolates or it could be used in combination with other MAbs.
It should be noted that there are many factors other than the epitopes that may influence the binding ability of the HRP2 MAbs used in a diagnostic test. The native conformation of HRP2 is yet unknown, and to what extent the structure would affect the binding of the MAbs has yet to be investigated. The effect of single amino acid substitutions or changes in the length of the epitopes on the binding affinity of the HRP2 MAbs to native protein in patient samples is also unclear.
Supplementary Material
ACKNOWLEDGMENTS
N.L. was supported by an NHMRC training scholarship. J.S.M. was supported by an NHMRC Practitioner Fellowship and a Government of Queensland Clinical Research Fellowship. M.L.G. was supported by an NHMRC Career Development Award. Funding for this project was provided in part by FIND.
We thank Allan McRae and Peter Visscher from the Queensland Statistical Genetics Laboratory, Queensland Institute of Medical Research, for assistance in writing the computer script to identify motifs.
Footnotes
Published ahead of print 18 January 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Baker J, Gatton ML, Peters J, Ho M-F, McCarthy JS. 2011. Transcription and expression of Plasmodium falciparum histidine-rich proteins in different stages and strains: implications for rapid diagnostic tests. PLoS One 6:e22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baker J, et al. 2010. Global sequence variation in the histidine-rich proteins 2 and 3 of Plasmodium falciparum: implications for the performance of malaria rapid diagnostic tests. Malar. J. 9:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baker J, et al. 2005. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. J. Infect. Dis. 192:870–877 [DOI] [PubMed] [Google Scholar]
- 4. Gamboa D, et al. 2010. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS One 5:e8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hayward RE, Sullivan DJ, Day KP. 2000. Plasmodium falciparum: histidine-rich protein II is expressed during gametocyte development. Exp. Parasitol. 96:139–146 [DOI] [PubMed] [Google Scholar]
- 6. Howard RJ, et al. 1986. Secretion of a malarial histidine-rich protein (Pf HRP II) from Plasmodium falciparum-infected erythrocytes. J. Cell Biol. 103:1269–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lambros C, Vanderberg JP. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65:418–420 [PubMed] [Google Scholar]
- 8. Lee N, et al. 2006. Effect of sequence variation in Plasmodium falciparum histidine-rich protein 2 on binding of specific monoclonal antibodies: implications for rapid diagnostic tests for malaria. J. Clin. Microbiol. 44:2773–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Panton LJ, et al. 1989. Purification and partial characterization of an unusual protein of Plasmodium falciparum: histidine-rich protein II. Mol. Biochem. Parasitol. 35:149–160 [DOI] [PubMed] [Google Scholar]
- 10. Rock EP, et al. 1987. Comparative analysis of the Plasmodium falciparum histidine-rich proteins HRP-I, HRP-II and HRP-III in malaria parasites of diverse origin. Parasitology 95:209–227 [DOI] [PubMed] [Google Scholar]
- 11. Trager W, Jensen JB. 1976. Human malaria parasites in continuous culture. Science 193:673–675 [DOI] [PubMed] [Google Scholar]
- 12. Wellems TE, Howard RJ. 1986. Homologous genes encode two distinct histidine-rich proteins in a cloned isolate of Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 83:6065–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.