Abstract
In November 2010, NDM-1-producing Klebsiella pneumoniae (NDMKP) was identified for the first time in South Korea from four patients with no history of traveling abroad who stayed for 21 to 205 days in a tertiary care hospital. All were sequence type (ST) 340 and had nearly identical XbaI pulsed-field gel electrophoresis (PFGE) patterns. The blaNDM-1-carrying plasmids were in the IncN group, with sizes ranging from 50 to 200 kb. These findings suggest that NDMKP had already been introduced into South Korea before this clustering was found.
TEXT
NDM-1 is a metallo-β-lactamase (MBL) first identified in carbapenem-resistant Klebsiella pneumoniae and Escherichia coli isolates from Swedish patients transferred from New Delhi, India, in 2008 (26). In a short period, NDM-1-producing Enterobacteriaceae have been reported in Africa, Asia, Australia, Canada, Europe, and the United States, starting in the United Kingdom, and many of those had links with India or Pakistan (15). Carbapenem-resistant Enterobacteriaceae (CRE) with acquired carbapenemases are rare in South Korean hospitals (8, 11).
In this study, we described four patients colonized by NDM-1-producing K. pneumoniae (NDMKP) who were hospitalized at a 2,700-bed tertiary care hospital in Seoul, South Korea. We characterized the genotype and phenotype of the strains. All isolates were identified as carbapenem-resistant K. pneumoniae by the MicroScan Neg Breakpoint Combo Panel type 44 (Siemens, West Sacramento, CA). The modified Hodge tests using ertapenem disks (4) were weakly positive, and a KPC-MBL Confirm ID kit (Rosco Diagnostica, Taastrup, Denmark) showed that only meropenem-dipicolinic acid tablets revealed an increase of the inhibition zone compared to that of meropenem tablets. Two NDMKP isolates were obtained from urine cultures, and the others were isolated from stool surveillance cultures (Table 1). Four patients were hospitalized for 21 to 205 days and received meropenem for 8 to 47 days before isolation of NDMKP. Three of them were admitted to the medical intensive care unit (MICU), but their periods in the MICU did not overlap. The remaining patient stayed at a surgical ward after a liver transplantation. There was no history of travel abroad found. None of the four patients was treated to eradicate NDMKP because all were considered to be colonizers. Stool surveillance cultures were performed on 71 patients who shared rooms or intensive care units with the patients carrying NDMKP, but only a carbapenemase-negative, carbapenem-resistant K. pneumoniae strain was isolated. The NDM-1-producing strains might have been introduced earlier in the hospital before this clustering was detected. Interestingly, three patients spontaneously decolonized, but one patient (case 4) carried NDMKP for more than 7 months. Prolonged colonization of NDM-1-producing Escherichia coli has been reported in a patient hospitalized for 13 months without exposure to carbapenems or overt infection by such bacteria (20). Prolonged carriage in the gut can be a factor facilitating the spread of NDM-1-producing Enterobacteriaceae.
Table 1.
Clinical features of the four patients with blaNDM-1-carrying Klebsiella pneumoniaea
| Patient | Age (yr)/gender and underlying disease | Admission date, date discharged or expired, and clinical outcome | Ward (dates of hospital stayb) | Period of isolation of NDMKP, specimen type, and pathogenicity | Carbapenem(s) administered before isolation of NDMKP (no. of days before isolation) |
|---|---|---|---|---|---|
| 1 | 71/F; pyogenic spondylitis | 3 June 2010; 21 January 2011; improved | GW124 (3 June–5 July), MICU1 (5 July–16 July), GW134 (16 July–30 July), MICU1 (30 July–3 August), GW134 (3 August–21 January 2011) | 5 November 2010–31 December 2010; stool; colonizer | Meropenem (8) |
| 2 | 52/M; dermatomyositis interstitial lung disease | 7 October 2010; 16 December 2010; expired from pneumothorax | MICU1 (7 October–12 October), MICU2 (12 October–16 December) | 8 November 2010–12 November 2010; urine; colonizer | Meropenem (16) |
| 3 | 71/M; vertebral osteomyelitis | 31 August 2010; 15 December 2010; improved | GW134 (31 August–2 September), MICU2 (2 September–17 September), GW134 (17 September–15 December) | 16 November 2010/stool/colonizer | Meropenem (22), ertapenem (13) |
| 4 | 59/M; liver transplantation due to HBV-associated liver cirrhosis | 28 July 2010; July 2011; improved | GW91 (28 July–3 August), MICU1 (3 August–12 August), SICU (12 August–3 September), GW102 (3 September–19 September), SICU (19 September–26 September), GW102/SICU (26 September to last follow-up on 30 June 2011) | 30 November 2010–6 December 2010/urine/colonizer; 15 December 2010–22 January 2011/stool/colonizer | Meropenem (47) |
NDMKP, NDM-1-producing Klebsiella pneumoniae; GW, general ward; MICU, medical intensive care unit; SICU, surgical intensive care unit; M, male; F, female.
In 2010 unless specified.
MICs of relevant antimicrobials were confirmed by the agar dilution method (Table 2). The interpretative breakpoints for tigecycline and colistin by the European Committee on Antimicrobial Susceptibility Testing and the breakpoints for other antimicrobials by the Clinical and Laboratory Standards Institute were used (4, 6). Three strains (F181, E1454, and F528) were highly resistant to imipenem and meropenem, but one from patient 4 (E5026) was susceptible to imipenem, with a MIC of 1 μg/ml, and intermediate to meropenem. Colistin, tigecycline, and gentamicin remained active for all strains, and tobramycin, amikacin, and aztreonam were variably active. The blaNDM-1 gene is usually associated with high-level MICs of carbapenems (10). Emergence of strains with low-level MICs suggested that additional mechanisms, such as an efflux pump or porin loss, may be involved in carbapenem resistance in NDM-1 producers. Detection of NDM-1 CRE is impeded when strains have lower-level imipenem and meropenem MICs (3).
Table 2.
Antimicrobial susceptibility profiles of the four NDM-1-producing isolatesa
| Antimicrobial, gene, or other characteristic | MIC (μg/ml) and susceptibility, gene presence, and plasmid size(s) of each strainb |
||||||
|---|---|---|---|---|---|---|---|
| F181 from patient 1 (wild type) | E1454 from patient 2 |
F528 from patient 3 |
E5026 from patient 4 |
||||
| Wild type | Transconjugant | Wild type | Transconjugant | Wild type | Transconjugant | ||
| Antimicrobials | |||||||
| Ampicillin | >128, R | >128, R | >128, R | >128, R | >128, R | >128, R | >128, R |
| Piperacillin | >128, R | >128, R | 32, I | >128, R | 64, I | >128, R | >128, R |
| Cephalothin | >128, R | >128, R | >128, R | >128, R | >128, R | >128, R | >128, R |
| Cefotaxime | >128, R | >128, R | 128,R | >128, R | >128, R | >128, R | >128, R |
| Ceftazidime | >128, R | >128, R | >128, R | >128, R | >128, R | >128, R | >128, R |
| Cefoxitin | >128, R | >128, R | >128, R | >128, R | >128, R | >128, R | >128, R |
| Aztreonam | 0.25, S | 32, R | 0.25, S | 64, R | 0.12, S | 64, R | 64, R |
| Cefepime | 128, R | 128, R | 8, S | >128, R | 32, R | 32, R | 32, R |
| Imipenem | 128, R | >128, R | 8, R | 16, R | 8, R | 1, S | 2, I |
| Meropenem | >128, R | >128, R | 2, I | 32, R | 1, S | 2, I | 2, I |
| Ciprofloxacin | 32, R | 64, R | 0.06, S | 128, R | 0.06, S | 64, R | 0.06, S |
| Tigecycline | 1, S | 2, S | 0.12, S | 2, S | 0.12, S | 0.5, S | 0.12, S |
| Colistin | 0.5, S | 0.5, S | 0.25, S | 0.5, S | 0.25, S | 0.5, S | 0.25, S |
| Gentamicin | 0.25, S | 1, S | 0.25, S | 0.5, S | 0.12, S | 0.25, S | 0.12, S |
| Tobramycin | 8, I | 16, R | 0.12, S | 8, I | 0.12, S | 0.25, S | 0.12, S |
| Amikacin | 4, S | 32, I | 0.25, S | 8, S | 0.25, S | 1, S | 0.25, S |
| Resistant genes other than blaNDM-1 | |||||||
| blaCTX-M-15 | − | − | − | − | − | + | +c |
| blaTEM/blaSHV | −/+ | +/+ | −/− | +/+ | −/− | +/+ | +d/− |
| armA | − | − | − | − | − | − | − |
| ISAba125 | + | + | + | + | + | + | + |
| Size of plasmid (kb) | 200 | 200, 60e, 50e | 50e | 200, 70e, 40 | 60e, 40 | 200, 50e | 100e |
Using the agar dilution method.
I, intermediate; R, resistant; S, susceptible; −, negative; +, positive.
blaCTX-M-15 type.
blaTEM-1 type.
blaNDM-1-carrying plasmid.
Extended-spectrum β-lactamase (ESBL) and plasmid-mediated AmpC β-lactamase (PABL) production was initially screened by the combined disk tests using cefotaxime-clavulanic acid, cefotaxime disks (4), and disks with boronic acid added (23). ESBL phenotypes were positive only in strain E5026 and its transconjugant, and PABL activity was detected in all isolates (Table 2). The presence of known MBLs (blaVIM, blaIMP, blaSPM-1, blaGIM-1, blaAIM-1, blaSIM-1, and blaNDM-1) (13, 24), ESBLs (blaTEM, blaSHV, and blaCTX-M) (25), and PABL genes (17) were determined using PCR. We also aimed to detect ISAba125 and 16S rRNA methylase (armA) by using the published primers (7). All PCR products were directly sequenced. PCR assays detecting MBL genes were positive only for blaNDM-1. E5026 and its transconjugant carried a blaCTX-M-15-type gene (Table 2). All of them carried blaDHA-1 genes. armA-specific PCRs were all negative. ISAba125, which is a common feature of blaNDM-1-positive Enterobacteriaceae, was located upstream of each blaNDM-1 gene (19). The absence of armA is related to susceptibility to gentamicin, a trait which is not found in NDM-1 producers of Indian origin (26).
Plate mating was performed using E. coli J53 (azide resistant) as a recipient, with transconjugants being selected on MacConkey agar containing 100 μg/ml of sodium azide and 8 μg/ml of ceftazidime. Plasmids in the transconjugants were typed as described previously (2). Plasmid sizes were determined by S1 nuclease restriction and pulsed-field gel electrophoresis (PFGE). The blaNDM-1-carrying plasmids were successfully transferred from the three index strains (E1454, F528, and E5026) to E. coli J53 at frequencies ranging from ca. 10−5 to 10−7 (transconjugant/donor). Hybridization of the membrane blot with a probe recognizing blaNDM-1 showed that the blaNDM-1-carrying plasmids varied in size, being ca. 200 kb in F181, ca. 60 and 50 kb in E1454, ca. 70 kb in F528, and ca. 50 kb in E5026 (Table 2). The blaNDM-1-carrying plasmids in this study all belonged to the IncN group. These results suggested that the plasmids may be quite unstable, as reported previously (14). This type of plasmid was previously known to be involved in the transmission of VIM-1, KPC-2, and CTX-M-1 as well as NDM-1 among K. pneumoniae isolates and has been also prevalent in E. coli and Salmonella species (1). Various replicon types are involved in blaNDM-1-carrying plasmids (19), but IncN has recently been found in only one NDM-1-positive E. coli strain (18). Highly efficient transmission of blaNDM-1-carrying plasmids (21) may explain the diversity and worldwide spread of blaNDM-1-carrying Enterobacteriaceae.
DNA sequences of housekeeping genes were uploaded to the multilocus sequence typing (MLST) database (http://pubmlst.org) (5). All 4 strains belonged to sequence type (ST) 340. Genomic DNA was digested with the restriction enzyme XbaI (Takara, Tokyo, Japan) and separated using the CHEF-DR II apparatus (Bio-Rad, Hercules, CA). The PFGE band patterns of the four strains were different by fewer than three bands, suggesting a clonal relationship among the strains (Fig. 1). Although no clear epidemiological linkage was found among the patients, the PFGE band patterns strongly suggest clonal dissemination of a strain within the hospital. All strains yielded ST 340, which is a single-locus variant of ST 258, the dominant ST of KPC3-producing K. pneumoniae worldwide (9) and the ST of one of two KPC2-producing K. pneumoniae found in South Korea (22), and this type, like the NDMKP of this study, has been shown to be gentamicin susceptible but tobramycin and amikacin resistant (12). ST 340 has already been found in NDMKP isolates from Oman, the United Kingdom, and Canada, of which two seem to be linked to India (16, 19). The clone of the NDMKP isolate of this study may possibly have been imported from regions of NDM endemicity, even though no direct epidemiologic links were found.
Fig 1.
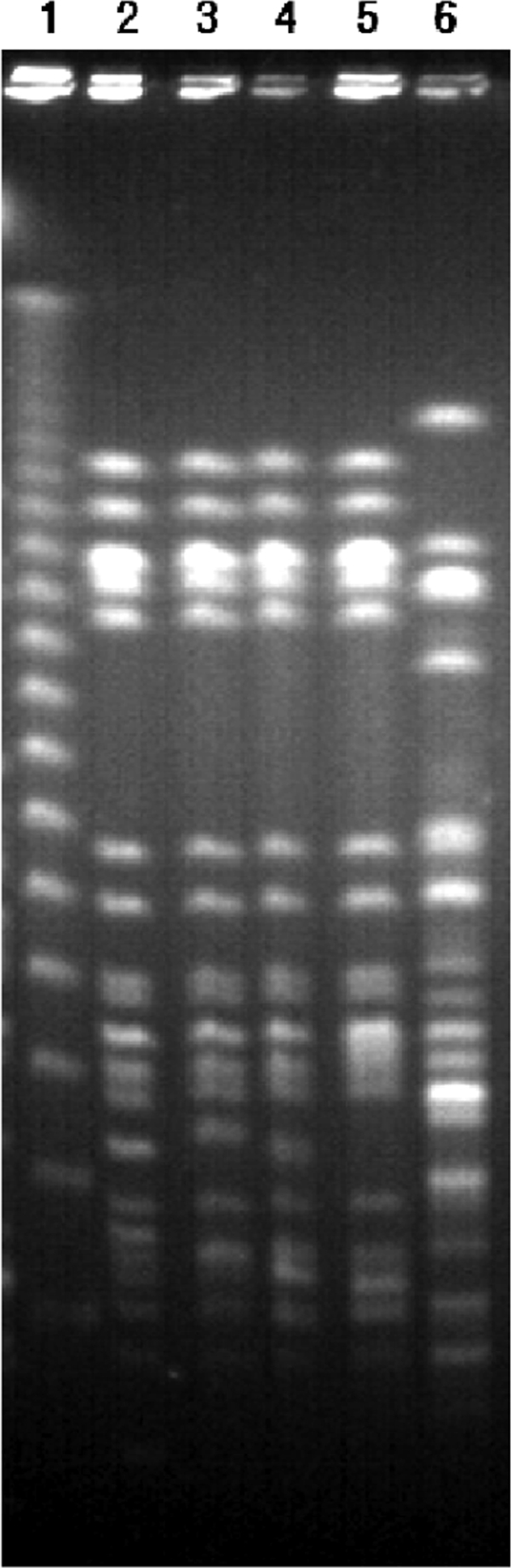
Comparison of PFGE patterns of XbaI-digested genomic DNA of four NDM-1-producing K. pneumoniae isolates. Lane 1, markers; lane 2, F181; lane 3, F528; lane 4, E1454; lane 5, E5026; lane 6, a clinical isolate of carbapenem-resistant K. pneumoniae without NDM-1.
In this study, the first emergence of NDMKP ST 340 strains in South Korea suggested that NDMKP strains might have been introduced earlier in the hospital and were already spread nosocomially. The one isolate revealing susceptible MICs to carbapenems implied the difficulty of detecting NDMKP, and resistance mechanism-based screening for CRE is required to prevent the spread of NDMKP.
Footnotes
Published ahead of print 18 January 2012
REFERENCES
- 1. Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carattoli A, et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 3. Castanheira M, et al. 2011. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006-2007. Antimicrob. Agents Chemother. 55:1274–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. CLSI 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI document M100-S21 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. EUCAST 2011. Breakpoint tables for interpretation of MICs and zone diameters, version 1.3. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/EUCAST_breakpoints_v1.3_pdf.pdf
- 7. Karthikeyan K, Thirunarayan MA, Krishnan P. 2010. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J. Antimicrob. Chemother. 65:2253–2254 [DOI] [PubMed] [Google Scholar]
- 8. Kim SY, et al. 2007. Prevalence and mechanisms of decreased susceptibility to carbapenems in Klebsiella pneumoniae isolates. Diagn. Microbiol. Infect. Dis. 57:85–91 [DOI] [PubMed] [Google Scholar]
- 9. Kitchel B, et al. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumarasamy KK, et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee HK, et al. 2005. Prevalence of decreased susceptibility to carbapenems among Serratia marcescens, Enterobacter cloacae, and Citrobacter freundii and investigation of carbapenemases. Diagn. Microbiol. Infect. Dis. 52:331–336 [DOI] [PubMed] [Google Scholar]
- 12. Livermore DM. 2009. Has the era of untreatable infections arrived? J. Antimicrob. Chemother. 64(Suppl 1):i29–i36 [DOI] [PubMed] [Google Scholar]
- 13. Nordmann P, Poirel L, Carrer A, Toleman MA, Walsh TR. 2011. How to detect NDM-1 producers. J. Clin. Microbiol. 49:718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nordmann P, Poirel L, Toleman MA, Walsh TR. 2011. Does broad-spectrum β-lactam resistance due to NDM-1 herald the end of the antibiotic era for treatment of infections caused by Gram-negative bacteria? J. Antimicrob. Chemother. 66:689–692 [DOI] [PubMed] [Google Scholar]
- 15. Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol. 19:588–595 [DOI] [PubMed] [Google Scholar]
- 16. Peirano G, Pillai DR, Pitondo-Silva A, Richardson D, Pitout JD. 2011. The characteristics of NDM-producing Klebsiella pneumoniae from Canada. Diagn. Microbiol. Infect. Dis. 71:106–109 [DOI] [PubMed] [Google Scholar]
- 17. Perez-Perez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poirel L, Bonnin RA, Nordmann P. 2011. Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob. Agents Chemother. 55:4224–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55:5403–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poirel L, Herve V, Hombrouck-Alet C, Nordmann P. 2011. Long-term carriage of NDM-1-producing Escherichia coli. J. Antimicrob. Chemother. 66:2185–2186 [DOI] [PubMed] [Google Scholar]
- 21. Potron A, Poirel L, Nordmann P. 2011. Plasmid-mediated transfer of the blaNDM-1 gene in Gram-negative rods. FEMS Microbiol. Lett. 324:111–116 [DOI] [PubMed] [Google Scholar]
- 22. Roh KH, et al. 2011. Isolation of a Klebsiella pneumoniae isolate of sequence type 258 producing KPC-2 carbapenemase in Korea. Korean J. Lab. Med. 31:298–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song W, et al. 2007. Detection of extended-spectrum β-lactamases by using boronic acid as an AmpC β-lactamase inhibitor in clinical isolates of Klebsiella spp. and Escherichia coli. J. Clin. Microbiol. 45:1180–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Woodford N. 2010. Rapid characterization of β-lactamases by multiplex PCR. Methods Mol. Biol. 642:181–192 [DOI] [PubMed] [Google Scholar]
- 25. Yong D, et al. 2005. Nosocomial outbreak of pediatric gastroenteritis caused by CTX-M-14-type extended-spectrum β-lactamase-producing strains of Salmonella enterica serovar London. J. Clin. Microbiol. 43:3519–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yong D, et al. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]


