Abstract
Optimal lipid storage and mobilization are essential for efficient adipose tissue. Nuclear receptor peroxisome proliferator-activated receptor γ (PPARγ) regulates adipocyte differentiation and lipid deposition, but its role in lipolysis and dysregulation in obesity is not well defined. This investigation aimed to understand the molecular impact of dysfunctional PPARγ on the lipolytic axis and to explore whether these defects are also confirmed in common forms of human obesity. For this purpose, we used the P465L PPARγ mouse as a model of dysfunctional PPARγ that recapitulates the human pparγ mutation (P467L). We demonstrated that defective PPARγ impairs catecholamine-induced lipolysis. This abnormal lipolytic response is exacerbated by a state of positive energy balance in leptin-deficient ob/ob mice. We identified the protein kinase A (PKA) network as a PPARγ-dependent regulatory node of the lipolytic response. Specifically, defective PPARγ is associated with decreased basal expression of prkaca (PKAcatα) and d-akap1, the lipase genes Pnplaz (ATGL) and Lipe (HSL), and lipid droplet protein genes fsp27 and adrp in vivo and in vitro. Our data indicate that PPARγ is required for activation of the lipolytic regulatory network, dysregulation of which is an important feature of obesity-induced insulin resistance in humans.
INTRODUCTION
Peroxisome proliferator-activated receptor γ (PPARγ) is an important regulator of adipogenesis and lipogenesis. However, accumulating evidence suggests that PPARγ may also coordinate the balance between fat deposition and mobilization through its effects on lipolysis. To elucidate the relevance of PPARγ for the lipolytic response, we took advantage of a rare pparγ mutation that causes lipodystrophy and severe metabolic disturbances in humans (34). According to the crystal structure, the P467L PPARγ mutation is situated in helix 12, an important region for ligand binding and PPAR transactivation (4). Characterization of patients with genetic defects in pparγ has revealed metabolic inflexibility in their adipose tissue, resulting from impaired fat sequestration to lipid droplets (LDs) and decreased nonesterified fatty acid mobilization (34, 37). However, the dissection of the PPARγ-dependent mechanisms that impair the flux of lipids in and out of adipose tissue has proven difficult, given the small number of patients affected by these mutations. In this respect, a humanized mouse model harboring a homologous mutation (P465L) offers clear experimental advantages.
Initial analysis of the humanized P465L PPARγ mice revealed that, unlike humans, these mice exhibited normal amounts of adipose tissue and were unexpectedly insulin sensitive (13). This apparent paradox was partially resolved when the P465L PPARγ mouse was crossed onto an ob/ob background. Under this condition of extreme positive energy balance, the P465L PPARγ mutant mouse recapitulated the human phenotype characterized by insulin resistance, reduced fat mass, hypertension, and dyslipidemia. In fact, the same deleterious, synergistic dysmetabolic effect of positive energy balance combined with a dominant-negative mutation of PPARγ has been observed in humans.
The lipolytic process involves tightly coordinated hormonal and biochemical signals (23) that ensure the appropriate release of fatty acids during fasting. Upon stimulation with catecholamines, HSL is activated and translocated to the lipid droplet surface, where it interacts with specific lipid droplet proteins, such as perilipin A, FSP27, caveolin 1 (CAV-1), or ADRP. Of note, HSL works in concert with other lipases, such as ATGL, maximizing the lipolytic output.
An explanation of how defects in PPARγ affect lipolysis remains elusive. The PPARγ-dependent molecular mechanisms leading to dysfunctional lipolytic responses and how a positive energy balance synergizes with the defects in PPARγ to dysregulate lipolysis are unclear. More importantly, these mechanisms may represent a vulnerable regulatory node that is perturbed in common states of obesity and insulin resistance. Here, we investigate the molecular mechanisms that cause this metabolic inflexibility by using the humanized P465L PPARγ knock-in mouse model and evaluating their potential relevance to common forms of human obesity. We provide evidence that PPARγ is necessary for an efficient lipolytic response and identify the protein kinase A (PKA)-AKAP1 complex as an important regulatory node dependent on PPARγ activation that contributes to the dysregulation of the lipolytic response observed in mutant mice. Furthermore, we examine whether PPARγ-dependent alterations in lipolysis contribute to obesity-induced metabolic disturbances in humans.
MATERIALS AND METHODS
Animals.
Heterozygous PPARγ P465L and PPARγ P465L × ob/ob (PLO) mutant mice were generated and bred as previously detailed (13). Body composition details are listed in File S1 in the supplemental material. The mice were housed in a temperature-controlled room (24°C) with a 12-h light/dark cycle. Chow diet and water were available ad libitum. All animal protocols were approved by the United Kingdom Home Office.
Lipolysis assay.
Adipocytes were isolated from the gonadal depot using the method described by Rodbell (32). A lipolysis assay was performed as described previously (29). The agonists used in this study were noradrenaline (NA) (β1-, β2-, β3-, and α2-AR), CGP12177A (partial β3-AR and β1, β2 antagonist), forskolin (adenylate cyclase), and dibutyryl cyclic AMP (DBcAMP) (a PKA activator), as well as insulin, and cilostamide and rolipram (PDE3 and PDE4 inhibitors, respectively). Chemicals for lipolysis were obtained from Sigma and Tocris.
Human studies. (i) Cohort 1.
The cohort 1 study included morbidly obese (MO) subjects who underwent bariatric surgery and 6 nonobese subjects as controls. Clinical details of patients included in the study are given in File S2 in the supplemental material. MO patients were classified according to their insulin sensitivity. Specifically, patients with homeostasis model assessment-insulin resistance (HOMA-IR) scores of <4 were considered low insulin resistant (MO-LowIR). The MO patients with HOMA-IR scores of >8 were considered high insulin resistant (MO-HighIR). Visceral adipose tissue biopsy specimens were obtained as described previously 3.
(ii) Cohort 2.
differentiation of human preadipocytes. Isolated subcutaneous preadipocytes from lean (body mass index [BMI] < 25) and obese (BMI > 30) subjects were cultured for a period of 14 days as described previously (11). Chronic rosiglitazone treatment refers to rosiglitazone incubation (0.1 and 1 μM) during differentiation. Acute rosiglitazone treatment refers to 48 h of incubation after full differentiation (after 14 days).
Real-time PCR.
Real-time PCR was performed as previously described (24). In rodent experiments, gene expression was corrected by the geometric average of 18S, β2-microglobulin, β-actin, and 36B4 (27). Primers for human samples were obtained from Sigma-Aldrich or prevalidated TaqMan primer-probe sets from Applied Biosystems.
Western blotting.
Protein lysates from adipocytes were subjected to SDS-10% PAGE, electrotransferred to a polyvinylidene difluoride (PVDF) membrane, and probed with the following rabbit anti-mouse antibodies: HSL, pHSL-Ser660, pHSL-Ser565 ATGL, PKAcat, AKT, and β-actin (Cell Signaling).
Cell transfection and reporter assays.
HEK293 cells were cultured and transfected as described previously (16) with plasmids expressing mouse PPARγ, the P465L mutant, and RXR. The Renilla luciferase plasmid pRL-TK (Promega) was also included in the transfection as an internal control, as well as a luciferase reporter plasmid containing the first 1,500 bp of the prkaca promoter cloned in a pgl4 basic vector (Promega). At day 2, the cells underwent 24 h of incubation with or without 1 μM rosiglitazone. On day 3, lysates were prepared and a luciferase assay was performed. Luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega).
EMSA.
Electrophoretic mobility shift assay (EMSA) and supershift assays using nuclear extract from HEK293T cells overexpressing RXR/PPAR were performed as described previously (16). The oligonucleotides used in the assay included mouse aP2 ppre (40). Oligonucleotide sequences for the PKAcatα subunit, PKAcatα subunit mutant, D-AKAP1, and D-AKAP1 mutant are available upon request.
ChIP assay.
For the ex vivo chromatin immunoprecipitation (ChIP) assay, mature adipocytes from gonadal white adipose tissue (WAT) were isolated with type II collagenase. The chromatin immunoprecipitation protocol used was adapted from that of Nelson et al. (26). Shearing DNA was incubated overnight at 4°C with PPARγ antibody (sc-7273 X). Total DNA concentrations in immunoprecipitates were quantified using nanodrop ND-1000 (thermo scientific). Primers used for ChIP assays were as follows: PKA_fw, 5′-CGGGCCTCCTTATGAATGACACCG-3′; PKA_rv, 5′-GCAGCGGCTGTCCACCTGCC-3′; akap1_fw, 5′-ACCCCCAGGTGGTGGAGACA-3′; and AKAP1_rv, 5′-ACCAGGCAGGGTAGGGCAGA-3′. As negative controls, we used primers not harboring any PPAR response element (PPRE), as follows: PKA_fw, 5′-CCCAAGGACACACAGAGCCACA-3′; PKA_rv, 5′-GCAGTCCCCGAGGGGGAAAGT-3′; akap1_fw, 5′-TCTGTTGCTGTGTATAGGATGCTGC-3′; and AKAP1_rv, 5′-TGAGCGTGAGTCACCACCCAGA-3′. Enrichment was calculated according to the following equation, where F is field change, E is efficiency of PCR, and CP is the crossing point of the fluorescent signal of the PCR over the threshold: F = [E1^(CPPPREseq target gene immunoprecipitated)/E2^(CPcontrolseq target gene immunoprecipitated)]/[E3^(CPPPREseq target gene input)/E4^(CPcontrolseq target gene input)], similar to what was described previously (1). The advantage of this equation is that it corrects for the difference in the efficiencies of the different amplicons in the PCR and corrects the enrichment in respect to a control sequence without any known PPRE.
Lipidomic analysis.
A total of 1.5 million adipocytes per sample were used (wild type [wt], n = 6; P465L, n = 6). Lipidomic analysis was performed as previously described (19, 25). The data obtained were converted into netCDF file format using D bridge software from MassLynx (Waters, Inc.). The data were processed using MZmine software version 0.60 (17).
RESULTS
Mutant P465L PPARγ impairs expression of the PKA catalytic and regulatory subunits.
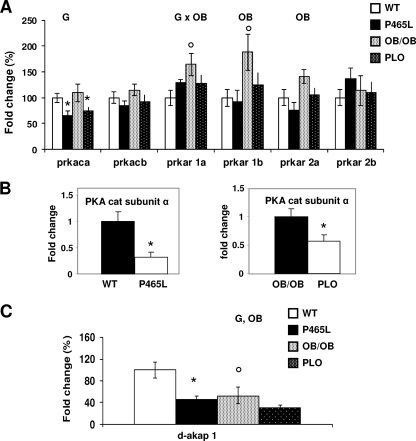
Based on a microarray data set from gonadal adipose tissue from P465L mice (data not shown), we identified and subsequently confirmed by real-time PCR a subset of genes suggestive of an impaired adrenergically mediated lipolytic response in P465L adipocytes. Isolated P465L adipocytes exhibited a selective decrease in the expression of the catalytic subunit alpha gene of PKA (prkaca) (Fig. 1A). Expression levels of the genes encoding PKA catalytic subunit beta and the regulatory proteins RI (prkar1a and prkar1b) and RII (prkar2a and prkar2b) were also measured. In agreement with the current literature, the expression levels of subunit RIα exceeded those of RIβ and showed that RIIβ was more highly expressed than RIIα. In our experimental model, the RII α and β subunits were not significantly dysregulated by the P465L mutation or by obesity, whereas the RI α and β subunits were upregulated by obesity but not by the P465L mutation. The downregulation of PKAcatα was confirmed at the protein expression level (Fig. 1B). Of note, the downregulation of PKAcatα and the lack of compensatory changes in other regulatory proteins in P465L are expected to diminish the functionality of PKA.
Fig 1.
PKAcatα and D-AKAP1 mRNA levels are decreased in P465L mutant carriers. (A) PKA subunit expression analysis of P465L mutant adipocytes from lean and obese mice versus mature adipocytes isolated from 12- to 16-week-old wt counterparts. (B) Protein levels of PKA subunits of P465L mutant adipocytes from lean and obese mice versus mature adipocytes isolated from wt counterparts. (C) akap-1 expression analysis of P465L mutant adipocytes from lean and obese mice versus mature adipocytes isolated from 12- to 16-week-old wt counterparts. The data represent means ± standard errors (SE) for 6 to 8 animals per group. Fold changes were established using the wt as 100. Differences in expression between groups were assessed by analysis of variance (ANOVA) (G, effect of P465L mutation; OB, effect of obesity; G × OB, interactive effect) and the Student t test. *, P465L versus wt or P465L/ob/ob versus ob/ob; °, wt versus ob/ob or P465L versus P465L/ob/ob. The level of probability was set at a P value of <0.05.
We also examined the expression levels of the scaffold protein AKAP1. AKAP1 is highly expressed in WAT (6) and has been suggested to create signaling platforms that function to anchor PKA regulatory subunits. It has recently been demonstrated that deletion of AKAP1 in adipocytes leads to impaired catecholamine-stimulated lipolysis (7).
The expression of d-akap1 was downregulated in P465L adipocytes from both lean P465L and obese PLO mice. This suggests d-akap1 may be a specific target of PPARγ action and identifies the PKA-AKAP1 complex as a potential PPARγ-dependent lipolytic regulatory node. Interestingly, the d-akap1 gene was also downregulated in the metabolically compromised and genetically obese ob/ob murine model (Fig. 1C).
Transcriptional regulation of prkaca and d-akap1 by PPARγ.
In silico analyses using NHR scan (33) and Patch 1.0 identified the presence of 3 candidate PPRE DR1 motifs in the first 1.5 kb upstream from the transcriptional start point of the prkaca promoter. The sequence for murine Dr1, located at bp −600, is similar to one reported in humans located at bp −781 (21) (Fig. 2). Analysis of the d-akap1 promoter also identified two DR1 motifs located 1 kb and >7 kb, respectively, upstream from the transcriptional start point. In addition, treatment of mouse adipose explants from wt mice with rosiglitazone resulted in increased mRNA levels of both genes (see File S3 in the supplemental material), suggesting that PPARγ activation may induce their transcriptional expression. Similarly, we investigated whether transcriptional regulation of these genes by PPARγ was also relevant in human adipocyte biology. Briefly, prkaca and d-akap1 mRNAs were upregulated after acute or chronic exposure to rosiglitazone in differentiated adipocytes (see Fig. 6B; see File S4 in the supplemental material).
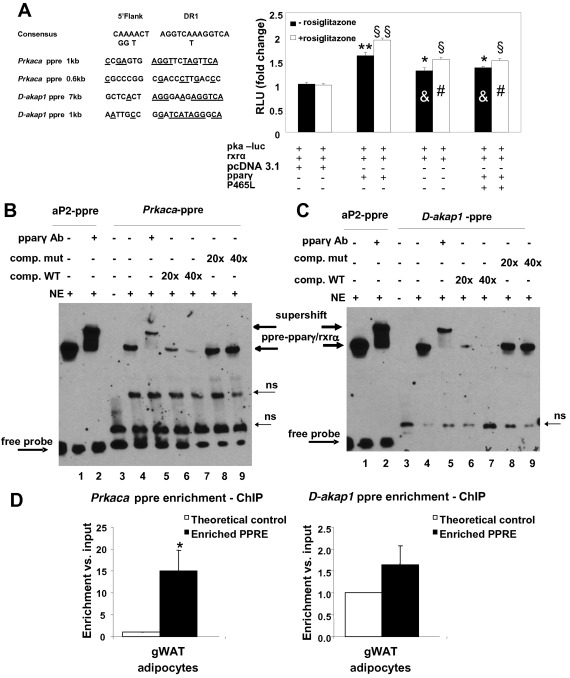
Fig 2.
Transactivation of the PKAcat subunit α promoter-luciferase reporter construct (pka-luc) by PPARγ and the PPARγ P465L mutant in HEK293T cells. (A) PPARγ transcriptionally activates prkaca. The pka-luc reporter plasmid was transiently cotransfected with a plasmid expressing RXRα together with PPARγ, the P465L mutant, or a combination of both into HEK293T cells. At 24 h posttransfection, the cells were incubated with 1 μM rosiglitazone (open bars) or vehicle (solid bars) and further incubated for 24 h before being harvested and assayed for luciferase activity. Here, we show a representative experiment out of 3 independent experiments (n = 12 for each experimental condition). The luciferase activity measured in cells transfected with pka-luc and RXRα with pcDNA 3.1 as a control for PPARγ and the P465L mutant (basal condition) in the absence of rosiglitazone was set at 1. **, P < 0.0001, and *, P < 0.001 compared to the basal condition. &, P < 0.001 compared to PPARγ transfection (− rosiglitazone). §§, P < 0.0001, and §, P < 0.001 compared to the basal condition (+ rosiglitazone). #, P < 0.0001 compared to PPARγ transfection (+ rosiglitazone). (B and C) PPARγ binds a new PPRE identified in prkaca and d-akap1 promoters. Shown are electrophoretic mobility shift and supershift assays on putative PPRE of the murine PKA (mPKA) catalytic subunit α promoter and the mAKAP1 promoter. The 3 biotin-labeled double-stranded oligonucleotide probes corresponding to the mouse PKAcat subunit α PPRE (prkaca-ppre) (B) and d-akap-PPRE (C) were incubated with 5 μg of nuclear extract (NE) of HEK293T cells overexpressing PPARγ and RXRα in the absence or presence of PPAR antibody. The gel was transferred to a nylon membrane, and the shifted bands were detected by incubating the membrane with streptavidin-horseradish peroxidase, followed by chemiluminescence detection. Lanes 1, biotin-labeled double-stranded aP2-ppre probe incubated with nuclear extract; lanes 2, biotin-labeled aP2-ppre probe incubated with nuclear extract in the presence of PPAR antibody; lanes 3, PKAcat α-ppre or D-AKAP1 biotinylated probes alone; lanes 4, PKAcat α-ppre or d-akap1 biotinated probes with nuclear extract; lane 5, PKAcat α-ppre or d-akap1 biotinated probes incubated with nuclear extract in the presence of PPAR antibody; lanes 6 and 7, for both PKAcatα and D-AKAP1, 20× and 40× excesses of unlabeled double-stranded wt oligonucleotides were included as competitors with nuclear extract and labeled wt probe; lanes 8 and 9, for both PKAcatα and D-AKAP1, 20× and 40× excesses of unlabeled double-stranded mutated oligonucleotides were included as competitors with nuclear extract and labeled wt probes. (D) PPARγ binds a new PPRE identified in the prkaca promoter in vivo. Shown is a chromatin immunoprecipitation assay of the putative PPRE of the mPKAcat subunit α promoter and mAKAP1 promoter. Isolated mature adipocytes from gonadal pads of wt mice were collected, and ChIP assays were performed, using an anti-PPARγ antibody to immunoprecipitate PPARγ-linked DNA. Quantification of PPRE sequences for both prkaca and d-akap1 promoters was performed by real-time PCR, considering the fold change with respect to a control sequence in the same gene and normalized to the input DNA as described in Materials and Methods. The data are expressed as means and standard errors of the mean (SEM) from six independent experiments. gWAT, gonadal WAT.
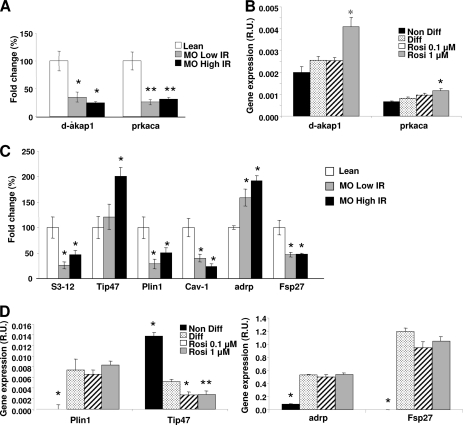
Fig 6.
(A to C) prkaca, d-akap1, and lipid droplet protein mRNA expression in adipose tissue from lean versus morbidly obese low-insulin-resistance and morbidly obese high-insulin-resistance subjects. (B and C) Adipocytes from obese subjects after acute exposure to rosiglitazone. (A) d-akap1 and prkaca mRNA expression is decreased in human obesity. (B)Rosiglitazone (Rosi) increases expression levels of d-akap1 and prkaca. *, differentiated (Diff) adipocytes. (C) Obesity severely affects the lipid droplet protein pool in human subjects. The data represent means ± SE. Differences in expression between groups were assessed by ANOVA and the Student t test. ★, P < 0.05 versus Diff; ★★, P < 0.005 versus Diff.
To investigate whether PKAcatα was a direct target of PPARγ, we performed analysis of its promoter using a luciferase assay. As shown in Fig. 2B, the luciferase activity was mildly increased (1.45-fold) relative to the basal condition after cotransfection of a pka-luc construct with pparγ and rxr plasmids. Moreover, we observed a small but robust increase after treatment with rosiglitazone, which was decreased in the presence of mutant PPARγ. We did not observe any induction when the construct was transfected with PPARα in the presence of its activator pirinix acid (WY14643) (see File S5 in the supplemental material), suggesting that PPARγ transcriptionally regulates prkaca with a degree of specificity.
EMSA confirmed that PPAR-RXR heterodimers were able to bind both the DR1 motif identified at bp −600 in the prkaca promoter and the one at >7 kb in the d-akap1 promoter, indicating that both are PPAR target genes (Fig. 2B and C, respectively). The specificity of PPARγ binding was further assessed by performing an EMSA using a set of oligonucleotides containing a mutated version of the PKAcatα subunit DR1 and another carrying the mutated version of the D-AKAP1 DR1 (see Fig. 5A) as competitors. In both cases, the presence of an excess of both mutated oligonucleotides did not interfere with the binding of PPARγ with the wt DR1.
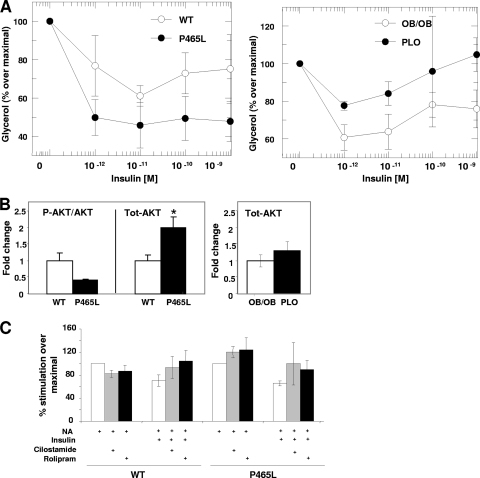
Fig 5.
(A) P465L carriers are more sensitive to the antilipolytic action of insulin. Dose-response curves show the inhibitory effect of insulin on the induced glycerol release in isolated mature white adipocytes from wt and P465L adipocytes in lean and obese mice treated with NA. (B) Increased levels of AKT in P465L carriers. Shown are the effects of the selective phosphodiesterase inhibitors cilostamide (PDE3) and rolipram (PDE4) on the lipolytic inhibition elicited by insulin in the presence of NA in lean mice. (C) PDE proteins are not responsible for the increased sensitivity to the antilipolytic action of insulin in P465L carriers. Shown are the results of Western blotting of Ser473-AKT and total AKT. Differences in expression between groups were assessed by the Student t test. The level of probability was set at a P value of <0.05.
In addition, we performed a ChIP assay to further confirm whether PPARγ was binding to the newly identified PPRE for prkaca and d-akap1 in vivo. Thus, a 15-fold enrichment for PKAcatα-immunoprecipitated DNA was observed in isolated mature adipocytes from gonadal WAT (Fig. 2D). This clearly implements the results obtained by EMSA and reinforces the idea of PPARγ as a transcriptional activator of prkaca. Unexpectedly, for d-akap1-immunoprecipitated DNA, we obtained only marginal enrichment (1.6-fold change), which suggests modest binding of PPARγ to the ppre sequence under in vivo conditions. Further research is needed to clarify this point.
Mutant P465L PPARγ impairs ATGL and HSL lipase, but not MGL, gene expression.
ATGL was downregulated in P465L adipocytes at both the mRNA and protein levels (Fig. 3A and B). Although Hsl mRNA remained unchanged, the protein levels were markedly downregulated, suggesting a posttranscriptional regulation of HSL, altered by the P465L mutation. No differences in the Ser-600 phosphorylation of HSL were observed, but Ser-565 phosphorylation, which prevents HSL activation by PKA (8), was increased under basal conditions (Fig. 3B). Interestingly, chemical inhibition of PKA signaling has been reported to increase phosphorylation of HSL at the AMPK activation site, Ser-565 (9), indicating this change in HSL may be related to dysfunctional PKA activation in P465L adipocytes. Interestingly, P465L adipocytes showed higher levels of AMPK than wt adipocytes and a small increase in phosphorylation at Thr-172, suggesting AMPK is activated in the context of dysfunctional PKA (see File S6 in the supplemental material). We extended our study to the characterization of CGI58, an ATGL-associated protein that has been shown to increase its lipolytic activity (20). abhl5 mRNA expression was decreased in obese adipocytes (ob/ob and PLO) (see File S7 in the supplemental material), suggesting the lipolytic capacity of ATGL may be further compromised in such models.
Fig 3.
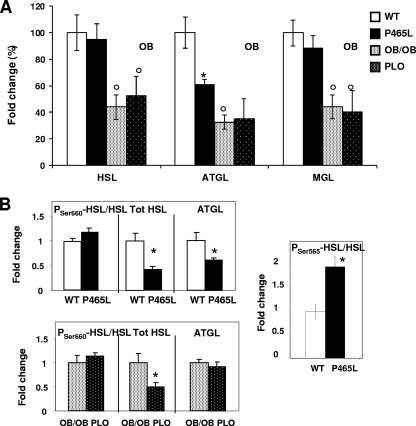
(A) atgl mRNA levels are decreased in P465L mutant carriers. Shown is lipase expression analysis of P465L mutant adipocytes from lean and obese mice versus mature adipocytes isolated from 12- to 16-week-old wt counterparts. The data represent means ± SE for 8 to 12 animals per group. Fold changes were established using the wt as 100. Differences in expression between groups were assessed by ANOVA and the Student t test. *, P465L versus wt or P465L/ob/ob versus ob/ob; °, wt versus ob/ob or P465L versus P465L/ob/ob. The level of probability was set at a P value of <0.05. (B) HSL and ATGL protein levels are decreased in P465L carriers. Shown are the results of Western blotting of ser660-HSL (Tot HSL), ser565-HSL HSL, and ATGL. Differences in expression between groups were assessed by ANOVA (G, OB, and G × OB) and the Student t test. *, P465L versus wt or P465L/ob/ob versus ob/ob. The level of probability was set at a P value of <0.05.
Mutant P465L PPARγ impairs the expression of the lipid droplet proteins ADRP and FSP27.
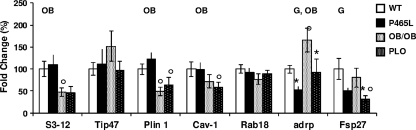
An important factor controlling the lipolytic response is the repertoire of lipid droplet-associated proteins (5, 10). adrp and fsp27 mRNA expression was downregulated in PPARγ mutant adipocytes of both lean and obese mice, indicating their strong PPARγ-dependent regulation (Fig. 4). These changes are compatible with increased lipolysis, facilitated by improved access of lipases to the lipid droplet surface (18, 30, 31). Interestingly, expression of other lipid droplet proteins, such as s3-12, perilipin A, and caveolin 1, was decreased in association with obesity and insulin resistance (ob/ob and PLO), but these changes were independent of the PPARγ mutation.
Fig 4.
Obesity and P465L PPARγ mutation modify the architecture of the lipid droplet protein pool. Shown is lipid droplet protein expression analysis of P465L mutant adipocytes from lean and obese mice versus mature adipocytes isolated from 12- to 16-week-old wt counterparts. The data represent means ± SE for 8 to 12 animals per group. Fold changes were established using the wt as 100. Differences in expression between groups were assessed by ANOVA (G, effect of P465L mutation; OB, effect of obesity; G, OB, interactive effect) and the Student t test. *, P465L versus wt or P465L/ob/ob versus ob/ob; °, wt versus ob/ob or P465L versus P465L/ob/ob. The level of probability was set at a P value of <0.05.
Impaired catecholamine-induced lipolytic response in PPARγ mutant white adipocytes.
Next, we investigated whether the changes in gene expression and protein levels translated a functional defect in the lipolytic response of PPARγ mutant mice. Our initial results indicated that the basal lipolytic response in isolated adipocytes was unaltered in PPARγ mutant adipocytes versus adipocytes from wild-type littermates in both lean and ob/ob genetic backgrounds. This suggests that in the absence of a metabolic challenge wild-type and mutant PPARγ genotypes show similar basal lipolytic responses despite changes in key elements of the lipolytic network.
However, the PPARγ mutant adipocytes exhibited an impaired lipolytic response to noradrenaline. This phenomenon was observed in adipocytes from lean PPARγ mutant mice and was associated with defective postreceptor signaling at the level of PKA response (Table 1) after dibutyril cAMP treatment. This functional defect is likely to result from the observed perturbations in the PKA–D-AKAP complex. Next, we studied whether a positive energy balance exacerbated the defect in the adrenergically stimulated lipolytic response. Obesity did impair NA-stimulated lipolysis, and this impairment was more evident in the PPARγ-defective PLO mice than in their ob/ob littermates (Table 1). As documented for the lean PPARγ mutant mice, PLO also exhibited a defect in PKA activity, further supporting a link between defective PPARγ function and altered activation of the PKA enzymatic complex. Moreover, in the context of positive energy balance (ob/ob background), the mutation for PPARγ was also associated with impaired maximal capacity of β3-AR (∼50%) and adenylyl cyclase response after forskolin treatment. In the context of these findings, our results show that the presence of the PPARγ mutation in the context of obesity further impairs catecholamine-stimulated lipolysis.
Table 1.
Effect of P465L mutation on catecholamine-induced lipolysis in isolated white mature adipocytes from lean and obese mice (females 12 to 16 weeks old)a
| Treatment | Maximal response (Emax) |
EC50 (nM) |
||||||
|---|---|---|---|---|---|---|---|---|
| Wt | P465L | ob/ob | P465L × ob/ob | Wt | P465L | Wt | P465L × ob/ob | |
| Noradrenaline | 302.5 ± 22.6 | 222.7 ± 25.8b | 103.5 ± 13.5 | 68.5 ± 12.2c | 9.8 ± 3.4 | 53.0 ± 29.8 | 10.8 ± 6.4 | 91.4 ± 88.5 |
| CGP 12177A | 113.6 ± 9.7 | 79.3 ± 8.0b | 93.4 ± 12.3 | 54.1 ± 9.9c | 509 ± 154 | 509 ± 154 | 122.3 ± 69.2 | 126.0 ± 107.9 |
| Forskolin | 123.5 ± 19.9 | 124.4 ± 16.5 | 242.2 ± 27.9 | 138.1 ± 14.5c | 8.1 ± 5.9 | 48.8 ± 33.1 | 1,330.0 ± 399.8 | 1,330.0 ± 399.8 |
| DBcAMP | 197.0 ± 19.3 | 197.0 ± 19.3 | 86.78 ± 11.3 | 86.78 ± 11.3 | 226.6 ± 118 | 53,078 ± 30,540b | 272.2 ± 212 | 3,801.2 ± 3,045.0c |
| Basal | 100 ± 8.5 | 85 ± 9.4 | 100 ± 10 | 83.8 ± 8.7 | ||||
The data are means ± standard errors. Lipolytic activity is expressed as a percentage of stimulation over basal lipolysis; a value of 100 was set up as a control for wt and ob/ob mice. Maximal action over basal lipolysis and significant differences were obtained using the GraFit computer program.
Versus the wt.
Versus ob/ob.
Increased antilipolytic response to insulin in PPARγ mutant adipocytes.
We found lean P465L mice to have increased insulin-mediated antilipolytic action compared to wild-type controls (Fig. 5A). These data were supported by increased total protein levels of AKT in PPARγ mutant adipocytes in the basal state (Fig. 5B). Next, we investigated whether this increased antilipolytic effect of insulin was due to the increased action of phosphodiesterases. The downregulation of pde3 and pde4 mRNAs in P465L adipocytes (see File S7 in the supplemental material) argues against that possibility. We did not observe differences between genotypes after treating adipocytes with PDE3 and PDE4 antagonists in the presence of insulin and NA (Fig. 5C). These data suggest that the higher antilipolytic action of insulin in P465L adipocytes is not fully mediated through phosphodiesterases. Other mechanisms explaining the antilipolitic action of insulin have been previously suggested (2, 14, 15). In the context of obesity, we found that PLO mice, unlike P465L mice, had impaired insulin-mediated antilipolytic action. These findings indicate that on a lean background, the P465L mutation decreased net lipolysis through combined effects of impairing adrenergically induced lipolysis and increasing the antilipolytic effect of insulin. However, the antilipolytic effect conferred by the PPARγ mutant in lean mice is offset by the lipolytic effect associated with obesity.
Dysregulation of PKAcatα and AKAP1 and remodeling of lipid droplet proteins are identified as a vulnerable regulatory metabolic network in human obesity.
We investigated whether the PPARγ-dependent lipolytic network comprising dysregulation of PKAcatα and AKAP1 and remodeling of lipid droplet proteins contributed to the decreased catecholamine-stimulated lipolysis typically associated with human obese and insulin-resistant states. We confirmed that prkaca and d-akap1 mRNA levels were downregulated in human obesity (Fig. 6A). Interestingly, no differences were observed between obese groups, suggesting no effect of the insulin resistance state. With respect to lipid droplet proteins, perilipin mRNA levels were downregulated in human obesity (Fig. 6C), similar to other studies (31). adrp and tip47 mRNA expression levels exhibited changes opposite those of perilipin, being upregulated in obese (LowIR and HighIR) versus lean subjects (Fig. 6C). Finally, caveolin 1 was greatly decreased in HighIR obese patients versus LowIR patients and controls. This finding suggests that defects in CAV-1 in obesity are related to the state of insulin resistance. Of note, global levels of PPARγ are decreased in morbidly obese subjects (35), supporting the concept of a direct association between PPARγ dysfunction and impairment in its lipolytic target genes in obesity.
Lipidomic analysis reveals remodeling in the adipose tissue profile of P465L adipocytes.
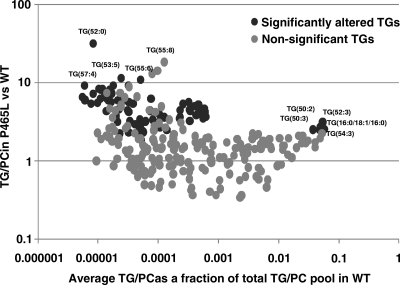
Based on the changes in the lipid droplet scaffold in PPARγ mutant adipocytes, we hypothesized that this may result in profound effects on lipid composition. We performed lipidomic analysis to assess the levels of triacylglycerides (TG) as a surrogate for fat content, as well as total phosphatidylcholine as a surrogate for membrane structures. We observed a substantial increase in the TG/phosphocholine (PC) ratios (Fig. 7). This is compatible with the existence of fewer but larger lipid droplets and defective lipolysis in PPARγ mutant adipocytes. These data also revealed substantial differences in specific sets of TG species whose pathophysiological relevance remains to be elucidated.
Fig 7.
P465L PPARγ mutation remodels the global lipidome of adipose tissue. Shown are fractional contributions to the total TG/PC pool. Significantly changed TG/PC values in P465L versus wt adipocytes are indicated.
DISCUSSION
We show that PPARγ regulates the lipolytic response through its modulatory effect on PKA-AKAP1, adipose tissue lipases, and lipid droplet proteins. We provide evidence that in obesity, a defect in PPARγ activity even further impairs the lipolytic response. Globally considered, our results indicate the presence of an important PPARγ-dependent functional regulatory node of the lipolytic axis in adipocytes. We also demonstrate that disturbances in the functionality of this node contribute to defective lipolytic responses associated with common forms of human obesity.
To determine the contribution of PPARγ to the lipolytic response, we took advantage of our P465L knock-in humanized mouse model that recreates the human P467L mutant. This mutation is known to cause a severe metabolic syndrome characterized by partial lipodystrophy, insulin resistance, and dyslipidemia in humans. However, the homologous mutation in rodents exhibited a relatively mild metabolic phenotype with only minor changes in adipose tissue morphology and distribution alongside maintained insulin sensitivity. This paradox was resolved after backcrossing the P465L knock-in into a genetically obese ob/ob background where the human metabolic phenotype was recapitulated. This indicated that despite the PPARγ defect being present, its expected pathophysiological effects become evident only when a surplus of nutrients exceeds the maximal storage and expansion of the adipose tissue allowed by the mutant PPARγ. Patients harboring the P467L mutation typically exhibit defects in triglyceride clearance in both the fasting and postprandial states. Thus, our initial hypothesis was that the metabolic phenotype of the P467L mutant was likely related to its inability to properly sequester free fatty acids (FFA) inside adipocytes, resulting in ectopic fat deposition and lipotoxicity-induced insulin resistance. More recently, Tan et al. (37) showed that functional defects in PPARγ lead to excessive release of FFA from triglyceride-rich lipoproteins, which contributes to the high systemic FFA levels exhibited by patients carrying the P467L mutation. Interestingly, in that study, adipose tissue lipolysis, measured as glycerol and NEFA outputs, was also decreased, suggesting the capacity of adipose tissue to mobilize lipids was severely compromised. Hence, the possibility that defects in PPARγ may directly contribute to an abnormal lipolytic response in adipose tissue remained. Thus, based on human data and our previous characterization of the PLO mouse, we hypothesized and confirmed that impaired lipid mobilization may balance the defects in lipid uptake, resulting in an apparently “normal” adipose tissue phenotype despite decreased adipocyte lipid turnover. Furthermore, we demonstrated that the deleterious metabolic effects typically associated with a positive energy balance and obesity are exacerbated in the presence of a defect in PPARγ.
We initially investigated whether the impaired lipolytic response in PLO mice was merely an exacerbation of the common metabolic defects associated with obesity and insulin resistance or, alternatively, the result of a primary defect related to the PPARγ mutation. In support of a PPARγ-mediated effect, our results have confirmed a direct role of PPARγ in the transcriptional control of specific functional nodes of the lipolytic axis, resulting in a defective catecholaminergic lipolytic response in PPARγ mutant adipocytes.
Interestingly, under nonstimulated conditions, PPARγ mutant adipocytes showed decreased expression of atgl mRNA and protein, as well as decreased HSL protein levels in comparison to wild types, suggesting that the functions of both lipases are affected by PPARγ dysfunction. This is in accordance with the presence of PPREs in these genes' promoters and their transcriptional regulation by rosiglitazone.
Dysregulation of lipid droplet proteins, biogenesis, and metabolism is progressively gathering momentum as a potential pathogenic mechanism mediating obesity-associated adipose tissue failure. In fact, defects in lipid droplet proteins have recently been described in models of insulin resistance and obesity (31). Here, we show that the expression of perilipin and s3-12 is significantly decreased in ob/ob mice, along with a similar tendency for fsp27 and caveolin-1. In contrast, adrp and tip47 mRNA expression levels were upregulated. It is relevant that this pattern of LD profile is fully consistent with the pattern of LD dysregulation observed in our obese cohorts and significantly differs from the natural replacement of lipid droplet proteins that occurs in normal differentiated adipocytes. The relevance of these changes in the expression of lipid droplet proteins to adipocytes is emphasized by recent in vivo and in vitro models, where the deficiency of one particular coating protein causes a hierarchical replacement by another set of proteins (36, 38, 41). However, while this allostatic remodeling in the scaffold of LD provides a certain degree of structural integrity, it also compromises the LD capacity to respond adequately to external stimuli, resulting in a net effect favoring the leakage of FFAs.
In this regard, the PPARγ mutation caused marked decreases in the expression of fsp27 and adrp in both lean and obese mice, in agreement with the presence of functional PPREs in their promoters (39). Additionally, the increase in tip47 mRNA expression observed in ob/ob adipocytes was suppressed in the presence of the PPARγ mutation. Altogether, these data suggest that defects in PPARγ affect the dynamics of the lipid droplet and prevent LDs from accumulating lipids in their cores, ultimately compromising both storage and mobilization of lipids.
Our results also indicate that the basal lipolytic response of the PPARγ mutant adipose tissue remains functional despite the decrease in the expression of lipases and the remodeling of lipid droplet proteins and the defects accumulated at different levels of the lipolytic axis. Indeed, glycerol release from isolated adipocytes appeared to be normal, suggesting that when the allostatic load is low under unstressed conditions, the deleterious metabolic effects of PPARγ dysfunction are masked or successfully compensated for. Only when the system is challenged by catecholamines do the PPARγ-dependent defects in the lipolytic machinery become pathophysiologically relevant.
In our study, we have identified the PKA complex as a vulnerable PPARγ-dependent regulatory lipolytic node. PKA is an enzyme constituted of catalytic and regulatory subunits that interact with AKAPs to form a regulatory complex that modulates the strength and responsiveness of PKA and helps to maintain the compartmentalization of cAMP pools. Interestingly, in the obese state, the sensitivity of PKA to catecholamines is reduced as a result of changes in the ratio of the regulatory subunits RI/RII (28). The increase in the expression of PKA regulatory subunits in ob/ob and PLO mice compared to their controls fully supports this finding. Interestingly, we observed that the PPARγ mutation caused a selective downregulation of the catalytic subunit alpha of PKA, which itself is enough to justify a functionally defective PKA. This appears to be induced by dysfunctional PPARγ, as it was observed not only in the PLO mice, but also in the P465L lean mice. Furthermore, our results indicate that PPARγ binds and activates the prkaca promoter and that the presence of the PPARγ mutation impairs such activation even in the presence of the PPARγ activator rosiglitazone. We observed a similar pattern of downregulation for d-akap1 in both the lean and obese P465L mutants versus wild types. We confirmed that PPARγ binds directly to the d-akap1 promoter, identifying for the first time free fatty acids (FFA) and d-akap1 genes as direct PPARγ target genes. The question of determining the strength of such transcriptional activation remains unresolved. Another interesting finding that requires further investigation is that the lean PPARγ mutants display increased sensitivity to the antilipolytic action of insulin but that this effect is lost in the context of obesity. We also show the relevance of the dysregulation of this PPARγ-dependent lipolytic regulatory node in common forms of obesity. In fact, adipose tissue from obese subjects exhibits specific downregulation of both prkaca and d-akap1 mRNAs (Fig. 6A). Our data are in agreement with previous data identifying D-AKAP1 as a potentially downregulated gene in the human obesity data set (12, 22).
In summary, we have shown, using the human P465L PPARγ mutant, that defective PPARγ causes impairment in catecholamine-induced lipolysis. Specifically, we have identified a dysfunction of the PKA-AKAP1 complex, as well as defects in the expression of major lipases and specific dysregulation of lipid droplet proteins associated with defective PPARγ. In the context of a positive energy balance, P465L mutation further impairs catecholamine-stimulated lipolysis through defects located at both the adrenoreceptor and postreceptor levels. It is relevant that the molecular mechanisms that characterize the impairment in lipolytic activity elicited by defective PPARγ were recapitulated in common forms of human obesity. Our data demonstrate that PPARγ coordinates both catabolic and anabolic processes in the adipocyte and that this effect is fundamental to maintaining the metabolic flexibility required for the efficient management of nutritional fluxes in and out of adipose tissue.
Supplementary Material
ACKNOWLEDGMENTS
We thank Erik Schoenmakers (Institute of Metabolic Science, University of Cambridge, Cambridge, United Kingdom) for providing us with the pgl4 vector and for his excellent technical advice and Andrew Whittle (Institute of Metabolic Science, University of Cambridge, Cambridge, United Kingdom) for his useful comments and suggestions. We also thank Janice Carter, Dan Hart, and Helen Westby for the excellent husbandry of the PPARγ-P465L mutant colony.
This work was funded by Diabetes UK, an MRC Programme grant, MITIN (Integration of the System Models of Mitochondrial Function and Insulin Signaling and its Application in the Study of Complex Diseases), and the Cambridge NIHR Biomedical Research Centre (A.V.-P.); FP6 HEPADIP (A.V.-P. and M.O.); the Human Frontier Science Programme (M.O.); Junta de Andalucia (P08-CTS-04369) and the Fondo de Investigacion Sanitaria (PS09/00997) (F.J.T.); and the Ministerio de Educacion y Ciencia (SAF2008-02073) and the Instituto de Salud Carlos III-CIBERobn (J.M.F.-R.).
S. R.-C. and S.C. performed the ex vivo experiments in mice (RNA and protein expression analysis and lipolysis, EMSA, ChIP, and luciferase assays) and collected and analyzed the data. V.R.V. and M.O. developed analytical platforms and performed and analyzed lipidomic experiments. N.B., J.M.M.-N., F.J.T., and J.M.F.-R. held and characterized the human cohorts, performed experiments and collected gene expression data from adipose depots, and analyzed the data. S.R.-C., S.C., and A.V.-P. designed the experiments. S.R.-C., S.C., N.B., J.M.M.-N., F.J.T., J.M.F.-R., V.R.V., M.O., and A.V.-P. discussed the manuscript. S.R.-C., S.C., and A.V.-P. coordinated and directed the project. S.R.-C. and A.V.-P. developed the hypothesis and wrote the manuscript.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print 6 March 2012
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1. Aparicio O, Geisberg JV, Struhl K. 2004. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Cell Biol. Chapter 17: Unit 17.7 [DOI] [PubMed] [Google Scholar]
- 2. Baltensperger K, et al. 1996. The beta-adrenergic receptor is a substrate for the insulin receptor tyrosine kinase. J. Biol. Chem. 271:1061–1064 [DOI] [PubMed] [Google Scholar]
- 3. Barbarroja N, et al. 2010. The obese healthy paradox: is inflammation the answer? Biochem. J. 430:141–149 [DOI] [PubMed] [Google Scholar]
- 4. Barroso I, et al. 1999. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature 402:880–883 [DOI] [PubMed] [Google Scholar]
- 5. Brasaemle DL. 2007. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 48:2547–2559 [DOI] [PubMed] [Google Scholar]
- 6. Bridges D, MacDonald JA, Wadzinski B, Moorhead GB. 2006. Identification and characterization of D-AKAP1 as a major adipocyte PKA and PP1 binding protein. Biochem. Biophys. Res. Commun. 346:351–357 [DOI] [PubMed] [Google Scholar]
- 7. Choi S. 2008. Insulin regulates adipocyte lipolysis via a compartmentalized AKT independent signaling pathway. Ph.D. dissertation. University of Pennsylvania, Philadelphia, PA: http://repository.upenn.edu/dissertations/AAI3345919. [Google Scholar]
- 8. Daval M, et al. 2005. Anti-lipolytic action of AMP-activated protein kinase in rodent adipocytes. J. Biol. Chem. 280:25250–25257 [DOI] [PubMed] [Google Scholar]
- 9. Djouder N, et al. 2010. PKA phosphorylates and inactivates AMPKalpha to promote efficient lipolysis. EMBO J. 29:469–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ducharme NA, Bickel PE. 2008. Lipid droplets in lipogenesis and lipolysis. Endocrinology 149:942–949 [DOI] [PubMed] [Google Scholar]
- 11. Fernandez-Real JM, et al. 2010. Study of caveolin-1 gene expression in whole adipose tissue and its subfractions and during differentiation of human adipocytes. Nutr. Metab. 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gomez-Ambrosi J, et al. 2004. Gene expression profile of omental adipose tissue in human obesity. FASEB J. 18:215–217 [DOI] [PubMed] [Google Scholar]
- 13. Gray SL, et al. 2006. Leptin deficiency unmasks the deleterious effects of impaired peroxisome proliferator-activated receptor gamma function (P465L PPARgamma) in mice. Diabetes 55:2669–2677 [DOI] [PubMed] [Google Scholar]
- 14. Hadcock JR, Port JD, Gelman MS, Malbon CC. 1992. Cross-talk between tyrosine kinase and G-protein-linked receptors. Phosphorylation of beta 2-adrenergic receptors in response to insulin. J. Biol. Chem. 267:26017–26022 [PubMed] [Google Scholar]
- 15. Houslay MD. 1998. Adaptation in cyclic AMP signalling processes: a central role for cyclic AMP phosphodiesterases. Semin. Cell Dev. Biol. 9:161–167 [DOI] [PubMed] [Google Scholar]
- 16. Jitrapakdee S, et al. 2005. The peroxisome proliferator-activated receptor-gamma regulates murine pyruvate carboxylase gene expression in vivo and in vitro. J. Biol. Chem. 280:27466–27476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Katajamaa M, Miettinen J, Oresic M. 2006. MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics 22:634–636 [DOI] [PubMed] [Google Scholar]
- 18. Keller P, et al. 2008. Fat-specific protein 27 regulates storage of triacylglycerol. J. Biol. Chem. 283:14355–14365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Laaksonen R, et al. 2006. A systems biology strategy reveals biological pathways and plasma biomarker candidates for potentially toxic statin-induced changes in muscle. PLoS One 1:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lass A, et al. 2006. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 3:309–319 [DOI] [PubMed] [Google Scholar]
- 21. Lemay DG, Hwang DH. 2006. Genome-wide identification of peroxisome proliferator response elements using integrated computational genomics. J. Lipid Res. 47:1583–1587 [DOI] [PubMed] [Google Scholar]
- 22. Marrades MP, Gonzalez-Muniesa P, Martinez JA, Moreno-Aliaga MJ. 2010. A dysregulation in CES1, APOE and other lipid metabolism-related genes is associated to cardiovascular risk factors linked to obesity. Obes. Facts 3:312–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin S, et al. 2009. Spatiotemporal regulation of early lipolytic signaling in adipocytes. J. Biol. Chem. 284:32097–32107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Medina-Gomez G, et al. 2005. The link between nutritional status and insulin sensitivity is dependent on the adipocyte-specific peroxisome proliferator-activated receptor-gamma2 isoform. Diabetes 54:1706–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Medina-Gomez G, et al. 2009. Adaptation and failure of pancreatic beta cells in murine models with different degrees of metabolic syndrome. Dis. Model Mech. 2:582–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nelson JD, Denisenko O, Bomsztyk K. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1:179–185 [DOI] [PubMed] [Google Scholar]
- 27. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. 2004. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26:509–515 [DOI] [PubMed] [Google Scholar]
- 28. Planas JV, Cummings DE, Idzerda RL, McKnight GS. 1999. Mutation of the RIIbeta subunit of protein kinase A differentially affects lipolysis but not gene induction in white adipose tissue. J. Biol. Chem. 274:36281–36287 [DOI] [PubMed] [Google Scholar]
- 29. Pujol E, et al. 2003. Gender- and site-related effects on lipolytic capacity of rat white adipose tissue. Cell. Mol. Life Sci. 60:1982–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Puri V, Czech MP. 2008. Lipid droplets: FSP27 knockout enhances their sizzle. J. Clin. Invest. 118:2693–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Puri V, et al. 2008. Cidea is associated with lipid droplets and insulin sensitivity in humans. Proc. Natl. Acad. Sci. U. S. A. 105:7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rodbell M. 1964. Metabolism of isolated fat cells. I. Effects of hormones on glucose metabolism and lipolysis. J. Biol. Chem. 239:375–380 [PubMed] [Google Scholar]
- 33. Sandelin A, Wasserman WW. 2005. Prediction of nuclear hormone receptor response elements. Mol. Endocrinol. 19:595–606 [DOI] [PubMed] [Google Scholar]
- 34. Savage DB, et al. 2003. Human metabolic syndrome resulting from dominant-negative mutations in the nuclear receptor peroxisome proliferator-activated receptor-gamma. Diabetes 52:910–917 [DOI] [PubMed] [Google Scholar]
- 35. Sewter C, Blows F, Considine R, Vidal-Puig A, O'Rahilly S. 2002. Differential effects of adiposity on peroxisomal proliferator-activated receptor gamma1 and gamma2 messenger ribonucleic acid expression in human adipocytes. J. Clin. Endocrinol. Metab. 87:4203–4207 [DOI] [PubMed] [Google Scholar]
- 36. Sztalryd C, et al. 2006. Functional compensation for adipose differentiation-related protein (ADFP) by Tip47 in an ADFP null embryonic cell line. J. Biol. Chem. 281:34341–34348 [DOI] [PubMed] [Google Scholar]
- 37. Tan GD, et al. 2008. Fatty acid metabolism in patients with PPARgamma mutations. J. Clin. Endocrinol. Metab. 93:4462–4470 [DOI] [PubMed] [Google Scholar]
- 38. Tansey JT, et al. 2001. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc. Natl. Acad. Sci. U. S. A. 98:6494–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Targett-Adams P, et al. 2005. A PPAR response element regulates transcription of the gene for human adipose differentiation-related protein. Biochim. Biophys. Acta 1728:95–104 [DOI] [PubMed] [Google Scholar]
- 40. Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. 1994. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 8:1224–1234 [DOI] [PubMed] [Google Scholar]
- 41. Wolins NE, et al. 2005. S3-12, adipophilin, and TIP47 package lipid in adipocytes. J. Biol. Chem. 280:19146–19155 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.