Abstract
The automation of DNA extraction and the use of commercial quantitative real-time PCR assays could help obtain more reliable results for the quantification of Epstein-Barr virus DNA loads (EBV VL). This study compared two automated extraction platforms and two commercial PCRs for measurement of EBV VL in 10 EBV specimens from Quality Control for Molecular Diagnostics (QCMD) and in 200 whole-blood (WB) specimens from transplant (n = 137) and nontransplant (n = 63) patients. The WB specimens were extracted using the QIAcube or MagNA Pure instrument; VL were quantified with the EBV R-gene quantification kit (Argene) or the artus EBV RG PCR kit (Qiagen) on the Rotor-Gene 6000 real-time analyzer; and the results were compared with those of a laboratory-developed PCR. DNA was extracted from the QCMD specimens by use of the QIAamp DNA minikit and was quantified by the three PCR assays. The extraction platforms and the PCR assays showed good correlation (R, >0.9; P, <0.0001), but as many as 10% discordant results were observed, mostly for low viral loads (<3 log10 copies/ml), and standard deviations reached as high as 0.49 log10 copy/ml. In WB but not in QCMD samples, Argene PCR tended to give higher VL values than artus PCR or the laboratory-developed PCR (mean difference for the 200 WB VL, −0.42 or −0.36, respectively). In conclusion, the two automated extraction platforms and the two PCRs provided reliable and comparable VL results, but differences greater than 0.5 log10 copy/ml remained between the two commercial PCRs after common DNA extraction.
INTRODUCTION
Primary Epstein-Barr virus (EBV) infection is the cause of the vast majority of cases of infectious mononucleosis, and the subsequent lifelong persistence of EBV in the infected host, although mostly asymptomatic, can lead to the development of several lymphoid and epithelial cancers in immunosuppressed and immunocompetent individuals (13, 18).
With the outstanding development of real-time quantitative PCR, measurement of EBV DNA loads during these EBV-associated diseases has been largely implemented in clinical practice (6, 11). The monitoring of EBV DNA loads in blood is required for transplant recipients at risk of posttransplantation lymphoproliferative disorders, and EBV DNA loads could also be a surrogate marker for the adjustment of the immunosuppressive regimen in these patients (7, 9). EBV DNA loads measured in plasma also appear to be a useful biomarker for the management of EBV-associated undifferentiated nasopharyngeal carcinoma (4). Although less clearly demonstrated, measurement of EBV DNA loads could also be helpful in other clinical situations, such as severe or atypical infectious mononucleosis and other EBV-associated malignancies in immunosuppressed or immunocompetent patients (6). Besides the debates on the clinical utility and the clinically relevant EBV DNA levels in various EBV-associated diseases, technical standardization of EBV loads has not yet been achieved (6, 7). There is still great variability among the different PCR methods at all steps of the analysis, resulting in great variability in interlaboratory results (1, 8, 16). This heterogeneity is a barrier to the optimal use of quantitative PCR assays. The automation of nucleic acid extraction procedures and the use of commercialized PCR assays could decrease this heterogeneity and improve the agreement and the clinical utility of EBV load measurements in routine settings (5, 15, 17).
The aims of this study were (i) to compare two automated platforms for the extraction of EBV DNA from whole-blood samples, the MagNA Pure LC system (Roche Applied Science, Meylan, France) (referred to below as MagNA) and the QIAcube instrument (Qiagen, Hilden, Germany) (referred to below as QIAcube), and (ii) to compare the results for EBV DNA loads in whole-blood samples, after DNA extraction with the QIAcube, obtained with two commercially available real-time PCR assays: the EBV R-gene quantification kit (provided free by Argene, Verniolle, France) (referred to below as the Argene PCR) and the artus EBV RG PCR kit (provided free by Qiagen, Hilden, Germany) (referred to below as the Artus PCR). The results obtained with the two commercially available PCR assays were also compared with those from a laboratory-developed EBV real-time quantitative PCR assay (Lab PCR). Additionally, the three PCR assays were tested with the 2009 proficiency panel from Quality Control for Molecular Diagnostics (QCMD).
MATERIALS AND METHODS
Clinical samples.
One hundred twenty-five patients sent to our institution for routine EBV load testing (86 transplant recipients with routine monitoring of EBV loads and 39 nontransplant patients with suspicion of an EBV-associated disease) were included in the study and gave 200 specimens of whole blood collected in EDTA tubes (Table 1). Eighty of these 200 samples were collected sequentially from eight patients (five transplant recipients, one patient with HIV infection, one patient with hypogammaglobulinemia, and one patient with EBV-associated encephalitis). The range of EBV loads assessed by the laboratory-developed PCR (see below) is presented in Table 1. All samples were collected between September and November 2008 and were stored at −80°C until use.
Table 1.
Whole-blood sample distribution
| Patient group | Range of viral loads (log10 copies/ml) | No. of whole-blood specimensa |
|---|---|---|
| Transplant recipients | ||
| Hematopoietic stem cells (n = 19) | 39 | |
| 0–3 | 15 | |
| 3–4 | 16 | |
| 4–5 | 7 | |
| >5 | 1 | |
| Solid organ (n = 67) | 98 | |
| 0–3 | 37 | |
| 3–4 | 20 | |
| 4–5 | 38 | |
| >5 | 3 | |
| Nontransplant patients | ||
| EBV primary infection (n = 2) | 4–5 | 2 |
| Other EBV-associated diseases (n = 37) | 61 | |
| 0–3 | 13 | |
| 3–4 | 9 | |
| 4–5 | 27 | |
| >5 | 12 |
The total for each patient group is given in boldface.
Study design.
First, all whole-blood specimens were extracted with the MagNA system or the QIAcube instrument and were amplified using the laboratory-developed PCR assay on the LightCycler platform, version 2.0, in order to compare the performances of the two extraction robots. DNA was isolated according to the manufacturer's instructions from 200 μl of whole blood with the DNA Isolation kit (Roche Applied Science, Meylan, France) for the MagNA instrument and with the QIAamp DNA Blood minikit (Qiagen, Hilden, Germany) for the QIAcube robot. The extracted DNA was eluted with 100 μl of elution buffer, aliquoted, and frozen at −80°C before use.
Second, the performances of the three EBV PCR assays were compared using aliquoted DNA extracts obtained with the QIAcube. The laboratory-developed PCR and the Argene PCR assays, described elsewhere (3, 5), targeted the BXLF1 thymidine kinase gene and were run on a LightCycler platform (version 2.0) and the Rotor-Gene 6000 platform, respectively. The Artus PCR targeted the EBNA1 gene and was run on the Rotor-Gene 6000 platform. All three PCR assays were multiplex PCRs for the simultaneous amplification of an internal control used to verify the efficacy of the extraction and the absence of PCR inhibitors in the amplification process. The characteristics of each PCR are summarized in Table 2.
Table 2.
Characteristics of real-time PCRs
| Characteristic | Argene PCR | Artus PCR | Laboratory-developed PCR |
|---|---|---|---|
| Target | BXLF1 | EBNA1 | BXLF1 |
| Probe technology | Hydrolysis | Hydrolysis | Hybridization |
| PCR cycle steps | 2 (95°C, 60°C) | 3 (95°C, 55°C, 72°C) | 3 (95°C, 58°C, 72°C) |
| Final vol (μl) in assay | 25 | 50 | 20 |
| Vol (μl) of extracted DNA | 10 | 20 | 10 |
| No., type of quantification standards | 4, plasmid | 4, plasmid | 5, Namalwa cells (2 EBV copies/cell) |
| Range of standard curve (copies/ml of whole blood) | 5,000–5,000,000 | 25,000–25,000,000 | 500–5,000,000 |
| Simultaneous amplification internal control | Yes | Yes | Yes |
In addition to the whole-blood samples, 10 lyophilized samples from the QCMD 2009 EBV proficiency panel were extracted manually (without the QIAcube robot) using the QIAamp DNA Blood minikit (Qiagen) and were quantified by the three PCR assays, as described above. The QCMD samples contain a lyophilized EBV strain quantified using electron microscopy. All QCMD samples were reconstituted using sterile water (nucleic acid amplification testing [NAT] quality).
Statistical analysis.
The EBV load measurements were expressed as log10 copies per milliliter. The correlation coefficients were calculated using a Spearman test, and the homogeneity of the variances was analyzed by the Fisher-Snedecor test (StatView, version 5.0; SAS Institute Inc., Cary, NC). The comparison of the viral loads obtained by the different technologies was represented on a Bland-Altman graph. Only viral loads positive by both assays compared were represented on the Bland-Altman graphs. Results could be defined as discordant either because the results of the Bland-Altman analysis fell outside the interval of the average ± 1.96 standard deviations (SD) (quantitatively discordant results) or because one EBV load measurement was positive by one method and negative by another (qualitatively discordant results).
RESULTS
Comparison of automated extractions.
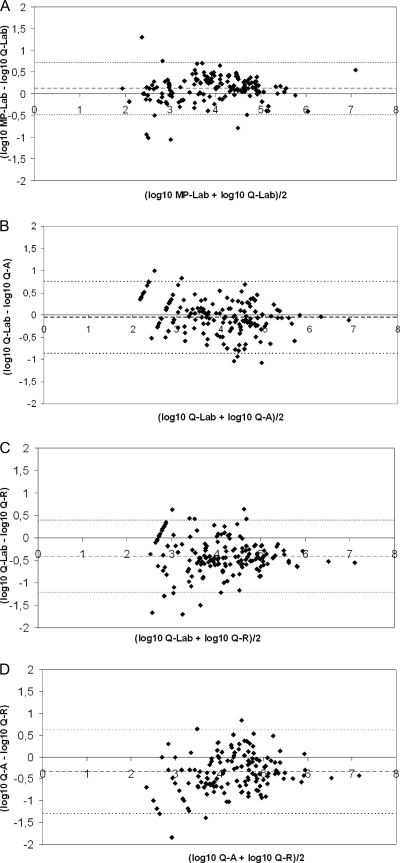
The analysis of EBV loads obtained after the two extraction procedures showed a linear correlation between the log10 EBV load and a Spearman correlation coefficient of 0.958 (P, <0.0001). Among the 200 samples, 159 were positive by both automated extraction technologies, 21 were negative by both, and 20 were qualitatively discordant. Five samples with low EBV loads (<3 log10 copies/ml) by QIAcube–Lab PCR were negative by MagNA–Lab PCR, and 15 samples positive by MagNA–Lab PCR (all but one with viral DNA loads [VL] of <3 log10 copies/ml) were negative by QIAcube–Lab PCR. Bland-Altman analysis of the 159 samples positive by both technologies showed that more than 95% of the samples were within the “mean ± 1.96 SD” (Fig. 1A). The mean difference between the EBV loads (log10 copies/ml) obtained by the two methods was 0.11, and the standard deviation was 0.31 (Table 3). The differences between viral loads were above 0.5 log10 for 7.6% of the samples and above 1 log10 for 1.9% of the samples.
Fig 1.
Bland-Altman analysis of EBV DNA loads positive by both technologies. (A) Comparison of extraction robots. MagNA the plus the laboratory-developed PCR on a LightCycler instrument (MP-Lab) was compared with QIAcube plus the laboratory-developed PCR on a LightCycler instrument (Q-Lab). A total of 159 samples were used. (B) Comparison of Q-Lab with Q-A (QIAcube plus the artus EBV RG PCR kit on a Rotor-Gene instrument). A total of 154 samples were used. (C) Comparison of Q-Lab with Q-R (QIAcube plus the Argene EBV R-gene quantification kit on a Rotor-Gene instrument). A total of 156 samples were used. (D) Comparison of Q-A and Q-R. A total of 151 samples were used. The bold line represents the mean differences; the thin lines represent the mean ± 1.96 standard deviation.
Table 3.
Mean differences (SD) between EBV DNA loads measured by the different methods
| Comparison of methodsa | Mean (SD) difference between EBV DNA loads (log10 copies/ml)b |
|---|---|
| MP-Lab PCR vs Q-Lab PCR | 0.11 (0.31) |
| Q-Lab PCR vs Q-Artus PCR | −0.06 (0.42)* |
| Q-Lab PCR vs Q-Argene PCR | −0.4 (0.41)* |
| Q-Artus PCR vs Q-Argene PCR | −0.34 (0.49)* |
MP, MagNA Pure extraction; Q, QIAcube extraction; Lab, laboratory-developed real-time PCR (on a LightCycler amplification platform); Artus PCR, Qiagen artus EBV RG PCR kit (on a Rotor-Gene amplification platform); Argene PCR, Argene EBV R-gene kit (on a Rotor-Gene amplification platform).
Asterisks indicate SD statistically different from those for MP-Lab PCR versus Q-Lab PCR (P <0.001 by the Fisher-Snedecor test).
No PCR inhibition occurred during these experiments.
Comparison of real-time PCRs after QIAcube extraction.
The Spearman correlation coefficients between the log10 EBV loads obtained by the three different PCRs ranged from 0.908 to 0.942 (P < 0.0001).
Eleven qualitatively discordant results were obtained between the laboratory-developed PCR and the Artus PCR. One sample was positive by the Artus PCR and negative by the laboratory-developed PCR, and 10 samples were negative by the Artus PCR and positive by the laboratory-developed PCR. Bland-Altman analysis of the 154 samples positive by both assays showed that 3.9% of the results were outside the “mean ± 1.96 SD” interval (Fig. 1B,). The most discordant results were measured at 5.69 log10 copies/ml and 3.42 log10 copies/ml by the Artus PCR and the laboratory-developed PCR, respectively. These results obtained from patient H are discussed below. The mean difference between EBV loads (log10 copies/ml) measured by Lab PCR and Artus PCR was −0.06, and the standard deviation was 0.42 (Table 3). The differences were above 0.5 log10 for 18.9% and above 1 log10 for 1.95% of the 154 samples positive by the two methods.
Among the 17 qualitatively discordant results between the laboratory-developed PCR and the Argene PCR, eight were negative by the Argene PCR and positive by the laboratory-developed PCR (all were below 3 log10 copies/ml). Nine samples were negative by the laboratory-developed PCR and positive by the Argene PCR (two were below 3 log10 copies/ml, and seven were between 3 and 4 log10 copies/ml). Bland-Altman analysis showed that 7.1% of the 156 results positive by the two methods were outside the “mean ± 1.96 SD” interval (Fig. 1C). The VL obtained for patient H, by the laboratory-developed PCR and the Argene PCR, were not discordant. The mean difference observed between the Lab PCR and the Argene PCR was −0.4, and the standard deviation was 0.41 (Table 3).
There were 18 qualitatively discordant results between the Artus and Argene PCRs. Four were negative by the Argene PCR and positive by the Artus PCR (all were below 3 log10 copies/ml), and 14 were negative by the Artus PCR and positive by the Argene PCR (five were below 3 log10 copies/ml, and eight were quantified between 3 and 5 log10 copies/ml). On the Bland-Altman graph, 6 of the 151 samples positive by both assays (4%) were discordant (Fig. 1D). One of them corresponded to patient H. The mean difference between the two methods was −0.34, and the standard deviation was 0.49 (Table 3).
The viral load differences measured between the Argene PCR and the laboratory-developed PCR or the Artus PCR were above 0.5 log10 for more than 40% of the samples positive by the two methods (66/156 and 62/151, respectively). Fewer than 6% of the samples showed a difference above 1 log10. In more than 92% of the samples with a difference above 0.5 log10, Argene PCR gave a higher viral load.
Follow-up of EBV VL in patients with various EBV-associated pathologies.
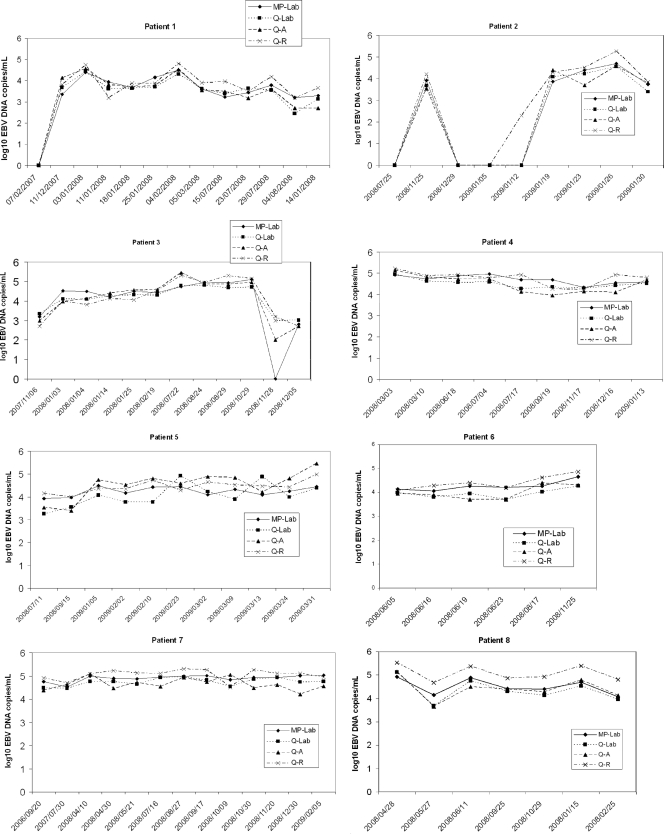
Figure 2 shows the longitudinal monitoring of EBV loads from eight patients (80 measurements, ranging from 6 to 13 samples/patient) by four methods: MagNA–laboratory-developed PCR, QIAcube–laboratory-developed PCR, QIAcube–Artus PCR, and QIAcube–Argene PCR. When the two extraction platforms were compared, the differences between viral loads were below 0.5 log for 86.3% of the samples and below 1 log for 96.3% of the samples. The differences between viral loads measured by the laboratory-developed and Artus PCRs were below 0.5 log for 90% of the samples and below 1 log for 98.8% of the samples. The viral load differences measured between the Argene PCR and the laboratory-developed or Artus PCR were below 0.5 log for 61.3% and 63.8% of the samples, respectively; 96.3% of samples showed a difference below 1 log when the Argene and laboratory-developed PCRs or the Argene and Artus PCRs were compared. Overall, EBV DNA load measurements were higher by the Argene PCR than by the laboratory-developed or Artus PCR.
Fig 2.
Follow-up, using four different EBV DNA load technologies, for eight patients with various pathologies. Patient 1 was a hematopoietic stem cell transplantation recipient with non-Hodgkin lymphoma; patient 2, a hematopoietic stem cell transplantation recipient (donor EBV serological status, positive; recipient serological status, negative [D+ R−]); patients 3, 4, and 5, kidney transplant recipients (D+ R−). Patient 6 had HIV; patient 7, hypogammaglobulinemia with primary EBV infection; patient 8, EBV encephalitis. MP, MagNA Pure extraction; Q, QIAcube extraction; Lab, laboratory-developed real-time PCR (on a LightCycler amplification platform); A, Qiagen artus EBV RG PCR kit (on a Rotor-Gene amplification platform); R, Argene EBV R-gene kit (on a Rotor-Gene amplification platform).
Results of the QCMD 2009 EBV proficiency panel.
Table 4 depicts the EBV load results for the 10 samples from the QCMD 2009 EBV proficiency panel after manual extraction (without the QIAcube or MagNA robot) and amplification by the three PCR assays (Lab, Artus, and Argene). The sample for which a negative result was expected was found negative by the three PCR assays. All nine positive results obtained by the laboratory-developed PCR were higher than the expected results, but the difference remained below 0.5 log10 copies/ml (mean Δlog10, +0.175). With the Argene PCR, eight of nine results were lower than the expected results (mean Δlog10 for the nine positive samples, −0.180). The maximum differences between the Argene PCR results and the expected results were −0.502 and +0.564. With the Artus PCR, seven of nine results were lower than the expected result (mean Δlog10 for the nine positive samples, −0.425) and four of them presented differences greater than 0.5 log10. The Spearman correlation coefficients for the laboratory-developed PCR, the Artus PCR, and the Argene PCR relative to the expected results were 0.999, 0.951, and 0.915, respectively.
Table 4.
Quantification of EBV DNA loads of QCMD 2009 proficiency panel specimens
| QCMD 2009 specimen no. | VL result (log10 copies/ml)a |
Difference from expected VL result (Δlog10 copies/ml) |
|||||
|---|---|---|---|---|---|---|---|
| Expected | Lab PCR | Artus PCR | Argene PCR | Lab PCR | Artus PCR | Argene PCR | |
| 1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 2 | 2.425 | 2.512 | 2.000 | 2.989 | 0.087 | −0.425 | 0.564 |
| 3 | 2.719 | 2.845 | 2.000 | 2.544 | 0.126 | −0.719 | −0.175 |
| 4 | 3.919 | 4.145 | 3.653 | 3.803 | 0.226 | −0.266 | −0.116 |
| 5 | 4.218 | 4.390 | 3.309 | 3.922 | 0.172 | −0.909 | −0.296 |
| 6 | 4.431 | 4.512 | 4.525 | 4.290 | 0.081 | 0.094 | −0.141 |
| 7 | 5.199 | 5.279 | 4.813 | 4.942 | 0.080 | −0.386 | −0.257 |
| 8 | 2.733 | 3.051 | 2.000 | 2.398 | 0.318 | −0.733 | −0.335 |
| 9 | 3.412 | 3.641 | 3.544 | 3.051 | 0.229 | 0.132 | −0.361 |
| 10 | 3.315 | 3.577 | 2.699 | 2.813 | 0.262 | −0.616 | −0.502 |
Lab PCR, laboratory-developed PCR (on a LightCycler amplification platform); Artus PCR, Qiagen artus EBV RG PCR kit (on a Rotor-Gene amplification platform); Argene PCR, Argene EBV R-gene kit (on a Rotor-Gene amplification platform). The Spearman correlation coefficients with regard to the Expected value (P ≤ 0.006) were 0.999 for the Lab PCR, 0.951 for the Artus PCR, and 0.915 for the Argene PCR.
DISCUSSION
The automation of nucleic acid extraction and the availability of commercial real-time quantitative PCR assays could improve the agreement and the clinical utility of EBV DNA load measurement in routine clinical settings. This study compared the EBV load results obtained by two automated extraction methods coupled to the same PCR assay and the EBV load results obtained by three EBV quantitative real-time PCR assays after the same extraction.
Few data have been published on the performance of automated extraction for EBV DNA quantification on whole blood (14), and to our knowledge, this is the first study of QIAcube extraction for EBV DNA load measurement. The comparison of the MagNA and QIAcube automated extraction platforms showed an excellent correlation of the EBV load results in 200 whole-blood specimens (R = 0.958; P < 0.0001), with fewer than 8% of VL differing by more than 0.5 log10, and with a low standard deviation of the differences (0.31). No PCR inhibition precluded the measurement of EBV load, since none of the samples inhibited the internal positive control in any of the PCR assays. All the qualitatively discordant VL (10%) concerned low EBV loads (below 3 log10 copies/ml): 15 of 20 were negative after QIAcube extraction and weakly positive after MagNA extraction, and 5 of 20 were negative after MagNA extraction and weakly positive after QIAcube extraction. This suggested a trend toward greater sensitivity of MagNA extraction than of QIAcube extraction, particularly for low viral loads. The two automated extraction platforms were equivalent regarding time spent and standardization. The QIAcube is not totally automated but offers the advantage of being more versatile, since almost all the manual extraction kits available from Qiagen can be adapted for the QIAcube with comparable results. No cross-contamination was observed with either the MagNA or the QIAcube, but it should be noted that the MagNA instrument includes a UV DNA decontamination process not included in the QIAcube.
The three PCR methods (Artus PCR, Argene PCR, and Lab PCR) were compared after a common extraction on the QIAcube platform. Overall, for the 200 blood specimens, the correlation between the different PCR assays was good (R > 0.9; P < 0.0001), with <10% quantitatively or qualitatively discordant results, but the standard deviation of the differences reached 0.49. In this study, the best correlation was observed for the laboratory-developed PCR and the Artus PCR, even though they were carried out with different amplification platforms (LightCycler versus Rotor-Gene) and even though they target distinct genes (thymidine kinase versus EBNA1).
The discordant results were observed mostly for EBV loads below 3 log10 copies/ml, as has been reported previously (1, 2, 5, 12, 16). In the transplantation setting, these discrepancies among low EBV loads in whole blood are most often irrelevant for the diagnosis of posttransplantation lymphoproliferative disorders (16). Nevertheless, greater sensitivity could become important if evidence emerges that low or medium EBV VL are related to other pathologies, such as acute rejection or graft dysfunction, or are useful for subtle immunosuppression monitoring (7). Although not explored in this study, the sensitivity of the quantitative PCR in matrices other than whole blood, such as serum or cerebrospinal fluid, could be important for the diagnosis of nasopharyngeal carcinoma, infectious mononucleosis, and EBV neurological disorders (6).
Despite the good correlation of EBV VL values measured using the three PCR methods, some major discrepancies were observed in this study. In one case (patient H), with VL above 3 log10 copies/ml, the quantification differed repeatedly by more than 2 log10 copies/ml between the Artus (5.69 log10 copies/ml) and Argene (3.45 log10 copies/ml) PCRs, despite the use of the same extraction process and the same PCR platform. Mutations involving the primers or the probe annealing site could account for this difference, since the Argene PCR and the laboratory-developed PCR target the EBV thymidine kinase gene, and the Artus PCR targets the EBNA1 gene, but the thymidine kinase gene was sequenced, and no mutations were observed (results not shown) (17). Targeting of genes present in multiple repeated copies may also cause this type of discrepancy, but to our knowledge, the thymidine kinase and EBNA1 genes are single-copy genes (15). This considerable difference between two commercial assays in a single laboratory has already been observed by others (2, 15, 17), suggesting that replacement of one method with another should be undertaken with caution. It is hoped that the ongoing implementation of a WHO EBV international standard for nucleic acid amplification-based assays will improve the standardization of EBV load measurement, making the clinical interpretation of EBV DNA loads less challenging.
The second type of discrepancy highlighted by this study was a trend for the Argene PCR to give higher EBV VL than the Artus PCR and the laboratory-developed PCR in whole blood (mean differences for the 200 WB VL, −0.42 and −0.36, respectively). Surprisingly, the QCMD panel quantified by the Argene PCR gave values below the expected results and below the results obtained by the laboratory-developed PCR, suggesting that EBV VL quantification depends on the matrix considered. This difference between the results for cell-free samples and cell-associated clinical samples has already been described (1, 5) and suggests that evaluation of a new commercial EBV DNA quantitative assay relies both on quality control and on clinical sample studies.
The main advantages of the commercial kit are that it provides standards and reagents produced using good manufacturing practices and is easy to develop for nonspecialized laboratories that cannot develop their own method. Some studies suggested that commercial assays and standardized reagents could help improve the comparability of assays between laboratories (2, 8, 10), but others observed no significant difference in interlaboratory quantitative precision when commercial reagents/assays rather than laboratory-developed assays were used (16).
In conclusion, these results demonstrated that the automated extraction systems, MagNA and QIAcube, and the two commercially available EBV real-time quantitative PCR assays, Argene and Artus, gave comparable results for EBV DNA measurement in whole blood. However, in this study, the two automated extraction techniques with the same PCR gave a higher correlation (P < 0.05) and a lower standard deviation (P, <0.001 by the Fisher-Snedecor test) than different PCR assays with the same extracted material. This work also underscored that 40% of VL measurements performed with two commercial assays in the same laboratory differed by more than 0.5 log10 copies/ml. This highlights the importance of using an international standard and reinforces the assumption that EBV VL should be monitored by the same assays and with the same specimen type, and should be interpreted for a given patient with regard to the clinical history, the risk factors for an EBV-associated disease, and the viral dynamics of the EBV DNA load.
ACKNOWLEDGMENTS
The authors declare no conflict of interest.
We thank José Labarere for his help in statistical analysis.
Footnotes
Published ahead of print 11 January 2012
REFERENCES
- 1. Abbate I, et al. 2011. Multicenter comparative study of Epstein-Barr virus DNA quantification for virological monitoring in transplanted patients. J. Clin. Virol. 50:224–229 [DOI] [PubMed] [Google Scholar]
- 2. Ahsanuddin AN, Standish MC, Caliendo AM, Hill CE, Nolte FS. 2008. Validation of an Epstein-Barr viral load assay using the QIAGEN Artus EBV TM PCR analyte-specific reagent. Am. J. Clin. Pathol. 130:865–869 [DOI] [PubMed] [Google Scholar]
- 3. Brengel-Pesce K, et al. 2002. Routine use of real-time quantitative PCR for laboratory diagnosis of Epstein-Barr virus infections. J. Med. Virol. 66:360–369 [DOI] [PubMed] [Google Scholar]
- 4. De Paoli P, Pratesi C, Bortolin MT. 2007. The Epstein Barr virus DNA levels as a tumor marker in EBV-associated cancers. J. Cancer Res. Clin. Oncol. 133:809–815 [DOI] [PubMed] [Google Scholar]
- 5. Fafi-Kremer S, et al. 2008. Evaluation of the Epstein-Barr virus R-gene quantification kit in whole blood with different extraction methods and PCR platforms. J. Mol. Diagn. 10:78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gärtner B, Preiksaitis JK. 2010. EBV viral load detection in clinical virology. J. Clin. Virol. 48:82–90 [DOI] [PubMed] [Google Scholar]
- 7. Gulley ML, Tang W. 2010. Using Epstein-Barr viral load assays to diagnose, monitor, and prevent posttransplant lymphoproliferative disorder. Clin. Microbiol. Rev. 23:350–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hayden RT, et al. 2008. Multicenter comparison of different real-time PCR assays for quantitative detection of Epstein-Barr virus. J. Clin. Microbiol. 46:157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heslop HE. 2009. How I treat EBV lymphoproliferation. Blood 114:4002–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ito Y, et al. 2010. Multicenter evaluation of prototype real-time PCR assays for Epstein-Barr virus and cytomegalovirus DNA in whole blood samples from transplant recipients. Microbiol. Immunol. 54:516–522 [DOI] [PubMed] [Google Scholar]
- 11. Kimura H, Ito Y, Suzuki R, Nishiyama Y. 2008. Measuring Epstein-Barr virus (EBV) load: the significance and application for each EBV-associated disease. Rev. Med. Virol. 18:305–319 [DOI] [PubMed] [Google Scholar]
- 12. Laus S, Kingsley LA, Green M, Wadowsky RM. 2011. Comparison of QIAsymphony automated and QIAamp manual DNA extraction systems for measuring Epstein-Barr virus DNA load in whole blood using real-time PCR. J. Mol. Diagn. 13:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luzuriaga K, Sullivan JL. 2010. Infectious mononucleosis. N. Engl. J. Med. 362:1993–2000 [DOI] [PubMed] [Google Scholar]
- 14. Mengelle C, et al. 2008. Comparison of two highly automated DNA extraction systems for quantifying Epstein-Barr virus in whole blood. J. Clin. Virol. 43:272–276 [DOI] [PubMed] [Google Scholar]
- 15. Perandin F, Cariani E, Pollara CP, Manca N. 2007. Comparison of commercial and in-house real-time PCR assays for quantification of Epstein-Barr virus (EBV) DNA in plasma. BMC Microbiol. 7:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Preiksaitis JK, et al. 2009. Interlaboratory comparison of Epstein-Barr virus viral load assays. Am. J. Transplant. 9:269–279 [DOI] [PubMed] [Google Scholar]
- 17. Ruiz G, Peña P, de Ory F, Echevarría JE. 2005. Comparison of commercial real-time PCR assays for quantification of Epstein-Barr virus DNA. J. Clin. Microbiol. 43:2053–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757–768 [DOI] [PubMed] [Google Scholar]