LETTER
Haemophilus haemolyticus is generally considered a human commensal and is found in the nasopharynx of a minority of individuals as well as in subgingival plaque (7). Most authors consider H. haemolyticus to be either nonpathogenic (2, 4, 7, 18) or a rare pathogen (1). The latter assessment appears to be based on two reports of endocarditis from the early part of the 20th century (3, 13); however, in neither case were requirements for the X and V factors reported, and thus, no species designation can be reliably made.
The only method of differentiation between Haemophilus influenzae and H. haemolyticus in clinical microbiology laboratories is based on the ability of H. haemolyticus to produce a zone of clear hemolysis on ovine, bovine, or equine blood agar plates (1, 6, 7, 9). However, the hemolytic activity may be lost on subculture (7), and it was recently shown that many isolates of H. haemolyticus are nonhemolytic even on primary isolation (11, 14).
Invasive H. influenzae disease is a reportable condition in Oklahoma, and all H. influenzae isolates from normally sterile sites are required to be submitted to the Oklahoma State Department of Health (OSDH) Public Health Laboratory (PHL) for confirmation and serotyping. For use in our ongoing studies of H. influenzae iron/heme acquisition, we obtained from the OSDH PHL all invasive H. influenzae isolates from 2003 and 2004. Seven of these isolates were subjected to multilocus sequence typing (MLST) as described by Meats et al. (12). We were consistently unable to amplify the gene fragment for the fucK gene from one strain, designated HI2028 and isolated from the blood of a 1-year-old male. Failure to amplify the fucK gene fragment from presumptive H. influenzae isolates has been considered an indicator of a misidentified strain (15). However, failure to detect the fucK gene cannot be considered conclusive since some strains of H. influenzae have recently been shown to lack the fucose operon (17, 19).
Based on the failure to amplify the fucK gene product, we considered that strain HI2028 may have been misidentified as H. influenzae and thus we proceeded to amplify 1,491 bp of the 16S rRNA gene from HI2028 using the primers UFPL and URPL (10). PCR amplicons were directly sequenced and yielded 1,448 bp of the 16S rRNA gene. Homology searches against the nucleotide collection using the MEGABLAST algorithm (http://blast.ncbi.nlm.nih.gov) with the determined 1,448 bp of the 16S rRNA gene from HI2028 indicated that the strain was H. haemolyticus but cannot be considered a definitive identification of the isolate as such.
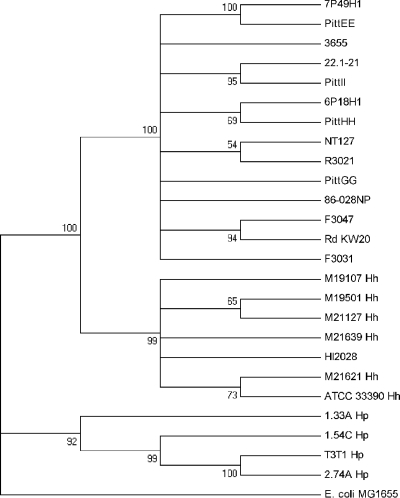
A multilocus sequence analysis system based on 5 gene loci (adk, pgi, recA, infB, and 16S rRNA) has been described to separate H. haemolyticus from H. influenzae (11, 18). The sequences of these genes from HI2028 were determined and concatenated as described by McCrea et al. (11). The partial gene sequences from the 200 Haemophilus strains in the original study (11) were retrieved and concatenated. A bootstrap consensus tree using 1,000 replicates was constructed, rooted to Escherichia coli, using the neighbor-joining method in MEGA 5.05 (20). The strain HI2028 clustered with strains designated H. haemolyticus (data not shown). Additionally, the 5 gene sequences were retrieved for 14 available H. influenzae genomic sequences, for 5 recently published H. haemolyticus genomic sequences (5), and for the H. haemolyticus type strain ATCC 33390. Figure 1 shows strain HI2028 clustering with the H. haemolyticus strains in a neighbor-joining tree generated as described above. Additional characteristics of HI2028 are shown in Table 1.
Fig 1.
Neighbor-joining dendrogram of Haemophilus strains. The tree, rooted to E. coli strain MG1655, is based on concatenated adk, pgi, recA, infB, and 16S rRNA gene sequences, with bootstrap values of greater than 50% of 1,000 bootstraps indicated. Strains are genome-sequenced H. influenzae strains unless indicated otherwise. Hh, H. haemolyticus strains, including the type strain ATCC 33390 and five recently sequenced isolates (5); Hp, H. parainfluenzae strains. HI2028 is the strain identified in this report.
Table 1.
Characteristics of strain HI2028 in comparison to H. haemolyticus, H. influenzae, and the Biotype IV cryptic genospecies of H. influenzae
| Characteristic | HI2028 | H. haemolyticus | H. influenzae | Biotype IV cryptic genospeciesa |
|---|---|---|---|---|
| X factor required | + | + | + | + |
| V factor required | + | + | + | + |
| H2s productionb | + | + | V | V |
| Indole productionb | + | Vc | V | − |
| Urease activityb | + | + | V | + |
| Orinthineb decarboxylase activity | − | − | V | + |
| Hemolysisd | − | V | − | − |
| Protein P6 amino acids at residues 33, 42, 59, 61, and 152e | G, S, N, E, A | G, S, N, E, A | A, A, D, T, A | G, S, N, E, S |
The cryptic Biotype IV genospecies causes urogenital and neonatal infections (16) and is closely related to H. haemolyticus (14).
Phenotype was determined as described by Kilian (6).
V, results are variable among strains (7).
Lack of hemolysis with strain HI2028 was determined on 5% defibrinated horse blood using BBL haemophilus identification quad plates (BD).
The amino acids at specific residues within the P6 protein have been used to differentiate H. influenzae and H. haemolyticus (14).
In summary, we have identified a strain of nonhemolytic H. haemolyticus, originally designated H. influenzae, from a patient with bacteremia. In reporting the H. haemolyticus genomic sequence, Jordan et al. stated that several presumptive H. influenzae isolates from invasive disease had been confirmed as H. haemolyticus, although no data were presented (5). Our finding supports the assertion of Jordan et al. and suggests that H. haemolyticus may occasionally be found among presumed H. influenzae clinical isolates and should be considered a more frequent cause of invasive disease than currently thought. Such misidentification of H. haemolyticus strains as H. influenzae may potentially lead to errors in national burden of invasive H. influenzae disease estimates as is the case with other clinical microbiology laboratory errors (8).
Nucleotide sequence accession numbers.
The partial gene sequences for adk, pgi, recA, infB, and the 16S rRNA from HI2028 have been deposited in GeneBank with the respective accession numbers JQ254883 to JQ254887. The GeneBank accession number for the gene encoding the P6 protein of HI2028 is JQ247079.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI29611 from the National Institute of Allergy and Infectious Disease. We gratefully acknowledge the support of the Children's Hospital Foundation.
We thank the Oklahoma State Department of Health Public Health Laboratory for furnishing strains.
Footnotes
Published ahead of print 1 February 2012
REFERENCES
- 1. Agrawal A, Murphy TF. 2011. Haemophilus influenzae infections in the H. influenzae type b conjugate vaccine era. J. Clin. Microbiol. 49:3728–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albritton WL. 1982. Infections due to Haemophilus species other than H. influenzae. Annu. Rev. Microbiol. 36:199–216 [DOI] [PubMed] [Google Scholar]
- 3. De Santo DA, White M. 1933. Hemophilus hemolyticus endocarditis. Am.J. Pathol. 9:381–392 [PMC free article] [PubMed] [Google Scholar]
- 4. Fink DL, St Geme JW. 2006. The genus Haemophilus, p 1034–1061 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes: a handbook on the biology of bacteria. Springer, New York, NY [Google Scholar]
- 5. Jordan IK, et al. 2011. Genome sequences for five strains of the emerging pathogen Haemophilus haemolyticus. J. Bacteriol. 193:5879–5880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kilian M. 1976. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J. Gen. Microbiol. 93:9–62 [DOI] [PubMed] [Google Scholar]
- 7. Kilian M. 2005. Genus III. Haemophilus Winslow, Broadhurst, Buchanan, Krumweide, Rogers and Smith 1917, 561, p 883–904 In Brenner DJ, Krieg NR, Staley JT. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2 Springer, New York, NY [Google Scholar]
- 8. LaClaire LL, et al. 2003. Identification of Haemophilus influenzae serotypes by standard slide agglutination serotyping and PCR-based capsule typing. J. Clin. Microbiol. 41:393–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ledeboer NA, Doern GV. 2012. Haemophilus. In Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW. (ed), Manual of clinical microbiology, 10th ed ASM Press, Washington, DC [Google Scholar]
- 10. LiPuma JJ, et al. 1999. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J. Clin. Microbiol. 37:3167–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McCrea KW, et al. 2008. Relationships of nontypeable Haemophilus influenzae strains to hemolytic and nonhemolytic Haemophilus haemolyticus strains. J. Clin. Microbiol. 46:406–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meats E, et al. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 41:1623–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miller CP, Branch A. 1923. Subacute bacterial endocarditis due to a hemolytic hemophilic bacillus. Arch. Intern. Med. 32:911–926 [Google Scholar]
- 14. Murphy TF, et al. 2007. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J. Infect. Dis. 195:81–89 [DOI] [PubMed] [Google Scholar]
- 15. Nørskov-Lauritsen N. 2009. Detection of cryptic genospecies misidentified as Haemophilus influenzae in routine clinical samples by assessment of marker genes fucK, hap, and sodC. J. Clin. Microbiol. 47:2590–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quentin R, Ruimy R, Rosenau A, Musser JM, Christen R. 1996. Genetic identification of cryptic genospecies of Haemophilus causing urogenital and neonatal infections by PCR using specific primers targeting genes coding for 16S rRNA. J. Clin. Microbiol. 34:1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ridderberg W, Fenger MG, Nørskov-Lauritsen N. 2010. Haemophilus influenzae may be untypable by the multilocus sequence typing scheme due to a complete deletion of the fucose operon. J. Med. Microbiol. 59:740–742 [DOI] [PubMed] [Google Scholar]
- 18. Sandstedt SA, et al. 2008. Comparison of laboratory-based and phylogenetic methods to distinguish between Haemophilus influenzae and H. haemolyticus. J. Microbiol. Methods 75:369–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shuel ML, Karlowsky KE, Law DK, Tsang RS. 2011. Nonencapsulated or nontypeable Haemophilus influenzae are more likely than their encapsulated or serotypeable counterparts to have mutations in their fucose operon. Can. J. Microbiol. 57:982–986 [DOI] [PubMed] [Google Scholar]
- 20. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]