Abstract
CASZ1 is a zinc finger (ZF) transcription factor that is critical for controlling the normal differentiation of subtypes of neural and cardiac muscle cells. In neuroblastoma tumors, loss of CASZ1 is associated with poor prognosis and restoration of CASZ1 function suppresses neuroblastoma tumorigenicity. However, the key domains by which CASZ1 transcription controls developmental processes and neuroblastoma tumorigenicity have yet to be elucidated. In this study, we show that loss of any one of ZF1 to ZF4 resulted in a 58 to 79% loss in transcriptional activity, as measured by induction of tyrosine hydroxylase promoter-luciferase activity, compared to that of wild-type (WT) CASZ1b. Mutation of ZF5 or deletion of the C-terminal sequence of amino acids (aa) 728 to 1166 (a truncation of 38% of the protein) does not significantly alter transcriptional function. A series of N-terminal truncations reveals a critical transcriptional activation domain at aa 31 to 185 and a nuclear localization signal at aa 23 to 29. Soft agar colony formation assays and xenograft studies show that WT CASZ1b is more active in suppressing neuroblastoma growth than CASZ1b with a ZF4 mutation or a deletion of aa 31 to 185. This study identifies key domains needed for CASZ1b to regulate gene transcription. Furthermore, we establish a link between loss of CASZ1b transcriptional activity and attenuation of CASZ1b-mediated inhibition of neuroblastoma growth and tumorigenicity.
INTRODUCTION
Neuroblastoma is a childhood cancer that arises from transformed embryonic neural crest cells that were destined to give rise to the developing sympathoadrenal nervous system and is responsible for the majority of extracranial solid tumors in childhood (22). As in other cancers, transcription factors involved in development play a key role in the transformation of normal progenitors into malignant neuroblastoma (3, 8, 10, 12, 17, 27). In humans, the transcription factor CASZ1 (also known as Castor or Cas), which localizes to chromosome 1p36, has been shown to be frequently deleted in neuroblastoma as well as other types of cancers, such as oligodendroglioma and breast cancer (1). Our previous studies show that CASZ1 functions as a tumor suppressor, inducing neuroblastoma cellular differentiation and suppressing neuroblastoma cell growth via regulation of specific target genes (18, 19).
CASZ1 is a transcription factor that has been studied in both invertebrates and vertebrates for its role in controlling differentiation of neural and cardiac progenitor cells. Originally characterized in Drosophila as a regulator of central nervous system (CNS) development, Drosophila Cas-regulated gene expression directs the appropriate timing of embryonic and postembryonic brain development in a subset of Drosophila neuroblasts (5, 7, 14, 25). In Xenopus laevis, silencing of CASZ1 results in an inability of a subset of cardiac precursor cells to arrest growth, migrate, and adopt the proper differentiated identity (6). Despite the critical role that CASZ1 plays in neural and cardiac developmental programs and tumorigenesis, the key structural elements that mediate CASZ1 function have yet to be elucidated in any species to date.
Human CASZ1 is structurally similar to Drosophila Cas. Both orthologs contain a centrally located, and highly conserved, series of four zinc finger (ZF) moieties defined by the structure Cys2-His2-Cys2-His2 (20). Human CASZ1 localizes to the nucleus and is expressed in two major, alternatively spliced isoforms: CASZ1a (with 11 ZFs and 1,759 amino acids [aa]) and CASZ1b (with 5 ZFs and 1,166 aa) (20). Like the longer CASZ1a isoform, CASZ1b plays a critical role in regulating gene expression and suppressing tumor cell growth (18). A BLAST analysis of the CASZ1 sequence shows that CASZ1b is the more evolutionarily retained isoform, with multiple species containing a CASZ1b ortholog but not a CASZ1a ortholog. In humans, the two isoforms are 66% identical, with all 1,166 amino acids of CASZ1b being identical to the first 1,166 amino acids of CASZ1a.
To obtain greater insight into the functions of CASZ1 and its role in neural and cardiac development, as well as tumorigenesis, it is important to obtain a better understanding of the mechanism of transcriptional regulation by CASZ1. This includes defining structural domains that mediate its translocation into the nucleus and its transcriptional activity. Moreover, it is essential to know whether these critical domains are also required for its biological functions, such as regulating cell proliferation. The determination of the critical domains of CASZ1 will be fundamental for elucidating its regulatory role during development and for future analyses of genomic changes in CASZ1 in neuroblastoma and other diseases.
In this report, we describe the first structure-function analysis conducted on CASZ1b in any species. We identified critical domains required for CASZ1b nuclear localization and transcriptional regulation. The loss of transcriptional function mutants of CASZ1b greatly decreased its ability to inhibit neuroblastoma cell growth, a result which further supports the hypothesis that CASZ1b transcriptional activity is required to suppress neuroblastoma cell growth.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney cells (HEK293T) were kindly provided by the Molecular Oncology Section of the Pediatric Oncology Branch of NCI. HEK293T was maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum as well as 100 μg/ml streptomycin, 100 U/ml penicillin, and l-glutamine. Neuroblastoma stable clones (described below) were maintained as described above but with medium containing 500 μg/ml G418 (Geneticin; Invitrogen) and 5 μg/ml blasticidin (Invitrogen). Cells were grown at 37°C with 5% CO2 and were passaged at 70 to 80% confluence 2 to 3 times per week.
Construction of missense, deletion, and fusion CASZ1b constructs and NLS-GFP constructs.
The QuikChange XL site-directed mutagenesis kit (Stratagene) was utilized to introduce various missense mutations into the wild-type (WT) pCMV-FLAG_CASZ1b cDNA plasmid (20) according to the manufacturer's protocol. Also, seven N-terminal deletion and two C-terminal deletion variants of CASZ1b were generated by PCR amplification of the appropriate region prior to cloning. The oligonucleotide primers used in the mutagenesis reactions will be provided upon request. Lastly, the double-stranded DNA (dsDNA) sequence for the first and second predicted nuclear localization signals was synthesized with an attached 5′ EcoRI site and a 3′ BamHI site and was cloned into the corresponding sites of the pmaxFP-Green-N vector (Lonza). All new constructs were fully sequence verified prior to analysis.
Transient transfections and generation of neuroblastoma stable clones.
For mRNA, protein, and subcellular localization experiments, plasmids were transiently transfected into HEK293T cells using the Lipofectamine 2000 cationic lipid reagent (Invitrogen) according to the manufacturer's protocol. Tyrosine hydroxylase (TH) is a known CASZ1 target (18, 19). In luciferase experiments, the TH promoter-pGL4.1-luc construct (TH-Luc), which contains the proximal 2 kb of the TH promoter, was generously provided by Gregory Wray (32). A cytomegalovirus (CMV)-driven β-galactosidase construct was cotransfected with TH-Luc and different CASZ1b mutants in order to provide an internal control for transfection efficiency. Luciferase activity was quantified after 24 h using the Dual-Luciferase reporter assay system (Promega), and β-galactosidase activity was measured concomitantly with the Luminescent β-galactosidase detection kit II (Clontech) to normalize the luciferase signals.
The neuroblastoma stable clone was made as reported previously (19). In brief, WT CASZ1b, the CASZ1b ZF4 mutant (ZF4m), the CASZ1b mutant with a deletion of aa 31 to 185 of the N terminus (ΔN31-185), and the CASZ1b mutant with a deletion of nuclear localization signal 1 (NLS1) (ΔNLS1) in pT-Rex-DEST30 were transfected into SH-SY5Ytet (SY5Ytet) cells that had been previously stably cloned with pcDNA6/TR (21). The transfected cells were cultured in selective medium containing both blasticidin S and G418. After 3 to 4 weeks of selection, individual colonies were selected and treated with 1 to 1,000 ng/ml of tetracycline in order to induce CASZ1b transcription. Stable clones expressing CASZ1b or CASZ1 mutations are labeled SY5YtetCASZ1b, SY5YtetCASZ1b ZF4m (ZF4m), SY5YtetCASZ1b ΔN31-185 (ΔN31-185), and SY5YtetCASZ1b ΔNLS1 (ΔNLS1). The stable clones with empty vector are labeled SY5Ytetemv.
RNA isolation and cDNA analysis by quantitative real-time PCR (qRT-PCR).
Total mRNA was collected 24 h after transfection using the RNeasy minikit (Qiagen) according to the manufacturer's protocol. Endogenous transcription of several known neural differentiation-associated CASZ1 target genes, including the TH gene, the nerve growth factor receptor (NGFR) gene, and the TrkA gene (19), was used to assess transcriptional activity of CASZ1b variants. Quantitative measurements of total β-actin, TH, TrkA, and NGFR levels were obtained using an ABI 7000 sequence detection system thermocycler, and the primer sequences will be provided upon request. Threshold cycle (CT) values were standardized to β-actin levels, and the fold change in mRNA compared to that of the pCMV-Tag2A (empty vector)-transfected control samples was calculated. This value was divided by the WT CASZ1b fold change in order to calculate the percentage of WT function that was retained by each variant using this assay system. All results are plotted as the mean plus or minus the standard deviation (SD) of at least three independent experiments, and Student's t test was used to assess statistical significance.
Protein isolation and Western blot analysis.
For assessment of protein levels, cells were lysed 24 h after transfection using the Qproteome mammalian protein prep kit (Qiagen), and 20 μg of total protein was separated and electroblotted as described previously (19). An anti-FLAG M2 monoclonal antibody (1 μg/ml; Sigma) and an anti-α-tubulin antibody (1 μg/ml; Santa Cruz Biotechnology) were used for detection of total FLAG-CASZ1b and α-tubulin levels, respectively. Protein bands were detected using a goat anti-mouse IgG-horseradish peroxidase (HRP)-conjugated secondary antibody (200 μg/ml; Santa Cruz Biotechnology) and visualized using enhanced chemiluminescence (Amersham Biosciences).
Analysis of nuclear localization by confocal microscopy.
HEK293T cells were transfected as described above in 8-well Lab-Tek chamber slides (catalog no. 177402). Cells were fixed, permeabilized, blocked, and stained as described previously (19). An anti-FLAG M2 monoclonal antibody (Sigma) and an Alexa Fluor 594-conjugated goat anti-mouse antibody (Molecular Probes) were used to detect FLAG-CASZ1b, and 4′,6′-diamidino-2-phenylindole (DAPI; Invitrogen) was used to stain chromatin within nuclei. Localization was analyzed and imaged using a Zeiss LSM 710 confocal microscope (Zeiss) at the NCI Confocal Microscopy Core Facility.
Cell proliferation and clonogenicity assays.
The proliferation of WT CASZ1b-expressing or CASZ1b variant-expressing SY5Ytet clones was analyzed as follows. Cells from each clone were plated in 96-well plates, and after 24 h, the medium was replaced with normal medium (for tetracycline-negative [Tet−] wells) or medium containing tetracycline (for Tet+ wells). This experiment was done in triplicate. After 3 or 6 days, a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay (Promega) was performed on the cells according to the manufacturer's protocol. To assess the effects of WT CASZ1b or CASZ1b variants on anchorage-independent cell growth, 1 × 104 cells were cultured in 0.7% top agarose in medium containing G418 and blasticidin S (with or without 100 ng/ml Tet) that was plated on a layer of 1.4% bottom agar/RPMI to prevent the adhesion of cells to the culture plates. This experiment was done in triplicate. The medium was changed twice per week, and visible colonies were counted after 3 weeks.
In vivo tumorigenesis.
Suspensions of SY5YtetCASZ1b, SY5YtetCASZ1b ZF4m, and SY5YCASZ1b ΔN31-185 (3 × 106) cells in Hanks' balanced salt solution (HBSS) were mixed with 3× the volume of Matrigel solution (Trevigen) and implanted subcutaneously in the dorsal flank of 20 SCID mice (6- to 8-week-old female mice) per cell line. The animal study was approved by the Animal Care and Use Committee of the National Cancer Institute (PB-023). The doxycycline-negative (Dox−) cohort of control mice received regular food, while the Dox+ cohort received food containing doxycycline for 1 week prior to tumor implantation and during the course of the experiment.
RESULTS
The ZF1 to ZF4 (ZF1-4) domain is critical for CASZ1b function.
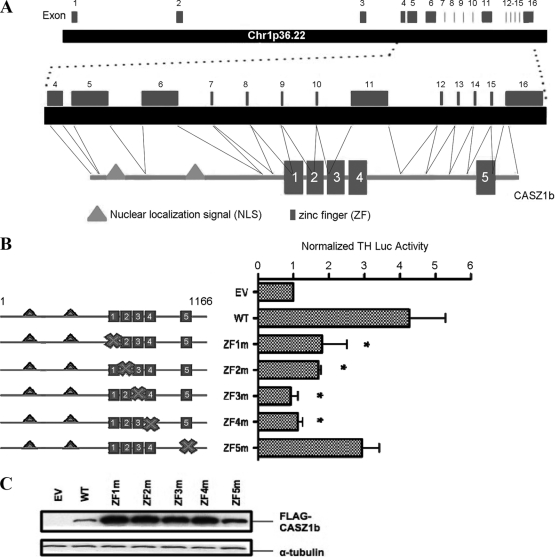
The primary RNA transcript of human CASZ1b has 16 exons and covers 149 kb of chromosome 1p36.22. Exons 1 to 3 comprise the 5′ untranslated region (UTR), while exons 4 to 16 encode the human CASZ1b protein-coding region (Fig. 1A). The protein contains a cluster of four predicted zinc fingers (ZFs) near the middle of the protein (ZF1 to ZF4) and a fifth predicted ZF (ZF5) located near the C terminus. Using the SMART domain prediction tool (http://smart.embl-heidelberg.de), the first four ZFs had low E values (ZF1, 5.34; ZF2, 0.81; ZF3, 0.93; and ZF4, 0.011), a result which indicates a high confidence in the presence of the motif and suggests they are functionally important. In contrast, ZF5 had the highest E value (22.9) among all five ZFs. Each of the five ZFs of CASZ1b contains a TFIIIA class C2H2 structural motif (the most common type of ZF among transcription factors), defined by a structural Cys2-His2-Cys2-His2 element. Loss of either of the two cysteines' sulfhydryl groups is classically used to abolish the proper folding of a ZF, thereby destroying its potential DNA- and/or protein-binding activity. In order to evaluate the relative functional importance of the ZFs, one of the C2H2 motif-defining cysteines in each ZF was individually mutated to an alanine in WT CASZ1b. Further, the tyrosine hydroxylase (TH) gene is a target gene of CASZ1b, and overexpression of WT CASZ1b in HEK293T cells strongly activated expression of a tyrosine hydroxylase promoter-luciferase (TH-Luc) reporter construct (Fig. 1B). Loss of any one of the first four ZFs resulted in a significant decrease in the ability to activate transcription of the TH-Luc reporter construct compared to that of WT CASZ1b (Fig. 1B). Loss of ZF1 or ZF2 resulted in an ∼60% decrease in induction of TH-Luc compared to that of the WT (P < 0.05). Despite their decrease in transcriptional activity, a ZF1 mutation or ZF2 mutation still stimulated induction of TH-Luc that was approximately 1.7-fold higher than that of the empty vector (EV) control (P < 0.05). Loss of ZF3 or ZF4 led to a nearly 80% decrease in induction of TH-Luc compared to that of the WT (P < 0.03), and the activity of these mutants was indistinguishable from that of the EV (P > 0.1). Mutation of ZF5 caused induction of TH-Luc activity that was approximately 3-fold higher than that of the EV (P < 0.05); it was slightly reduced compared to the transcriptional induction of the WT CASZ1b, although this difference was not statistically significant (P = 0.14) (Fig. 1B). Western blot analysis and immunocytochemical staining indicated that the reduced transcriptional activity for the ZF1 to ZF4 mutants was not due to decreased levels of CASZ1 protein levels (Fig. 1C) or an alteration in subcellular localization, as CASZ1 ZF mutant proteins retained their nuclear localization (data not shown). These data demonstrate that alteration of ZF1 to ZF4 significantly decreases the ability of CASZ1b to activate transcription of a target promoter.
Fig 1.
The ZF1-4 domain is critical for CASZ1b function. (A) Genomic structure and schematic of the CASZ1b protein with the two nuclear localization signals and the five ZFs. (B) Activation of the TH-luciferase construct 24 h after transfection by the CASZ1b ZF mutants. The mutated ZF is indicated by an “X” in the diagram next to each sample (*, P <0.05). (C) Western blot showing the steady-state protein levels in 293T cells transfected with plasmids encoding the CASZ1b ZF mutants and WT CASZ1b 24 h after transfection.
The ZF1-4 domain is highly conserved.
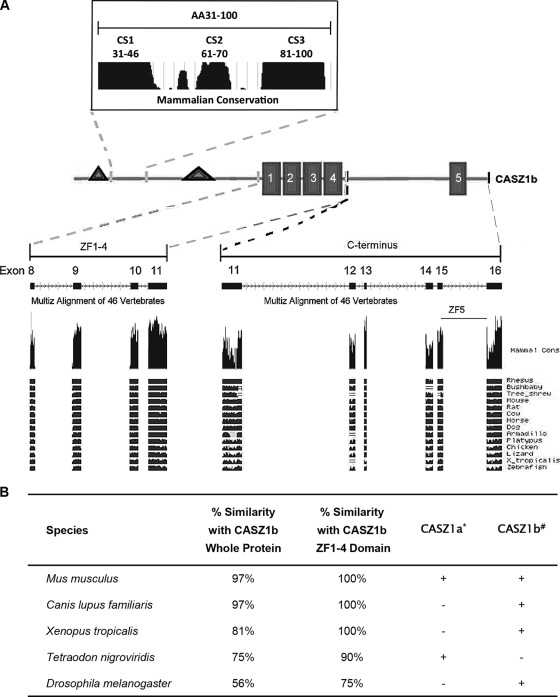
Drosophila Cas contains only a single set of four ZFs, which are highly homologous to ZF1 to ZF4 in human CASZ1b. Exons 8 to 11 encode the human protein sequence comprising ZF1 to ZF4, and this sequence (including the intervening amino acids between the ZF domains) was entered into the BLAT program of the UCSC Genome Bioinformatics website (http://genome.ucsc.edu/) in order to determine the levels of conservation among multiple species. Figure 2A shows the results of the multispecies alignment and illustrates the high level of evolutionary conservation of the ZF1-4 domain. Figure 2B shows the enrichment of sequence conservation in the ZF1-4 domain compared to that of the full-length protein in multiple species, including Xenopus tropicalis (100% versus 81%, respectively) and Drosophila melanogaster (75% versus 56%, respectively), a finding which demonstrates that the ZF1-4 domain has been highly conserved throughout evolution and is consistent with the observed functional importance of this domain.
Fig 2.
High conservation of the ZF1-4 domain and N-terminal domain of CASZ1b among vertebrates. (A) Screenshot of CASZ1 conservation among different species (from http://www.genome.ucsc.edu/). (B) Conservation of the ZF1-4 domain across multiple species. *, a plus sign indicates the existence of the CASZ1a isoform; #, a plus sign indicates the existence of CASZ1b in different species.
The C terminus does not contribute significantly to CASZ1b function.
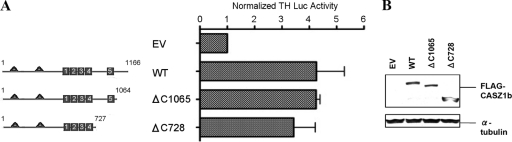
The left panel of Fig. 3A shows a graphical representation of the two C-terminal mutants that were generated from the region between ZF4 and the C terminus of the WT CASZ1b construct: ΔC1065 represents a deletion of the region between ZF5 and the C terminus, and ΔC728 deletes the region between ZF4 and the C terminus, which includes ZF5 and represents a truncation of 38% of CASZ1b. Both of the C-terminal mutants significantly induce TH-Luc expression compared to that of the EV (P < 0.05), with ΔC1065 and ΔC728 retaining 100% ± 3% and 80% ± 18% of WT function, respectively (Fig. 3A, right panel). Total CASZ1b protein levels were not appreciably altered by the C-terminal truncations (Fig. 3B). This result indicates that critical transcriptional regulatory domains localize to the N terminus as well as to the ZF1-4 domain of the CASZ1b protein but not to the C terminus.
Fig 3.
The C terminus does not significantly contribute to CASZ1b transcriptional activity. (A) Activation of the TH-luciferase construct 24 h after transfection with CASZ1b containing the ΔC728 and ΔC1065 plasmids. ΔC728 is a deletion of aa 728 to 1166 of CASZ1b (a 38% truncation of the full-length protein). (B) Western blot showing that steady-state protein levels are comparable to those of WT CASZ1b 24 h after transfection.
NLS1 is critical for nuclear localization.
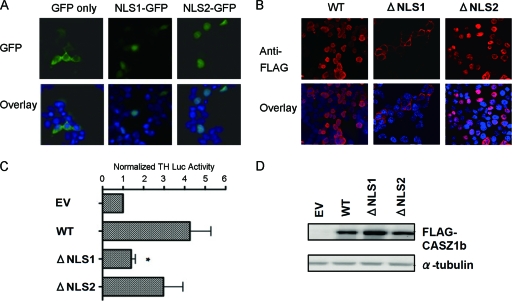
We next focused on the N terminus of CASZ1b. Using the WoLF PSORT protein subcellular localization prediction tool (http://wolfpsort.org), we identified a predicted nuclear localization signal (NLS) at aa 23 to 29 (denoted NLS1) (Fig. 1A). Previously, we identified a predicted bipartite nuclear localization signal at aa 232 to 248 (20) (denoted NLS2) (Fig. 1A). In order to determine the functions of NLS1 and NLS2, the two sequences were individually fused to the N terminus of green fluorescent protein (GFP), and the ability of each sequence to localize GFP to the nucleus was monitored. As shown in Fig. 4A, both NLS1 and NLS2 were able to independently localize GFP to the nucleus, while the vector containing GFP alone was distributed primarily in the cytoplasm. This confirmed that both NLS1 and NLS2 are functionally capable of directing nuclear import. However, deletion of NLS1 (ΔNLS1) in WT CASZ1b demonstrated that it is necessary for nuclear localization in the full-length protein, while deletion of NLS2 (ΔNLS2) produced only a slight decrease in nuclear localization (Fig. 4B). As expected, loss of NLS1 produced a complete loss of activation of TH-Luc (P < 0.05), while loss of NLS2 did not significantly alter TH-Luc activation compared to that of WT CASZ1b (Fig. 4C).
Fig 4.
NLS1 is required for nuclear localization. (A) Colocalization of GFP, NLS1-GFP, and NLS2-GFP with the nucleus. Chromatin is stained with DAPI. Magnification, ×63. (B) Assessment of ΔNLS1 and ΔNLS2 CASZ1b cellular localization. FLAG-tagged WT CASZ1, ΔNLS1 CASZ1b, and ΔNLS2 CASZ1b are stained with an anti-FLAG antibody, and chromatin is stained with DAPI. Magnification, ×63. (C) Activation of the TH-luciferase construct by WT, ΔNLS1, and ΔNLS2 CASZ1b 24 h after transfection. Data are plotted as the means ± SDs of two independent experiments (*, P < 0.05; **, P < 0.001). (D) Western blot showing steady-state levels of WT, ΔNLS1, and ΔNLS2 CASZ1b 24 h after transfection.
CASZ1b transcriptional activity depends upon aa 31 to 185.
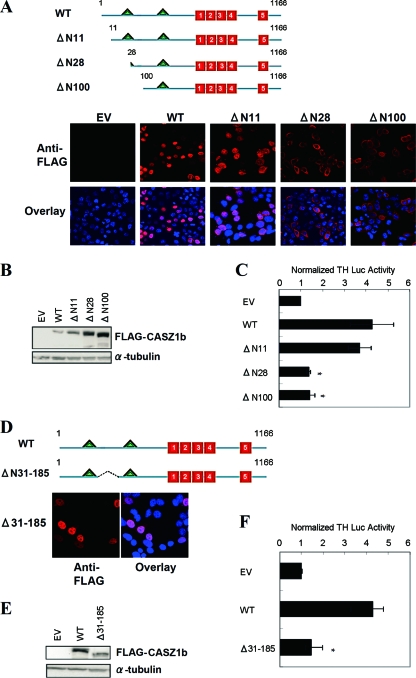
In order to identify functionally important domains at the N terminus, a series of progressive deletions prior to NLS2 were generated from the WT CASZ1b construct (Fig. 5A, top). The ability of the N-terminal mutants to localize to the nucleus was analyzed by immunocytochemical staining. Nuclear localization was abolished in the constructs containing partial or complete NLS1 deletion, further supporting that NLS1 is required for nuclear localization (Fig. 5A, bottom). Deletion of the first 11 amino acids of the N terminus of CASZ1b (CASZ1b ΔN11) did not significantly alter induction of TH-Luc expression (87% ± 12% of that of the WT) (Fig. 5A), indicating that this region is not required for transcriptional regulation in this assay. Each of the progressively larger N-terminal truncations did not localize to the nucleus, and as expected, these variants showed a complete loss of induction of TH-Luc (P < 0.05) (Fig. 5C). To further define the critical region located within the N terminus of CASZ1b, a deletion was introduced between NLS1 and NLS2, and it was assayed for its ability to induce TH-Luc transcription. Notably, deletion of aa 31 to 185 caused a complete loss of CASZ1b function without affecting protein levels or nuclear localization (Fig. 5D to F). These results indicate that CASZ1b transcriptional activity depends upon the region defined by aa 31 to 185.
Fig 5.
The CASZ1b transcriptional activation domain is defined by aa 31 to 185. (A) Schematic of the full-length CASZ1b protein with the two NLSs (triangles) and the five ZFs (boxes) indicated. The three N-terminal deletions are also indicated. Colocalization of WT FLAG-CASZ1b and the three N-terminal variants with the nucleus is shown underneath. Chromatin is stained with DAPI. Magnification, ×63. (B) Western blot showing steady-state WT and variant CASZ1b protein levels 24 h after transfection. (C) Activation of the TH-luciferase construct by WT CASZ1b and the three N-terminal deletions 24 h after transfection. Two of the mutants lose complete function, while only ΔN11 retains function (*, P < 0.001). (D) Cellular localization of the ΔN31-185 CASZ1b mutant protein after staining with anti-FLAG antibody. Chromatin is stained with DAPI. Magnification, ×63. (E) Western blot showing steady-state WT and ΔN31-185 CASZ1b protein levels 24 h after transfection. (F) Activation of the TH-luciferase construct 24 h after transfection of 293T cells with WT CASZ1b and ΔN31-185 CASZ1b plasmids (*, P < 0.001).
Contribution of additional predicted domains to transcriptional activation.
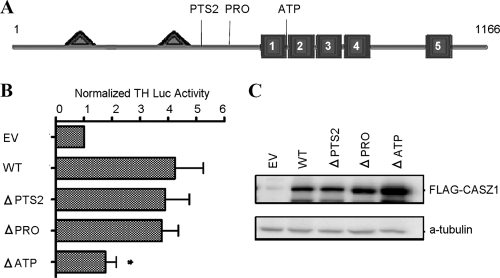
A motif scan of the CASZ1b protein sequence uncovered a novel peroxisome-targeting sequence type 2 (PTS2) at aa 277 to 285, a predicted proline-rich region located at aa 384 to 417, and a predicted ATP/GTP-binding motif at aa 541 to 548, which were designated PTS2, PRO, and ATP, respectively (Fig. 6A). In order to determine the contribution of these domains to CASZ1b transcriptional activation, each of the predicted domains was individually deleted (ΔPTS2, ΔPRO, and ΔATP), and the ability to induce TH-Luc activity was analyzed. Figure 6B shows that only ΔATP significantly decreased CASZ1b function, retaining 41% ± 9% of WT function (P < 0.05), yet this decrease in function cannot be separated from a potential destabilization of the ZF1-4 domain caused by the deletion of a portion of this domain. None of the deletions decreased CASZ1b protein levels (Fig. 6C) or nuclear localization (data not shown).
Fig 6.
Contribution of various predicted domains to CASZ1b transcriptional activity. (A) Schematic of full-length CASZ1b showing the locations of the predicted peroxisome-targeting sequence (PTS2), proline-rich domain (PRO), and ATP/GTP-binding motif (ATP). (B) Activation of the TH-luciferase construct 24 h after transfection by ΔPTS2, ΔPRO, and ΔATP CASZ1b variants (*, P < 0.001). (C) Western blot showing that steady-state protein levels are comparable to those of WT CASZ1b 24 h after transfection.
Reduced CASZ1b function alters endogenous induction of genes important in neural differentiation.
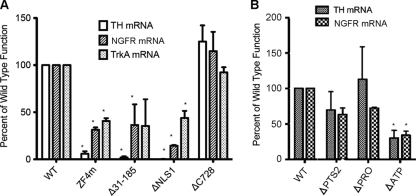
High expression of TrkA and NGFR is associated with a good prognosis in neuroblastoma, and restoration of either TrkA or NGFR expression in neuroblastoma cell lines suppresses tumor cell growth (9, 13, 29). These genes have been shown to be upregulated by CASZ1 (19). To determine whether the domains critical for CASZ1b activation of the TH-Luc construct were also important for endogenous transcriptional activation of CASZ1 target genes, four representative variants, including ZF4m, Δ31-185, ΔNLS1, and ΔC728 CASZ1b, were transfected into 293T cells. Consistent with the TH-Luc data, CASZ1b's transcriptional regulation of TH, TrkA, or NGFR mRNA levels was significantly attenuated by a ZF4m, Δ31-185, or ΔNLS1 mutation, while the ΔC728 mutation did not have any appreciable effect on any of the evaluated genes (Fig. 7A). Moreover, we investigated the transcriptional activity of other CASZ1b variants that contain predicted functional domains for regulating endogenous TH and NGFR mRNA levels in 293T cells. Consistent with the TH-Luc data (Fig. 6B), CASZ1b's transcriptional regulation of TH or NGFR mRNA levels was significantly attenuated by the ΔATP mutation but not by the ΔPTS2 and ΔPRO mutations (Fig. 7B).
Fig 7.
CASZ1b regulates endogenous gene transcription. (A) Quantitative PCR was used to assess relative levels of TH, NGFR, and trkA mRNA 24 h after transfection of 293T cells with either WT CASZ1b, ZF4m, Δ31-185, ΔNLS1, or ΔC728 plasmids. (B) Quantitative RT-PCR was used to assess relative levels of TH and NGFR mRNA 24 h after transfection of 293T cells with WT CASZ1b or with ΔPTS2, ΔPRO, or ΔATP. Results are the means ± SDs of three independent experiments (*, P < 0.05).
CASZ1b transcriptional activity is required for growth suppression in neuroblastoma cells.
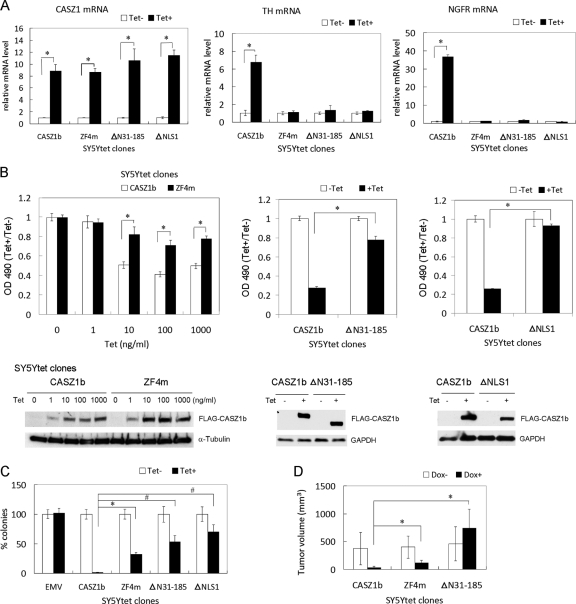
CASZ1b is a neuroblastoma tumor suppressor gene. To determine whether the transcription regulation domains of CASZ1b discovered in the 293T cells are also critical in neuroblastoma cells, three representative CASZ1b variants, including ZF4m, Δ31-185, and ΔNLS1 in the pT-Rex-DEST30 vector, were stably transfected into SY5Ytet neuroblastoma cells. Endogenous transcriptional activation of CASZ1 target genes was evaluated by quantitative real-time PCR. In this tetracycline-inducible (Tet+) system, after 24 h of Tet treatment, WT CASZ1b and the CASZ1b variants were induced to comparable levels of mRNA (Fig. 8A, left panel) and protein (Fig. 8B, bottom). Consistent with the 293T data, CASZ1b's transcriptional regulation of TH or NGFR mRNA levels was significantly attenuated by a ZF4m, Δ31-185, or ΔNLS1 mutation in the SY5Y cells (Fig. 8A, middle and right panels). Lastly, the effect on biological function of the transcriptionally inactive CASZ1b variants was compared to that of the transcriptionally active WT CASZ1b. Utilizing a tetracycline-inducible WT CASZ1b in SY5Y neuroblastoma cells, we found that after 3 days of Tet (Tet+) treatment, there was an ∼60% suppression of cellular proliferation compared to that of noninduced (Tet−) cells (Fig. 8A), whereas upon CASZ1b ZF4m induction, there was an ∼30% suppression of cellular proliferation compared to that of the noninduced cells (Fig. 8B, left). We found that after 3 days of Tet treatment, upon CASZ1b Δ31-185 induction, there is no suppression of cellular proliferation compared to that of the noninduced cells (data not shown); however, after 6 days of Tet treatment, CASZ1b Δ31-185 induction caused ∼20% suppression of cellular proliferation compared to that of the noninduced cells. This suppression is not comparable to that of WT CASZ1b because there was 80% suppression of cell proliferation upon WT CASZ1 induction after 6 days of Tet treatment (Fig. 8B, middle). CASZ1 ΔNLS1 did not suppress cellular proliferation after 6 days of Tet treatment compared to that of noninduced cells (Fig. 8B, right panel). To test whether CASZ1b variants inhibit tumorigenicity of neuroblastoma cells, anchorage-independent growth was assessed by soft agar clonogenicity, and visible clones were counted after 3 weeks. The CASZ1b ZF4m-, CASZ1b Δ31-185-, or CASZ1b ΔNLS1-expressing SY5Y cells showed a decrease in soft agar clonogenicity compared to that of noninduced control cells, but they were ∼30-fold to ∼70-fold less effective than WT CASZ1b in decreasing soft agar clonogenicity (Fig. 8C). To assess in vivo tumorigenicity, SY5YtetCASZ1b, CASZ1b ZF4m, or CASZ1b Δ31-185 cells were implanted in a subcutaneous site in mice that had received normal food or doxycycline-containing food during the prior week (9 or 10 mice per group). Doxycycline-treated animals showed 90% decreases in tumor growth for SY5YtetCASZ1b cells, 70% decreases for SY5YtetCASZ1b ZF4m cells, and no decrease for SY5YtetCASZ1b Δ31-185 cells at 4.5 weeks compared to the growth in placebo-treated mice (Fig. 8D). Consistent with the transcription activity data, this indicates that these CASZ1b critical domains are involved in both transcriptional regulation and the ability of CASZ1b to inhibit neuroblastoma tumor growth.
Fig 8.
CASZ1b variants are less efficient than WT CASZ1b at inhibiting neuroblastoma cell proliferation and tumor growth. (A) Quantitative PCR was used to assess relative levels of CASZ1b, TH, and NGFR mRNA after 24 h of Tet (1 μg/ml) treatment in SY5Ytet cells that have been stably transfected with Tet-inducible WT, ZF4m, Δ31-185, or ΔNLS1 CASZ1b. The mRNA levels of WT CASZ1b and the variants were significantly upregulated compared to those of noninduced control cells (left panel; *, P < 0.02). TH and NGFR mRNA levels were significantly upregulated by WT CASZ1b but not by ZF4m, Δ31-185, or ΔNLS1 CASZ1b (middle and right panels, respectively; *, P < 0.005). (B) The left panel shows the MTS assay of SY5YtetCASZ1b and ZF4m cells treated with different amounts of Tet for 3 days. ZF4m is about 2-fold less effective than WT CASZ1b at suppressing cell proliferation (*, P < 0.0001). The middle panel shows the MTS assay of SY5YtetCASZ1b and Δ31-185 cells treated with Tet (1 μg/ml) for 6 days. Δ31-185 CASZ1b is less effective than WT CASZ1b at suppressing cell proliferation (*, P < 0.0001). The right panel shows the MTS assay of SY5YtetCASZ1b and ΔNLS1 cells treated with Tet (1 μg/ml) for 6 days. ΔNLS1 CASZ1b is less effective than WT CASZ1b at suppressing cell proliferation. *, P < 0.0001. A Western blot analysis of steady-state proteins from SY5YtetCASZ1b and CASZ1b mutant cells after 24 h of treatment with Tet is shown at the bottom of panel B. (C) Soft agar assay showing a 30-fold to 70-fold decrease in the ability of CASZ1b variants to inhibit the clonogenicity of SY5Y cells compared to that of WT CASZ1 (*, P < 0.003; #, P < 0.02). (D) SY5YtetCASZ1b, ZF4m, or Δ31-185 cells were subcutaneously injected into nude mice for 4.5 weeks. ZF4m and Δ31-185 CASZ1b are less effective than WT CASZ1b at suppressing tumor growth in the xenografts (n = 9 or 10 mice per group; volume = [length × width2]/4; *, P < 0.0005). Data are the means ± SDs.
DISCUSSION
During neural differentiation in Drosophila, progenitor cells withdraw from the cell cycle and begin expressing a particular set of genes that is specific to the subtype of tissue into which they are developing. The temporospatial program of gene expression in these cells is initiated by a distinct class of neural fate determination genes which includes the transcription factor CASZ1 (5, 11, 15). In mammals, CASZ1 suppresses neuroblastoma growth, induces cell differentiation, and inhibits cell migration, indicating that CASZ1 regulates a transcriptome enriched in genes associated with developmental processes (19). CASZ1a and CASZ1b are the two major isoforms of the CASZ1 gene, and CASZ1b has activity that is similar, if not identical, to that of CASZ1a for regulating gene transcription and suppressing tumor growth (18). In this report, we identified for the first time the domains within CASZ1b that mediate transcriptional control of a set of CASZ1b target genes. We assessed whether alteration of CASZ1b transcriptional activity altered a known CASZ1b biofunction. We found that the N terminus of CASZ1b (aa 31 to 185) and the ZF1-4 domain are required for CASZ1b transcriptional activity. In addition, we report that NLS1 is necessary and sufficient for CASZ1b nuclear localization but that NLS2 is not required. Moreover, loss of transcriptional activity of CASZ1b led to the decrease in its capability to inhibit neuroblastoma cell proliferation.
By using representative loss-of-function mutants, we showed that CASZ1b transcriptional activity is regulated by the ZF1-4 domain, suggesting a role for this domain in protein or DNA binding. A study in Drosophila supports a DNA-binding role for this domain (16). The DNA-binding sequence of Cas was determined from a population of dsDNA fragments with degenerate sequences, and surprisingly, the binding sequence shared 9 of 10 bases with another neural fate determination gene, Hunchback (hb) (16). Also, Cas was shown to specifically bind a known endogenous hb-binding site from Drosophila genomic DNA (16). Our data expand upon the results of earlier studies (16) by identifying that ZF3 and ZF4, in particular, most highly contribute to transcriptional regulation and to the putative DNA-binding function of Cas as well. The main purpose of our study was limited to defining critical domains required for nuclear localization and transcriptional activity of the CASZ1b gene, but our future goal is to determine whether the ZF domains of CASZ1b are critical for DNA binding. Drosophila Cas contains only ZF1 to ZF4, and these domains remain highly evolutionarily conserved. Our finding that the C-terminal region after ZF4, which includes ZF5, did not appear to contribute to transcriptional activity is consistent with evolutionary sequence conservation, as Drosophila Cas does not contain this region. Moreover, among other species, the C-terminal region after ZF4 has less conservation of homology than the cross-species homology seen in ZF1 to ZF4 (Fig. 2A).
Bioinformatic analysis showed that there are two NLSs located at the N terminus of CASZ1b which we designated NLS1 (aa 23 to 29) and NLS2 (aa 232 to 248). Both NLS1 and NLS2 are able to translocate enhanced green fluorescent protein (EGFP) into the nucleus (Fig. 3C). Unexpectedly, the results of the NLS1 and NLS2 deletions demonstrated the requirement of only NLS1 for CASZ1b nuclear localization. In addition, we report that the N terminus of CASZ1b is critical for transcriptional regulation. This domain, defined by aa 31 to 185, contains two highly conserved regions and multiple predicted phosphorylation sites (data not shown), suggesting a role in protein-protein interaction. It will be important for future studies to identify the presence and identity of coactivators or corepressors that interact with CASZ1 at this site in order to further dissect the molecular means by which CASZ1 mediates its transcriptional functions.
Neuroblastoma is an embryonic tumor of the sympathetic nervous system that is thought to arise from a defective neuronal differentiation program within neural crest cells (4, 22, 24, 26), and increased expression of CASZ1 is observed in neuroblastoma cell lines upon induction of differentiation by all-trans retinoic acid (19). Restoration of CASZ1 expression in neuroblastoma induces cell differentiation, inhibits cell movement, and suppresses cell proliferation by regulating specific gene expression patterns (19). The data presented here support a similar conclusion. Overexpression of CASZ1b induces expression of the differentiation-associated TH, NGFR, and trkA genes. These genes are important neural differentiation genes, and the loss of the NGFR gene or trkA in neuroblastoma tumors portends a poor prognosis (9, 13, 23, 28–31). The ability of various CASZ1b structural variants to activate endogenous expression of these genes that is found in 293T cells is also found in SY5Y neuroblastoma cells (Fig. 7A and B and 8A), suggesting that the critical transcription regulation domains of CASZ1b defined by using 293T cells are also critical in other cell lines. We found that the loss of transcriptional function in these variants (CASZ1b ZF4m, Δ31-185, and ΔNLS1) not only fails to induce expression of these genes but also decreases their ability to inhibit tumor growth compared to that of WT CASZ1b (Fig. 8B to D). Interestingly, based on a soft agar assay, none of these variants completely lost their function of suppressing clonogenicity. It is possible that CASZ1b utilizes different domains to recruit different cofactors and regulate distinct transcription programs. Thus, the CASZ1b variants we assessed may lose only a part of the transcriptional program regulated by WT CASZ1 transcription programs, a result which would be consistent with a partial attenuation of tumor suppression activity. This is not unusual; for example, others' studies revealed that different p53 transcriptional activation requirements, associated with different target gene expression programs, are important in the settings of acute genotoxic stress and oncogenic stimuli (2). In the future, we will test this hypothesis by assessing the different transcriptional programs regulated by CASZ1 variants. The induction of CASZ1b ΔNLS1 in SY5Y cells also slightly suppresses clonogenicity compared to that of noninduced control cells; this might be due to the 10% CASZ1b ΔNLS1 that retains nuclear localization. Although the CASZ1b Δ31-185 mutant retains some tumor suppressor activity, as evidenced by the 50% decrease in anchorage-independent growth, in vivo tumor suppressor activity was completely lost (Fig. 8). We hypothesized that CASZ1b Δ31-185 might turn on some genes that are suppressed by WT CASZ1 and that these genes might stimulate angiogenesis or stimulate cell growth only under certain microenvironments so that although the cells grow slower in soft agar, growth will not be the same in the xenografts. Our study provides the first direct evidence that the previously described CASZ1-mediated growth suppression in neuroblastoma is mediated by its transcriptional program. Given the intensive investigations of gene structure coming with next-generation sequencing approaches and the emerging sequence variations in the human genome, structure-function analyses such as our study are critical for the ultimate understanding of benign or pathological alterations in the CASZ1 gene.
By identifying and analyzing the critical domains within CASZ1b, we begin to unravel the molecular mechanisms by which neural and cardiomyocyte differentiation is regulated and move one step closer to understanding the impact of this gene on neuroblastoma pathogenesis. Collectively, these data are consistent with a model whereby CASZ1b-mediated growth suppression and gene regulation are transmitted through the transcriptional activity of the ZF1-4 domain and the N terminus of the protein, which are critical for its function.
ACKNOWLEDGMENTS
We thank Susan Garfield, head of the NIH CCR Confocal Facility, for her kind assistance and advice with the confocal microscopy images, Gregory Wray from the Institute for Genome Sciences & Policy, Duke University, for generously providing the tyrosine hydroxylase promoter-pGL4.1-luc (TH-Luc) construct, Chris Redfern and Quentin Campbell Hewson of the Northern Institute for Cancer Research, Newcastle University, United Kingdom, for providing the SY5Ytet (SY5Ytet12) cell line, Choh Yeung of the Molecular Oncology Section at the POB, NCI, for providing the HEK293T cells, and Chunxi Wang and Stanley He of the Cellular and Molecular section at the POB, NCI, for help with animal studies. We appreciate the critical review and discussion of Dinah Singer and David Levens of the CCR.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, and Center for Cancer Research.
Footnotes
Published ahead of print 13 February 2012
REFERENCES
- 1.Bagchi A, Mills AA. 2008. The quest for the 1p36 tumor suppressor. Cancer Res. 68:2551–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brady CA, et al. 2011. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145:571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan P, Donev R, Hewamana S. 2008. Targeting transcription factors for therapeutic benefit. Mol. Biosyst. 4:909–919 [DOI] [PubMed] [Google Scholar]
- 4.Brodeur GM. 2003. Neuroblastoma: biological insights into a clinical enigma. Nat. Rev. Cancer 3:203–216 [DOI] [PubMed] [Google Scholar]
- 5.Brody T, Odenwald WF. 2002. Cellular diversity in the developing nervous system: a temporal view from Drosophila. Development 129:3763–3770 [DOI] [PubMed] [Google Scholar]
- 6.Christine KS, Conlon FL. 2008. Vertebrate CASTOR is required for differentiation of cardiac precursor cells at the ventral midline. Dev. Cell 14:616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui X, Doe CQ. 1992. ming is expressed in neuroblast sublineages and regulates gene expression in the Drosophila central nervous system. Development 116:943–952 [DOI] [PubMed] [Google Scholar]
- 8.De Laurenzi V, et al. 1998. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J. Exp. Med. 188:1763–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eggert A, et al. 2000. Molecular dissection of TrkA signal transduction pathways mediating differentiation in human neuroblastoma cells. Oncogene 19:2043–2051 [DOI] [PubMed] [Google Scholar]
- 10.Grimmer MR, Weiss WA. 2006. Childhood tumors of the nervous system as disorders of normal development. Curr. Opin. Pediatr. 18:634–638 [DOI] [PubMed] [Google Scholar]
- 11.Grosskortenhaus R, Robinson KJ, Doe CQ. 2006. Pdm and Castor specify late-born motor neuron identity in the NB7-1 lineage. Genes Dev. 20:2618–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grotzer MA, Castelletti D, Fiaschetti G, Shalaby T, Arcaro A. 2009. Targeting Myc in pediatric malignancies of the central and peripheral nervous system. Curr. Cancer Drug Targets 9:176–188 [DOI] [PubMed] [Google Scholar]
- 13.Harel L, Costa B, Fainzilber M. 2010. On the death Trk. Dev. Neurobiol. 70:298–303 [DOI] [PubMed] [Google Scholar]
- 14.Hitier R, Chaminade M, Preat T. 2001. The Drosophila castor gene is involved in postembryonic brain development. Mech. Dev. 103:3–11 [DOI] [PubMed] [Google Scholar]
- 15.Isshiki T, Pearson B, Holbrook S, Doe CQ. 2001. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106:511–521 [DOI] [PubMed] [Google Scholar]
- 16.Kambadur R, et al. 1998. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 12:246–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Killick R, et al. 2011. p73: a multifunctional protein in neurobiology. Mol. Neurobiol. 43:139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, Naranjo A, Thiele CJ. 2011. CASZ1b, the short isoform of CASZ1 gene, coexpresses with CASZ1a during neurogenesis and suppresses neuroblastoma cell growth. PLoS One 6:e18557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, et al. 2011. CASZ1, a candidate tumor-suppressor gene, suppresses neuroblastoma tumor growth through reprogramming gene expression. Cell Death Differ. 18:1174–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Yang X, Tan F, Cullion K, Thiele CJ. 2006. Molecular cloning and characterization of human Castor, a novel human gene upregulated during cell differentiation. Biochem. Biophys. Res. Commun. 344:834–844 [DOI] [PubMed] [Google Scholar]
- 21.Lovat PE, et al. 2002. GADD153 and 12-lipoxygenase mediate fenretinide-induced apoptosis of neuroblastoma. Cancer Res. 62:5158–5167 [PubMed] [Google Scholar]
- 22.Maris JM. 2010. Recent advances in neuroblastoma. N. Engl. J. Med. 362:2202–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsushima H, Bogenmann E. 1993. Expression of trkA cDNA in neuroblastomas mediates differentiation in vitro and in vivo. Mol. Cell. Biol. 13:7447–7456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McConville CM, Forsyth J. 2003. Neuroblastoma—a developmental perspective. Cancer Lett. 197:3–9 [DOI] [PubMed] [Google Scholar]
- 25.Mellerick DM, Kassis JA, Zhang SD, Odenwald WF. 1992. castor encodes a novel zinc finger protein required for the development of a subset of CNS neurons in Drosophila. Neuron 9:789–803 [DOI] [PubMed] [Google Scholar]
- 26.Nakagawara A. 2004. Neural crest development and neuroblastoma: the genetic and biological link. Prog. Brain Res. 146:233–242 [DOI] [PubMed] [Google Scholar]
- 27.Rothschild G, Zhao X, Iavarone A, Lasorella A. 2006. E proteins and Id2 converge on p57Kip2 to regulate cell cycle in neural cells. Mol. Cell. Biol. 26:4351–4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schramm A, et al. 2005. Biological effects of TrkA and TrkB receptor signaling in neuroblastoma. Cancer Lett. 228:143–153 [DOI] [PubMed] [Google Scholar]
- 29.Schulte JH, et al. 2009. The low-affinity neurotrophin receptor, p75, is upregulated in ganglioneuroblastoma/ganglioneuroma and reduces tumorigenicity of neuroblastoma cells in vivo. Int. J. Cancer 124:2488–2494 [DOI] [PubMed] [Google Scholar]
- 30.Summerhill EM, Wood K, Fishman MC. 1987. Regulation of tyrosine hydroxylase gene expression during differentiation of neuroblastoma cells. Brain Res. 388:99–103 [DOI] [PubMed] [Google Scholar]
- 31.Thiele CJ, Li Z, McKee AE. 2009. On Trk—the TrkB signal transduction pathway is an increasingly important target in cancer biology. Clin. Cancer Res. 15:5962–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warner LR, et al. 2009. Functional consequences of genetic variation in primates on tyrosine hydroxylase (TH) expression in vitro. Brain Res. 1288:1–8 [DOI] [PubMed] [Google Scholar]