Abstract
Mitochondrial DNA sequences offer major advantages over the more usual nuclear targets for loop-mediated isothermal amplification approaches (mito-LAMP) because multiple copies occur in every cell. Four LAMP primers [F3, FIP(F1c+F2), BIP(B1c+B2), and B3] were designed based on the mitochondrial nad1 sequence of Opisthorchis viverrini and used for a highly specific assay (mito-OvLAMP) to distinguish DNA of O. viverrini from that of another opisthorchiid (Clonorchis sinensis) and other trematodes (Haplorchis pumilio, Haplorchis taichui, Fasciola hepatica, and Fasciola gigantica). Conventional PCR was applied using F3/B3 primer pairs to verify the specificity of the primers for O. viverrini DNA templates. All LAMP-positive samples could be detected with the naked eye in sunlight, by gel electrophoresis (stained with ethidium bromide), and by addition of SYBR green I to the product in sunlight or under UV light. Only DNA from O. viverrini yielded amplification products by LAMP (and by PCR verification), and the LAMP limit of detection was as little as 100 fg (10−4 ng DNA), indicating that this assay is 10 to 100 times more sensitive than PCR. Field testing was done using representative egg and metacercarial samples collected from localities where the fluke is endemic. With the advantages of simplicity, rapidity, sensitivity, and cost effectiveness, mito-OvLAMP is a good tool for molecular detection and epidemiology studies in regions or countries where O. viverrini is endemic, which can lead to more effective control of opisthorchiasis and trematodiasis.
INTRODUCTION
Opisthorchiasis caused by the fish-borne zoonotic liver fluke Opisthorchis viverrini Poirier, 1886 (Southeast Asian liver fluke), is a major disease in Thailand, Laos, Cambodia, and southern Vietnam (7, 13, 23, 24, 26, 27). This species can induce cholangitis and cholangiocarcinoma, the latter being the most severe consequence of chronic opisthorchiasis (22, 24, 26). The consumption of raw or improperly cooked freshwater fish containing infective metacercariae of O. viverrini remains a traditional habit in Vietnam, Thailand, and Laos, and it can lead to opisthorchiasis and its potentially fatal sequelae (5, 21, 24). Typically, infections by O. viverrini go unnoticed until severe symptoms and/or pathological lesions of the hepatic and biliary system are detected in patients admitted to medical stations and hospitals (3, 25, 26).
Recently, a simple, cost-effective, easily performed technique, known as loop-mediated isothermal amplification (LAMP) (15, 17), was developed that permits the rapid and highly specific amplification of a trace amount of DNA under isothermal conditions (normally at 63°C). The LAMP reaction is highly specific due to its use of four LAMP primers [namely, F3, FIP(F1c+F2), BIP(B1c+B2), and B3] binding to six independent regions in the target sequence (15, 19). Successful LAMP depends on the appropriate selection of the target gene, the optimal design of turn-back primers (10), and the generation of visible turbidity derived from the production of magnesium pyrophosphate (14). More importantly, the LAMP reaction is performed in inexpensive equipment such as a regular water bath or a heating block, and its products are easily observed with the naked eye or visualized by the addition of fluorescent dyes such as SYBR green I, with or without using a UV lamp (15). LAMP has been successfully developed for rapid detection of a range of zoonotic platyhelminths, such as Fasciola hepatica and Fasciola gigantica (1), Clonorchis sinensis (4), Schistosoma japonicum (29), Taenia spp. (16), Paragonimus westermani (6), and, recently, O. viverrini (2). For these LAMP reactions, nuclear genes were mainly chosen as the target regions.
Greater sensitivity will be possible if the target nucleotide sequence is present in high copy numbers and is highly specific and widely conserved within a particular pathogen species or group (10, 15, 19). For this reason, mitochondrial sequences have major advantages over the more usual nuclear targets. The animal mitochondrial genome is a double-stranded DNA circle (normally 13.5 to 20 kb) containing 36 or 37 genes (2 rRNA, 22 tRNA, and 12 or 13 protein-coding genes) (8, 11). Each cell contains many mitochondria, providing many copies of any mitochondrial DNA (mtDNA) target region. Furthermore, the circular mitochondrial DNA molecules are relatively resistant to degradation. Complete or near-complete mtDNA genomes for various zoonotic parasitic pathogens of public health concern are becoming available in the GOBASE database (http://gobase.bcm.umontreal.ca/) (18), including many helminths obtained by us and our collaborators (8, 11). Additionally, updated information is available from our own growing collecting of mitochondrial sequences from O. viverrini (T. H. Le et al., unpublished data). Using these resources, we have developed a range of mtDNA-based LAMP assays (i.e., mito-LAMP) for detection of common helminthic zoonoses of public medical and veterinary importance. The mito-LAMP strategy could also be developed for detection of agents of other neglected tropical diseases. Here, we chose a mitochondrial protein-coding gene, nad1, as a LAMP target for O. viverrini. This gene encodes one of the subunits of nicotinamide dehydrogenase. Many nad1 sequences are available from O. viverrini strains of different geographic origins and from other species of trematode, including some closely related to O. viverrini. This makes it possible to design primers for O. viverrini-specific mito-LAMP (mito-OvLAMP).
In this paper, we present the development of a mito-LAMP strategy for O. viverrini targeting nad1 and of experimental assays for evaluation of sensitivity and specificity in order to provide a method for rapid detection of any life stage of this zoonotic trematode in any country where the organism is endemic.
MATERIALS AND METHODS
Parasite samples.
Samples of O. viverrini from Vietnam were collected from humans in the central part of the country (Binh Dinh, Phu Yen, Dac Lak, Quang Nam, and Thua Thien-Hue provinces) (7, 13), and adults and eggs of Thai origin (Khon Kaen province) were provided by Paiboon Sithithaworn of Khon Kaen University. Other samples used as controls for specificity were collected by us, including a related small liver fluke (Clonorchis sinensis), large liver flukes (Fasciola hepatica and F. gigantica), a giant intestinal fluke (Fasciolopsis buski), and minute intestinal flukes (Haplorchis pumilio and H. taichui) (Table 1).
Table 1.
List of samples and organisms providing nad1 sequences for designing primers and DNA template used in the assessment of the specificity and sensitivity of LAMP
| Species | Sample | Accession no.a | Origin | Purpose |
|
|---|---|---|---|---|---|
| Primer design | Specificity/sensitivity | ||||
| O. viverrini | OvBD1 | EU443831 | Binh Dinh, Vietnam | ✓ | ✓ |
| OvBD | DQ882172 | Binh Dinh, Vietnam | ✓ | ✓ | |
| OvDL3 | DQ882174 | Dac Lak, Vietnam | ✓ | ✓ | |
| OvPY3 | EU443833 | Phu Yen, Vietnam | ✓ | ||
| OvQN | EU443832 | Quang Nam, Vietnam | ✓ | ||
| OvHU | Hue City, Vietnam | ✓ | |||
| OvL | DQ882175 | Vientiane, Laos | ✓ | ✓ | |
| OvKK | Khon Kaen, Thailand | ✓ | ✓ | ||
| OvKO | Khon Kaen, Thailand | ✓ | ✓ | ||
| OvMH | Thailand | ✓ | |||
| OvVT1 | GQ401025 | Vientiane, Laos | ✓ | ||
| OvCP1 | GQ401064 | Champasak, Laos | ✓ | ||
| OvKM7 | GQ401046 | Khammouane, Laos | ✓ | ||
| OvSV12 | GQ401060 | Savannakhet, Laos | ✓ | ||
| OvKD1 | GQ401082 | Kandal, Cambodia | ✓ | ||
| OvSK1 | GQ401096 | Sakaeo, Thailand | ✓ | ||
| OvKS | EU022343 | Kalasin, Thailand | ✓ | ||
| OvLP | EU022346 | Lampang, Thailand | ✓ | ||
| OvSK | EU022348 | Sakon Nakhon, Thailand | ✓ | ||
| OvMS | EU022350 | Mahasarakham, Thailand | ✓ | ||
| C. sinensis | CsAmur-RU | FJ381664 | Amur River, Russia | ✓ | |
| O. felineus | OfUT-RU | EU921260 | Novosibirsk, Russia | ✓ | |
| F. gigantica | FgIN-IN | Indonesia | ✓ | ||
| FgNB-VN | EU260072 | Ninh Binh, Vietnam | ✓ | ✓ | |
| F. hepatica | FhAU-AU | AF216697 | Geelong, Australia | ✓ | ✓ |
| C. sinensis | CsND-VN | Nam Dinh, Vietnam | ✓ | ✓ | |
| F. buski | FbN-VN | Nghe An, Vietnam | ✓ | ✓ | |
| H. pumilio | HpuD2-VN | Nghe An, Vietnam | ✓ | ||
| H. taichui | HtaTL-TH | Bangkok, Thailand | ✓ | ||
Samples with no accession number were obtained in this study.
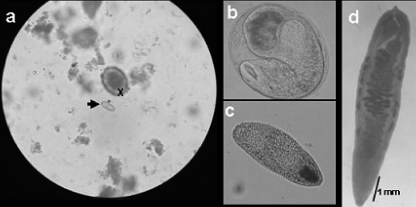
Stool specimens were collected from men over 45 years old who lived in an area where O. viverrini is endemic (a village in Phu Yen province of central Vietnam). Stools from individual donors were screened for parasite eggs using the Kato-Katz method (9); the eggs were collected for microscopic examination and identification (Fig. 1), and stool samples were subjected to DNA extraction.
Fig 1.
Photomicrographs of samples collected from fish and people living in the village where O. viverrini is endemic used in this study. (a) Egg of O. viverrini (arrow) and an egg of Ascaris lumbricoides (X) (magnification, ×400); (b and c) metacercarial cyst and a metacercaria from fish (magnification, ×100); (d) unstained adult O. viverrini worm from a patient (magnification, ×40).
Metacercariae and/or metacercarial cysts of any trematode species present were pooled from muscle samples of freshwater fish (Carassius auratus gibelio), a silver carp weighing up to 30 or 40 g and commonly eaten by habitual consumers of raw fish. These were sampled randomly from ponds. A single fish in this area commonly contains metacercariae of a number of trematode species. Entire fish or large portions of fish muscle were digested using 2% pepsin at pH 2. The digested material was passed through a 1-mm-mesh sieve and allowed to settle until the sediment was clearly visible. Several rounds of washing with physiological saline (i.e., 0.85% [wt/vol] NaCl), settling, and decanting the supernatant followed until metacercarial cysts could be seen in the sediment. All samples (adult worms, pooled trematode eggs from stools, and metacercariae and metacercarial cysts from fish) were stored in 70% ethanol and kept at −20°C prior to DNA extraction.
Genomic DNA preparation.
Genomic DNA was extracted from individual O. viverrini metacercariae and metacercarial cysts and from adult O. viverrini and worms of other species using a commercial QIAamp DNA extraction kit (Qiagen Inc.) as described previously (13). Additionally, for stools known to contain trematode eggs, an AccuPrep stool DNA extraction kit (Bioneer, Daejeon, South Korea) was used. Briefly, after the addition of 400 μl SL buffer to 100 mg stool, the tube was vortexed for 30 s and then incubated at 60°C for 10 min. Following centrifugation at 12,000 rpm for 5 min, the supernatant was transferred to a new tube, and 400 μl binding buffer added to this tube, which was incubated again at 60°C for 10 min. After the addition of 100 μl isopropanol, the tube was vortexed for 5 s and briefly centrifuged. The supernatant liquid was transferred into the binding column fitted onto a collection tube, and the combination was then centrifuged at 8,000 rpm for 1 min. The column was fitted to a new tube, 500 μl washing buffer 1 was added, and the tube was centrifuged again at 8,000 rpm for 1 min. This step was repeated using washing buffer 2, followed by centrifugation at 13,000 rpm for 1 min. Finally, the column was fitted onto a collection tube, 100 μl elution buffer was added, and after standing at room temperature for 5 min, the combination was centrifuged at 8,000 rpm for 1 min. The eluted genomic DNA from stool samples was stored at −20°C until use.
The identity of each trematode DNA sample was confirmed by species-specific PCR as previously described for C. sinensis (13), for heterophyids (28), and for Fasciola spp. (12). DNA concentration was estimated using a GBC UV/visible 911A spectrophotometer (GBC Scientific Equipment Pty. Ltd., Australia). Each sample was diluted to 100 ng/μl, and 1 μl was used as the template in a normal LAMP or PCR in a 25-μl volume.
Design of LAMP primers for O. viverrini.
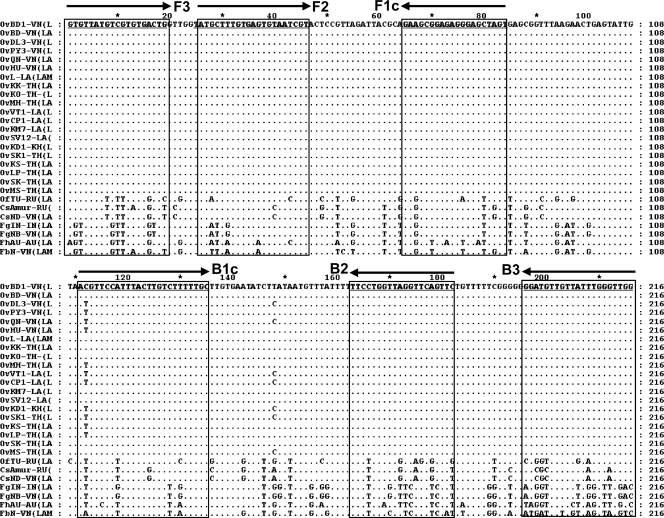
LAMP primers specific for O. viverrini were designed following inspection of the multiple alignment of 27 nad1 sequences, including 20 of O. viverrini (Table 1). Four LAMP primers, F3, FIP(F1c+F2), BIP(B1c+B2), and B3 (Table 2), were designed based on regions conserved in O. viverrini but differing in other species of trematode (Fig. 2). PrimerExplorer V4 software (http://primerexplorer.jp/elamp4.0.0/index.html) was used to determine the primer sequences. The F1c and F2 sequences of the forward inner primer (FIP) as well as the B1c and B2 sequences of the reverse inner primer (BIP) are joined by TTTT spacers.
Table 2.
Sequence of primers for LAMP reaction
| Primera | Sequence (5′–3′) | Length (bp) |
|---|---|---|
| F3 | GTGTTATGTCGTGTGACTG | 19 |
| B3 | CCAACCCAAATAACAACATCC | 21 |
| FIP(F1c+F2) | ACTAGCTCCCTCTCCGCTTCTTTTTATGCTTTGTGAGTGTAATCGT | 46 |
| BIP(B1c+B2) | ACGTTCCATTTACTTGTCTTTTTGCTTTTGAACTGAACCTAACCAGGAA | 49 |
F3, forward outer primer; FIP, forward inner primer (comprising F1c and F2 sequences with a TTTT link [italics] added between them); BIP, reverse inner primer (comprising B1c and B2 sequences, with the TTTT link); and B3, reverse outer primer.
Fig 2.
Comparative alignment of 27 nad1 sequences from O. viverrini and related trematodes for demonstration of variability between species in regions chosen for the primers for mito-OvLAMP (see Table 1 for a list of samples and strains from which nad1 sequences were obtained). The top sequence is the corresponding OvBD1 nad1 sequence (Vietnam); dots in the sequences below indicate identity. The horizontal arrows indicate the directions (sense and antisense) of the LAMP primers (underlined), including F3 (forward outer primer), F2 and F1c (forward inner primers); B1c and B2 (reverse inner primers), and B3 (reverse outer primer). Regions used for OvLAMP primers are highly conserved among O. viverrini samples but differ in other trematodes.
PCR assays using outer primers F3 and B3 to confirm specificity and sensitivity.
Conventional PCR using the external primers F3 and B3 was performed on each DNA template of O. viverrini and on a range of other trematodes (listed in Table 1), to test specificity of the primers. An amplicon of 216 bp was expected for O. viverrini. PCR was conducted in a 25-μl reaction volume containing 12.5 μl PCR master mix (Fermentas Inc.), 2 μl (5 pmol/μl) each primers F3 and B3, 1 μl DNA template (100 ng/μl), and 7.5 μl water. A negative (no DNA) control was included. The reaction conditions were an initial step at 94°C for 3 min, then 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 5 min.
The sensitivity of the PCR was also assayed to establish the detection limit of O. viverrini DNA. A serial dilution of genomic DNA of a Vietnamese sample (OvBD1) was used. The quantities of template used were 10, 1, 10−1, 10−2, 10−3, 10−4, and 10−5 ng DNA. The assay was performed with 1 μl of the diluted template in each case. A negative (no DNA) control was included. The PCR products (10 μl of each) were examined on a 1% agarose gel, stained with ethidium bromide, and visualized under UV light (Wealtec).
LAMP performance and assays.
The LAMP reaction was performed in a total volume of 25 μl containing 12.5 μl of 2× LAMP reaction buffer [40 mM Tris-HCl (pH 8.8), 20 mM KCl, 16 mM MgSO4, 20 mM (NH4)2SO4, 0.2% Tween 20, 1.6 M betaine, 2.8 mM each deoxynucleoside triphosphate], 2.5 μl 10× Bsm DNA polymerase buffer, 1 μl of primer mix (20 pmol FIP, 20 pmol BIP, 2.5 pmol F3 primers, and 2.5 pmol B3 primers), 1 μl of target genomic DNA, and 7 μl of diethyl pyrocarbonate (DEPC)-treated water. First, the mixture was heated at 95°C for 3 min and then chilled on ice. Next, 1 μl (8 units) of the Bsm DNA polymerase large fragment (Fermentas Inc.) was added. The mixture was incubated at 63°C for 70 min in a water bath and then heated to 80°C for 5 min to terminate the reaction. The LAMP products were inspected to determine positivity by the naked eye (i) directly in sunlight, (ii) after the addition of SYBR green I (Invitrogen Inc.) in sunlight, (iii) under UV light after the addition of SYBR green I, and (iv) by electrophoresis in 1.5% agarose gel with ethidium bromide staining under UV light (Wealtec).
The specificity of LAMP was examined using 1 μl of genomic DNA (100 ng/μl) in each case, from six samples of O. viverrini (Vietnam, Laos, Thailand origin); one sample of C. sinensis (Vietnam); three fasciolids, F. hepatica (Australia), F. gigantica (Vietnam), and Fasciolopsis buski (Vietnam); and two heterophyids, H. pumilio (Vietnam) and H. taichui (Thailand) (Table 1). A negative control (no DNA) was included.
The sensitivity of the mito-OvLAMP was assayed with the 10-fold dilution series of DNA as described above (i.e., 10, 1, 10−1, 10−2, 10−3, 10−4, and 10−5 ng). The assay was performed with 1 μl of template in each case. A negative control (no DNA) was included. The LAMP products were subjected to electrophoresis on a 1.5% agarose gel stained with ethidium bromide, followed by visualization after addition of SYBR green I by the naked eye in sunlight and under UV in a Dolphin apparatus (Wealtec).
RESULTS
LAMP and PCR performance for confirmation of the O. viverrini template.
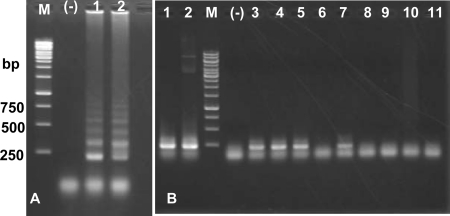
To confirm the identity of DNA from O. viverrini, two DNA templates (OvBD1, from Binh Dinh, Vietnam, and OvKK, from Khon Kaen, Thailand) were used for a LAMP and a PCR experiment. The temperature for LAMP was set to 63°C for 70 min (1 to 2°C lower or higher than 63°C was tested with less positive results [data not shown]). Each LAMP reaction yielded a typical ladder-like pattern on an agarose gel stained with ethidium bromide, while the negative control did not (Fig. 3A). When SYBR green I was added to the products, either under UV light or in sunlight, clearly positive results were seen with the naked eye. Both OvBD1 and OvKK and other O. viverrini templates generated the expected 216-bp PCR product, while samples from other trematodes yielded no amplicons (Fig. 3B).
Fig 3.
Confirmation of the identity of O. viverrini genomic DNA (OvBD1, Vietnam; OvKK, Thailand) by loop-mediated isothermal amplification (LAMP) and PCR. A total of 100 ng of template was used in each case. (A) Products of mito-OvLAMP on 1.5% agarose stained with ethidium bromide. Lanes: M, 1-kb ladder marker (numbers on the left are sizes in base pairs); (-), negative control (no DNA); 1 and 2, LAMP products from templates of OvBD1 and OvKK, respectively. (B) Electrophoresis of PCR products of 216 bp (using primers F3 and B3) on 1% agarose. Lanes: M, 1-kb ladder marker; (-), negative control (no DNA) for templates, see Table 1): 1, OvBD1 (O. viverrini); 2, OvKK (O. viverrini); 3, OvDL3 (O. viverrini); 4, OvPY3 (O. viverrini); 5, OvQN (O. viverrini); 6, CsND (Clonorchis sinensis); 7, OvL (O. viverrini); 8, FhAU (Fasciola hepatica); 9, FgNB (F. gigantica); 10, HpuD2 (Haplorchis pumilio); 11, HtaTL (H. taichui).
LAMP specificity for O. viverrini.
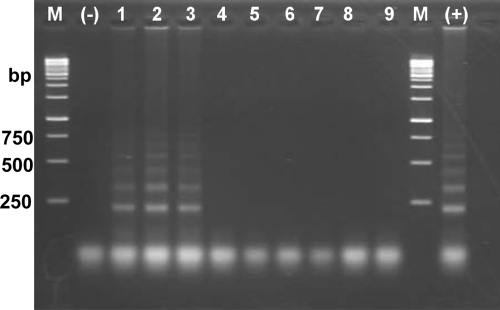
To determine the specificity of the mito-OvLAMP assay, genomic DNA samples from different life stages of O. viverrini and other trematodes, including C. sinensis (Vietnam isolate), F. hepatica (Australia), F. gigantica (Vietnam), Fasciolopsis buski (Vietnam), H. pumilio (Vietnam), and H. taichui (Thailand), were used. LAMP products were amplified only from DNA samples of O. viverrini eggs, adults, and metacercariae, and no amplification occurred with other trematode samples (Fig. 4).
Fig 4.
Specificity assessment of the mito-OvLAMP assay by visualization on 1.5% agarose gel stained with ethidium bromide. Lanes: M, 1-kb ladder marker (numbers on the left are sizes in base pairs); (-), negative control (no DNA); (+), positive control (100 ng DNA template of O. viverrini); 1 through 9, LAMP reaction with DNA template (100 ng) from O. viverrini eggs, O. viverrini adult worm, O. viverrini single metacercaria, C. sinensis (CsND), Fasciolopsis buski (FbN), F. hepatica (FhAU), F. gigantica (FgNB), H. pumilio (HpuD2), and H. taichui (HtaTL).
Analytical sensitivity of LAMP and PCR for O. viverrini.
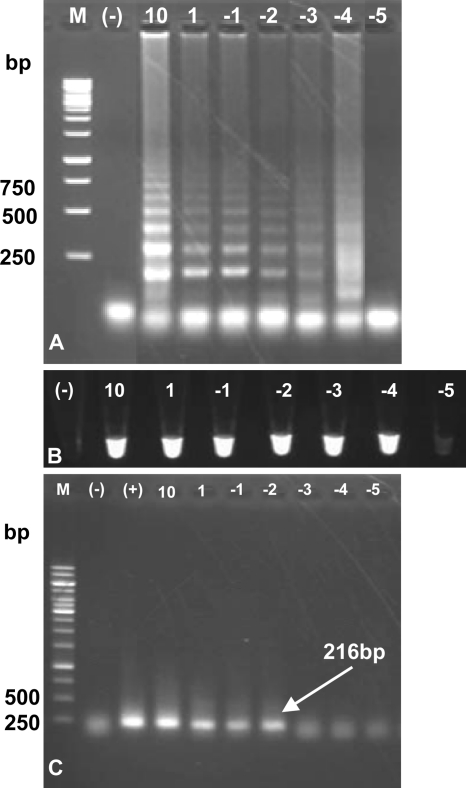
Tenfold serial dilutions of O. viverrini genomic DNA of a Vietnamese isolate (OvBD1) were used to compare the sensitivity between the mito-OvLAMP and the PCR tests. Results indicated that the detection limit for the LAMP reaction was between 10−3 ng (1 pg) and 10−4 ng (100 fg) of template (Fig. 5A and B), while it was 10−2 ng (10 pg) for the conventional PCR assay (Fig. 5C). This suggested that the LAMP is 100 times more sensitive than the PCR assay.
Fig 5.
Sensitivity assessment of the LAMP assay for O. viverrini using serial dilutions of genomic DNA template and comparison with PCR (performed using outer primers F3 and B3). (A and B) Sensitivity analysis of the LAMP assay by visualization on 1.5% agarose stained with ethidium bromide (A) or under UV light after the addition of SYBR green I (B). (C) Sensitivity analysis of the conventional PCR revealed on 1% agarose (arrow indicates a 216-bp DNA fragment at the highest template dilution for PCR performance). Lanes: M, 1-kb ladder marker (numbers on the left are sizes in base pairs); (-), no-DNA template (water added) as a negative control; (+), 100 ng template (positive control), 10, 10 ng; 1, 1 ng; −1: 100 pg; −2, 10 pg; −3, 1 pg; −4, 100 fg; −5, 10 fg.
LAMP test with field samples collected from fish and humans.
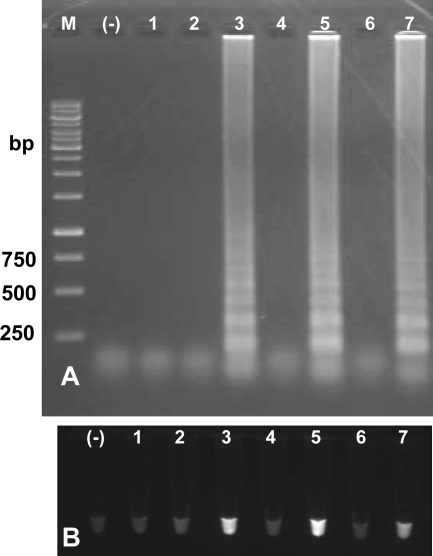
The specific LAMP assay was used in attempts to detect O. viverrini metacercariae and cysts from silver carp and to detect eggs from feces of humans in an area where O. viverrini is endemic. Most of the inhabitants (mainly males) frequently eat raw fish in the traditional style (7, 13). Three pools of mixed metacercariae from fish muscle samples and four samples of mixed eggs collected from human feces (Fig. 1) were used for evaluation of mito-OvLAMP. One of three metacercarial samples and two of four fecal egg samples produced a positive LAMP reaction; these results were verified by visualization after addition of SYBR green I under UV light (Fig. 6). The positive result in the mito-OvLAMP assay was in agreement with the microscopic examinations of the O. viverrini eggs and metacercariae (Fig. 1) and with PCR using specific F3-B3 primer pairs (data not shown). Neither the number of metacercariae per fish nor egg counts (eggs per gram of feces) were calculated in this study.
Fig 6.
Results of specific LAMP assays for detection of O. viverrini using DNA material from mixed metacercariae extracted from freshwater fish and eggs from feces of humans. (A) Visualization on 1.5% agarose stained with ethidium bromide; (B) with SYBR green I under UV light. Lanes: M, 1-kb ladder marker (numbers on the left are sizes in base pairs); (-), LAMP negative control (no DNA template); 1 through 3, DNA of possibly mixed metacercariae from muscles of fish caught in ponds where O. viverrini is endemic; 4 through 7, LAMP products from DNA of eggs, possibly of more than one species, extracted from individual stool samples from humans (after raw fish consumption) in a village where O. viverrini is endemic.
DISCUSSION
The problem of O. viverrini-contaminated fish and the increasing rate of cholangiocarcinoma in the community is a great concern. More effective methods to detect every life stage of this dangerous worm are required. Until now, a variety of microscopic, immunological, and PCR (and real-time PCR) approaches have been used to detect metacercariae in fish and to detect eggs and adult worms as well as antibodies in late-stage opisthorchiasis patients (24), including a LAMP assay using the internal transcribed spacer 1 (ITS-1) in the nuclear ribosomal gene cluster as a target (2). However, it would be more reasonable to use a mitochondrial target for a LAMP assay, due to its stability and the likely higher copy number for provision of DNA template. A typical animal cell is likely to contain hundreds of copies of the mitochondrial genome (20).
In this study, a mito-OvLAMP method targeting the nad1 gene of O. viverrini was successfully developed. The selection of the mitochondrial nad1 gene was appropriate because it contained highly conserved regions for all geographical strains of this species, forming an ideal target for the design of LAMP primers. This was specifically tested with a range of O. viverrini life stages, including eggs, metacercariae, and adults of different geographical origins. The mito-OvLAMP assay in this study is more sensitive than the conventional PCR, capable of producing LAMP ladder-like pattern products from as little as 100 fg of DNA template. The Bsm DNA polymerase large fragment worked well, generating a large amount of DNA products which could be visualized by adding fluorescent SYBR green I for inspection under UV light or with the naked eye. In this study, we have used four methods of determining positive reactions by visual inspection of the LAMP tubes: (i) with the naked eye in sunlight without any additions (turbidity determined from the production of magnesium pyrophosphate); (ii) by gel electrophoresis (with ethidium bromide staining) to inspect the DNA generated; (iii) with the naked eye in sunlight after the addition of SYBR green I to the product; (iv) under UV light after the addition of SYBR green I to the product. The last three of these methods employ a sensitive fluorescent dye for detecting double-stranded DNA.
In fact, in our study, the first method (magnesium pyrophosphate turbidity) is not emphasized, since in sunlight it was difficult to determine sensitivity and variability between the tubes. Interestingly, all LAMP-positive reaction tubes examined under UV light after the addition of SYBR green I appear similar in intensity, although the amounts of product evident via LAMP on the agarose gel stained with ethidium bromide are clearly different. However, the distinction between the positive result at the lowest detection level and the negative at the next dilution is very sharp in sunlight or under UV light. We suggest that visualization using SYBR green I is most practical due to its ease of application in a basic laboratory.
In conclusion, the mito-LAMP method targeting mitochondrial nad1, developed in this study, demonstrated successful identification of any life stage of O. viverrini in diverse hosts in several areas where opisthorchiasis is endemic.
ACKNOWLEDGMENTS
This work received financial support from the National Foundation for Science and Technology Development (NAFOSTED) in Vietnam (grant no. 106.16-2010.60) to Thanh Hoa Le. We express thanks to colleagues for their kind provision of materials used in this study and to international and national collaborators (provincial stations) for their assistance in the field and laboratory work. We extend our thanks to David Blair of School of Marine and Tropical Biology, James Cook University (Townsville, Australia), for the invaluable review of the manuscript.
Footnotes
Published ahead of print 8 February 2012
REFERENCES
- 1. Ai L, et al. 2010. Rapid identification and differentiation of Fasciola hepatica and Fasciola gigantica by a loop-mediated isothermal amplification (LAMP) assay. Vet. Parasitol. 174:228–233 [DOI] [PubMed] [Google Scholar]
- 2. Arimatsu Y, Kaewkes S, Laha T, Hong SJ, Sripa B. 2012. Rapid detection of Opisthorchis viverrini copro-DNA using loop-mediated isothermal amplification (LAMP). Parasitol. Int. 61:178–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Braconi C, Patel T. 2010. Cholangiocarcinoma: new insights into disease pathogenesis and biology. Infect. Dis. Clin. North. Am. 24:871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cai XQ, et al. 2010. Sensitive and rapid detection of Clonorchis sinensis infection in fish by loop-mediated isothermal amplification (LAMP). Parasitol. Res. 106:1379–1383 [DOI] [PubMed] [Google Scholar]
- 5. Chai JY, Shin EH, Lee SH, Rim HJ. 2009. Foodborne intestinal flukes in Southeast Asia. Korean J. Parasitol. 47(Suppl.):S69–S102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen MX, et al. 2011. Sensitive and rapid detection of Paragonimus westermani infection in humans and animals by loop-mediated isothermal amplification (LAMP). Parasitol. Res. 108:1193–1198 [DOI] [PubMed] [Google Scholar]
- 7. De NV, et al. 2003. The food-borne trematode zoonoses of Vietnam. The current status of parasitic diseases in Vietnam. SE Asian J. Trop. Med. 34:12–34 [PubMed] [Google Scholar]
- 8. Hu M, Gasser RB. 2006. Mitochondrial genomes of parasitic nematodes—progress and perspectives. Trends Parasitol. 22:78–84 [DOI] [PubMed] [Google Scholar]
- 9. Katz N, Chaves A, Pellegrino J. 1972. A simple device for quantitative stool thick smear technique in schistosomiasis mansoni. Rev. Inst. Med. Trop. São Paulo 14:397–400 [PubMed] [Google Scholar]
- 10. Kimura Y, et al. 2011. Optimization of turn-back primers in isothermal amplification. Nucleic Acids Res. 39:e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Le TH, Blair D, McManus DP. 2002. Mitochondrial genomes of parasitic flatworms. Trends Parasitol. 18:206–213 [DOI] [PubMed] [Google Scholar]
- 12. Le TH, De NV, Blair D, Sithithaworn P, McManus DP. 2006. Clonorchis sinensis and Opisthorchis viverrini: development of a mitochondrial-based multiplex PCR for their identification and discrimination. Exp. Parasitol. 112:109–114 [DOI] [PubMed] [Google Scholar]
- 13. Le TH, et al. 2008. Human fascioliasis and the presence of hybrid/introgressed forms of Fasciola hepatica and Fasciola gigantica in Vietnam. Int. J. Parasitol. 38:725–730 [DOI] [PubMed] [Google Scholar]
- 14. Mori Y, Notomi T. 2009. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 15:62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mori Y, Nagamine K, Tomita N, Notomi T. 2001. Detection of loop-mediated isothermal reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150–154 [DOI] [PubMed] [Google Scholar]
- 16. Nkouawa A, Sako Y, Nakao M, Nakaya K, Ito A. 2009. Loop-mediated isothermal amplification method for differentiation and rapid detection of Taenia species. J. Clin. Microbiol. 47:168–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Notomi T, et al. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O'Brien EA, et al. 2009. GOBASE: an organelle genome database. Nucleic Acids Res. 37:D946–D950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parida MM, Sannarangaiah S, Dash PK, Rao PV, Morita K. 2008. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 18:407–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robin ED, Wong R. 1988. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J. Cell Physiol. 136:507–513 [DOI] [PubMed] [Google Scholar]
- 21. Sithithaworn P, et al. 2007. Genetic variation in Opisthorchis viverrini (Trematoda: Opisthorchiidae) from northeast Thailand and Laos PDR based on random amplified polymorphic DNA analyses. Parasitol. Res. 100:613–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smout MJ, et al. 2011. Infection with the carcinogenic human liver fluke, Opisthorchis viverrini. Mol. Biosyst. 7:1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sohn WM, et al. 2011. Adult Opisthorchis viverrini flukes in Humans, Takeo, Cambodia. Emerg. Infect. Dis. 17:1302–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sripa B, et al. 2011. Opisthorchiasis and Opisthorchis-associated cholangiocarcinoma in Thailand and Laos. Acta Trop. 120(Suppl. 1):S158–S168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sripa B, Kaewkes S, Intapan PM, Maleewong W, Brindley PJ. 2010. Food-borne trematodiases in Southeast Asia: epidemiology, pathology, clinical manifestation and control. Adv. Parasitol. 72:305–350 [DOI] [PubMed] [Google Scholar]
- 26. Sripa B, et al. 2007. Liver fluke induces cholangiocarcinoma. PLoS Med. 4:e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Touch S, Komalamisra C, Radomyos P, Waikagul J. 2009. Discovery of Opisthorchis viverrini metacercariae in freshwater fish in southern Cambodia. Acta Trop. 111:108–113 [DOI] [PubMed] [Google Scholar]
- 28. Van Van K, Dalsgaard A, Blair D, Le TH. 2009. Haplorchis pumilio and H. taichui in Vietnam discriminated using ITS-2 DNA sequence data from adults and larvae. Exp. Parasitol. 123:146–151 [DOI] [PubMed] [Google Scholar]
- 29. Xu J, et al. 2010. Sensitive and rapid detection of Schistosoma japonicum DNA by loop-mediated isothermal amplification (LAMP). Int. J. Parasitol. 40:327–331 [DOI] [PubMed] [Google Scholar]