Abstract
Tissue-specific alternative splicing is achieved through the coordinated assembly of RNA binding proteins at specific sites to enhance or silence splicing at nearby splice sites. We used high-throughput sequencing (RNA-Seq) to investigate the complete spectrum of alternative splicing events that are regulated by the epithelium-specific splicing regulatory proteins ESRP1 and ESRP2. We also combined this analysis with direct RNA sequencing (DRS) to reveal ESRP-mediated regulation of alternative polyadenylation. To define binding motifs that mediate direct regulation of splicing and polyadenylation by ESRP, SELEX-Seq analysis was performed, coupling traditional SELEX with high-throughput sequencing. Identification and scoring of high-affinity ESRP1 binding motifs within ESRP target genes allowed the generation of RNA maps that define the position-dependent activity of the ESRPs in regulating cassette exons and alternative 3′ ends. These extensive analyses provide a comprehensive picture of the functions of the ESRPs in an epithelial posttranscriptional gene expression program.
INTRODUCTION
Nearly all human multiexon gene transcripts undergo alternative splicing, and most also undergo multiple alternative splicing events within the same transcript (29, 46). As a result, alternative splicing provides a mechanism that broadens gene expression through a nearly exponential expansion of the number of distinct gene products that can be produced from the rather modest number of ∼20,000 human genes. The regulation of alternative splicing is mediated by RNA binding proteins (RBPs) that interact with exonic and intronic sequences and function as splicing enhancers or silencers, depending on the regulator and/or binding context (27). This regulation is combinatorial, with the splicing outcome being determined by the net activities of RBPs that bind within or near alternative exons (1, 20). While many of the regulatory factors, such as the well-characterized hnRNP and SR families of proteins, are generally ubiquitously expressed, the number of known regulators with cell or context-specific expression is growing (6). Technological advances in the past several years have led to the identification of the genome-wide targets for several well-characterized splicing regulators, such as Nova1/2, the Fox family proteins, and PTBP1/PTBP2 (reviewed in reference 12). Most of the targets described to date consist of simple cassette exons, and these regulators can induce either splicing or skipping of different exons. A recurring theme has been that splicing regulators induce either exon splicing or skipping in a position-dependent manner. These observations have given rise to the concept of “RNA maps,” whereby the binding position of the protein relative to a regulated alternative exon determines whether it promotes or represses splicing. These RNA maps along with other experimental and bioinformatics data suggest a broader “splicing code,” wherein the collective identification of the expression levels and binding sites for all splicing regulators can predict and determine the splicing patterns in different cell types and cellular milieus (1). However, the catalog of known splicing factors is incomplete and the binding sites and RNA maps have thus far been determined for only a limited number of splicing regulators. Therefore, the definition of the broader splicing code requires a more complete characterization of the genome-wide activities of an expanded set of splicing factors, including those with more restricted expression patterns.
We identified ESRP1 and ESRP2 as paralogous epithelial cell-type-specific splicing proteins that play a role in the epithelial-to-mesenchymal transition (EMT), an important developmental process that also can contribute to cancer metastasis (38, 47–49). Previous analyses used splicing-sensitive exon and human exon junction (HJAY) microarrays to define a set of alternative cassette exons and simple 3′ or 5′ splice sites that they regulate (47, 49). However, existing microarray platforms are generally limited to the detection of simple changes in splicing for which they contain representative probe sets. To more broadly define the ESRP-regulated splicing network, as well as ESRP-mediated regulation of alternative polyadenylation, we used high-throughput sequencing (RNA-Seq) and direct RNA sequencing (DRS) of mRNA 3′ ends. We also carried out an unbiased determination of the ESRP1 binding motif by harnessing the power of systematic evolution of ligands by exponential enrichment (SELEX) in conjunction with RNA-Seq (SELEX-Seq). Mapping of these motifs within the broader genome-wide ESRP-regulated splicing network yielded an RNA map that describes the position-dependent functions of the ESRPs. Together our results define a network of ESRP targets that encode proteins that function in pathways and protein interaction networks that are likely to have important roles in epithelial mesenchymal transitions in development and disease.
MATERIALS AND METHODS
Cell culture, transfection, and transduction.
PNT2, MDA-MB-231, and 293T cells were maintained, transfected, and transduced as described previously (48).
Library preparation and sequencing.
Sequencing libraries were prepared using mRNA-Seq sample preparation kits (Illumina) according to the manufacturer's instructions. Total RNA (10 μg) was used to prepare poly(A) RNA for fragmentation, followed by cDNA synthesis with random hexamers and ligation to Illumina adaptor sequences. The samples were quantified using an Agilent 2100 Bioanalyzer, loaded onto flow cells for cluster generation, and sequenced on an Illumina IIx genome analyzer using a single-read protocol to generate 76-bp reads (Illumina).
Splice junction database.
We constructed a database of splice junctions in human genes using the Ensembl transcript annotations (release 57) (9). The database includes all known splice junctions observed in Ensembl transcripts, as well as hypothetical splice junctions obtained by all possible pairwise fusions of exons within genes. In total, the database contains ∼3.5 million splice junctions.
Mapping of RNA-Seq reads.
During quality assessment of the 76-bp single-end reads obtained by RNA-Seq, we found that the first two 25-bp segments of these reads had a high rate of mapping to the human genome, while the third 25-bp segment had a much lower mapping rate due to increased sequencing error near the 3′ ends of RNA-Seq reads. Thus, we decided to use the first 50 bp of each read for mapping and subsequent analysis. We mapped RNA-Seq reads to the human genome (hg19) and the splice junction database, using the software Bowtie (18) and allowing up to three mismatches. Each mapped splice junction read required at least 8 bp from both sides of the splice junction. To obtain unique junction mapping reads, we removed splice junction reads that mapped to either the human genome (hg19) or multiple splice junctions. Each junction sequence is 84 bp long and is composed of the last 42 bp of the 5′-end exon and the first 42 bp of the 3′-end exon, thereby ensuring that at least 8 bp of a 50-bp read can be mapped across the junction.
Detection of differential alternative splicing using RNA-Seq data.
For each alternatively spliced cassette exon detected by the RNA-Seq data, we calculated its exon inclusion level (ψ) in any given sample, using the counts of reads that uniquely mapped to its upstream splice junction (UJC), downstream splice junction (DJC), and skipping splice junctions (SJC), as follows: [(UJC + DJC)/2]/[(UJC + DJC)/2 + SJC].
To detect differential alternative splicing events between two samples, we developed a multivariate Bayesian algorithm called MATS (multivariate analysis of transcript splicing). MATS uses a multivariate uniform prior to model the between-sample correlation in exon splicing patterns and a Markov chain Monte Carlo (MCMC) method coupled with a simulation-based adaptive sampling procedure to calculate the P value and false discovery rate (FDR) of differential alternative splicing. The MATS approach provides the flexibility to identify differential alternative splicing events that match a given user-defined threshold. Suppose that ψ1 and ψ2 are the estimated exon inclusion levels of an exon in two conditions; in this work we used MATS to calculate the P value and FDR for a value of | ψ1 − ψ2 | that is <0.05, representing a difference of at least 5% in exon inclusion levels between the two conditions. The same approach was also used to identify differential alternative 5′ and 3′ splice site usage. In this case we calculated the percentage of alternative 3′ and 5′ splice sites using long junction counts (LJC) (i.e., the number of junctions reads mapping to the isoform that results in a longer exon) divided by the sum of long junction counts and short junction counts (SJC). The MATS software can be downloaded from http://intron.healthcare.uiowa.edu/mats/, and further details are provided elsewhere (36).
RT-PCR validations.
RNA-Seq-predicted ESRP-regulated splicing events that passed the 5% FDR threshold were selected for validation based primarily on an unbiased identification of all cassette exons that were readily amenable to unambiguous reverse transcription-PCR (RT-PCR)-based interpretation. Thus, nearly all simple cassette exons with exon sizes under 300 nucleotides (nt) that were flanked by constitutive exons were tested. However, a limited number of events that occurred in transcripts that encoded functionally relevant proteins were also chosen. In addition, given the substantial false-negative rate and to also test a limited subset of predicted events that did not meet our stringent 5% FDR cutoff, we also selected an additional 33 cassette exons for validation (see Table S3 in the supplemental material). Quantification of alternative splicing was performed using standard RT-PCR incorporating radiolabeled dCTP or high-throughput (HT) RT-PCR at the Université de Sherbrooke as described previously (47). Complete HT-RT-PCR data can be accessed at http://palace.lgfus.ca.
Antibodies and immunoblotting.
Cell lysates were resolved on 4 to 12% NuPAGE bis-Tris gels (Invitrogen) and immunoblotted as described previously (48). Antibodies used were anti-β-actin (1:1,000; AC-15; Sigma), anti-Flag (1:1,000; Stratagene), and anti-Rbm9 (1:2,000; Bethyl Laboratories).
SELEX-Seq.
Random 20-mer RNA sequences were generated from a 103-base DNA oligonucleotide containing a T7 promoter, restriction sites, 20 random bases, and an SP6 promoter sequence. The full sequence (with N standing for randomized bases) is AATTTATAATACGACTCACTATAGGGAGAAAGTTGGCCGCAGTATCGATANNNNNNNNNNNNNNNNNNNNCTCGAGTTCTATAGTGTCACCTAAATCAAGCTT.
This oligonucleotide was made into a double-stranded DNA library using Klenow Exo Minus (Promega). This randomized double-stranded DNA pool was blunt cloned into the pCR-Blunt vector and used as a template for T7 transcription per the manufacturer's instructions (Ambion).
A recombinant glutathione S-transferase (GST)-Esrp1 fusion protein was produced and purified as previously described (47). GST-Esrp1 was bound to GST beads rotating at 4°C for 30 min, and RNA was also precleared against GST beads. The randomized RNA pool was incubated with GST-Esrp1 at a 30:1 to 40:1 molar ratio of RNA to protein in a 200-μl binding reaction mixture (16 mM HEPES [pH 7.9], 95 mM KCl, 2 mM MgCl2, 60 μM EDTA, 1 mM dithiothreitol [DTT], 6% glycerol, 0.1 mM heparin [Sigma H-3393], and 0.15 mM phenylmethylsulfonyl fluoride [PMSF]). Binding reaction mixtures were incubated with rotation at room temperature for 30 min and washed five times with the same binding buffer. Bound RNA was isolated from beads using TRIzol reagent according to the manufacturer's instructions (Invitrogen) and used for reverse transcription with an SP6-specific primer to create a cDNA library from the bound RNA. This cDNA was then used as a template for a T7 transcription. This cycle was repeated for a total of seven rounds. In the seventh round, the KCl concentration for binding and washes was increased to 300 mM.
Sequencing libraries were prepared from round 0, 2, 3, 6, and 7 cDNA pools using a PCR strategy with modified Illumina adaptor sequences containing four nucleotide barcodes specific to each round and sequenced with an Illumina II genome analyzer to generate 44-bp single-end reads.
SELEX-Seq data analysis.
The SELEX-Seq sequences are 44 nt in length. Each sequence starts with a 4-nt barcode, followed by a 16-nt constant region that includes a ClaI site and then by a 20-nt region of random sequences (the sequences selected by SELEX), and ends with a 4-nt constant region that marks the start of the XhoI site. From the total 32.4 million reads, we removed reads with ambiguous barcodes, reads with undetermined nucleotides (N) within the 20-nt random sequence region, and reads with mutated ClaI or XhoI sites. To avoid PCR amplification bias, we also removed redundant 20-nt random sequences from each round and kept unique 20-nt sequences for ESRP motif analysis. These filtering steps resulted in a final set of 19.8 million reads from rounds 0, 2, 3, 6, and 7 of SELEX. (We note that the motif analysis was not significantly different when the same analysis was performed without removing redundant sequences.) We then enumerated all 4,096 possible 6-mers and calculated the frequency of each 6-mer in the 20-nt random sequence region from each round of SELEX, defined as the total occurrence of a 6-mer divided by the total number of unique 6-mer positions within all 20-nt random sequence regions from a given round. We ranked all possible 6-mers in each round based on their relative frequencies. We noted that the top 12 6-mers from the final round (round 7) (see Table S4 in the supplemental material) all exhibited progressive increases in relative frequencies from early to late rounds of SELEX, including a few that matched previous bioinformatically predicted ESRP binding sites (47). Thus, these top 12 6-mers were selected to derive an ESRP motif score in the RNA map analysis (see below). We also calculated a position weight matrix (PWM) for 6-mers and 8-mers using an approach previously applied to SELEX-Seq data for transcription factor binding sites (15).
RNA electrophoretic mobility shift assay (EMSA) analysis.
Selected sequences for in vitro transcription were inserted into the ClaI-Xho restriction sites in plasmid pDP19RC-ΔEE, and transcription by T7 polymerase (Ambion) after XhoI digestion was carried out as described previously (13). [32P]UTP-radiolabeled RNAs were used at a specific activity of 2.4 × 108 cpm/μg, and 50,000 cpm (∼8 to 13 fmol) of radiolabeled RNA and GST-Esrp1 were incubated in a 10-μl binding reaction mixture (16 mM HEPES [pH 7.9], 95 mM KCl, 2 mM MgCL2, 60 μM EDTA, 1 mM DTT, 6% glycerol, 0.1 mM heparin [Sigma H-3393], and 0.15 mM PMSF). Binding reaction mixtures were incubated at 30°C for 30 min, loaded on 4% nondenaturing Tris-borate acrylamide gels, and electrophoresed at 4°C for 1.5 h at 200 V.
Minigene analysis of alternative 5′ and 3′ splice sites.
LPPR2 and HNRNPH3 minigene sequences were PCR amplified from genomic DNA isolated from PNT2 cells. The PCR products were cloned into the NotI and EcoRV sites of the pI-11(-H3)-PL adenovirus based splicing minigene (14). Point mutations were introduced via site-directed mutagenesis with a QuikChange kit (Stratagene). Cotransfections of minigene plasmids and EV and ESRP expression plasmids were performed as described previously (47).
RNA map analysis.
We used the top 12 6-mers identified in the SELEX-Seq data to define an ESRP “binding score” using an approach similar to that previously used to define predicted Nova binding sites (41). We assigned the score based on the overall percentage of nucleotides covered by any of these top 6-mers within a 45-nt window and slid this window in 1-nt increments across the ESRP-regulated cassette exons and the 250 nt of intronic sequences flanking these exons as well as the upstream and downstream exons. The complete set of ESRP-regulated exons identified either in previous Affymetrix exon 1.0/HJAY analyses or by RNA-Seq, with validated switches in splicing of at least 10% in either experimental system, comprised a total of 276 exons (see Table S3 in the supplemental material). Of these, 103 were ESRP-enhanced exons and 173 were ESRP-silenced exons. As a control data set we used 3,508 alternatively spliced exons that are present on HJAY arrays and in genes expressed in the cell lines used here but for which there was no evidence of ESRP-mediated regulation. For each position of the RNA map, we calculated the P value of the Wilcoxon rank sum test for ESRP-enhanced versus background values and ESRP-silenced versus background values, respectively. RNA map analysis for alternative polyadenylation was similarly performed using the ESRP binding motif score and evaluating enrichment relative to a background set of 9,016 alternative poly(A) sites from DRS analysis that were not predicted to be regulated by ESRPs (with FDR > 0.5) at each position 250 nt upstream and downstream of the poly(A) site.
Identification of ESRP-regulated changes in polyadenylation.
We used a recently described method for direct RNA sequencing (DRS) of 3′ ends of poly(A) RNA and sequenced one channel for control MDA-MB-231 cells and another for cells ectopically expressing ESRP1 (28). The sequencing was performed by Helicos Biosciences and we aligned direct RNA sequencing reads to human genome assembly 19 (hg19) using the indexDPgenomic tool in Helisphere at http://open.helicosbio.com/mwiki/index.php/Releases. This analysis yielded 3,338,956 and 3,537,072 uniquely mapped reads for the control and ESRP expressing samples, respectively. The uniquely mapped reads with a minimal mapped length of 25 and alignment score of 4.0 were kept for further analysis. We first identified the 5′ ends of mapped reads as individual poly(A) sites. We next filtered all reads for those arising from internal poly(A) priming using a previously described approach (11). To construct a consensus poly(A) annotation for downstream analysis, we used pooled data from both EV and ESRP experiments to iteratively cluster all individual poly(A) sites within 40 nt to its nearest poly(A) site on the same chromosome strand. The weighted coordinate, which was calculated as the sum of the product of the coordinate of an individual poly(A) and its percentage of usage in the whole cluster, was taken as the representative coordinate of the corresponding poly(A) cluster. The frequencies of poly(A) clusters in the different samples were calculated according to the above consensus coordinates of poly(A) clusters in the pooled data. Next, the poly(A)s residing in the whole gene region, including exons, introns, and the downstream 500-nt region of the terminal exon, were collected as possible poly(A)s of a certain gene. The Fisher exact test was conducted on all possible pairs of poly(A)s of one gene in two different experiments (EV and ESRP) to test whether there is a change in relative usage of two poly(A)s, and the Benjamini-Hochberg method was used to calculate FDR. The pairs of poly(A)s with FDRs less than 0.05 were defined as statistically significant events. We used single-end RNA-Seq reads for EV and ESRP experiments to infer the exon-exon junctions, which are used to classify APA events [alternative poly(A) sites on the same terminal exon or 3′ untranslated region (UTR)]. The junction prediction was done in TopHat (39). The predicted junctions and known gene annotation were taken together to do the APA type classification. Three categories are assigned: APA events (alternative polyadenylation within the same 3′ UTR, APA3 events [alternative poly(A) sites coupled with alternative 3′ splice site choices], and APA5 events [alternative poly(A) sites coupled with alternative 5′ splice site choices]; events falling outside the above three categories are classified as “other”. To investigate the agreement between DRS and RNA-Seq data and to validate the DRS predictions, we counted RNA-Seq reads within 300-nt upstream regions of poly(A)s and conducted a one-sided Fisher exact test based on RNA-Seq reads. We defined consensus validated events as those with an FDR of less than 0.05 from DRS and a P value of less than 0.01 from RNA-Seq with the same direction of change in both data sets. We also filtered using a cutoff of at least a 10% change in poly(A) site use from DRS and discarded events that could not be classified as APA, APA3, or APA5. For the purposes of investigating ESRP binding motifs within the events we also removed significant APA genes with more than two poly(A)s for drawing an RNA map. We also noted that a number of the APA3 and APA5 type events corresponded to comparison of two or more closely approximated poly(A) sites with a single alternative poly(A), and we therefore retained only the most representative comparison within that gene with the most significant P value.
Functional interaction network analysis.
All network analysis was performed with the Reactome FI Cytoscape plugin (33, 50). The extended 209,988 protein functional interactions (FIs) were obtained from (50). The pathway enrichment analysis was performed for each module that contains both ESRP targets and linker genes, and we calculated the enriched pathways from six pathway databases: Reactome (R) (43), Panther (P) (24), CellMap (C) (http://cancer.cellmap.org), NCI-Nature (N) (http://pid.nci.nih.gov), NCI-BioCarta (B) (http://pid.nci.nih.gov), and KEGG (K) (16). FDR values were calculated based on 1,000 permutations on all genes in the PPI network.
RESULTS
Extending the spectrum of ESRP-regulated splicing using RNA-Seq analysis.
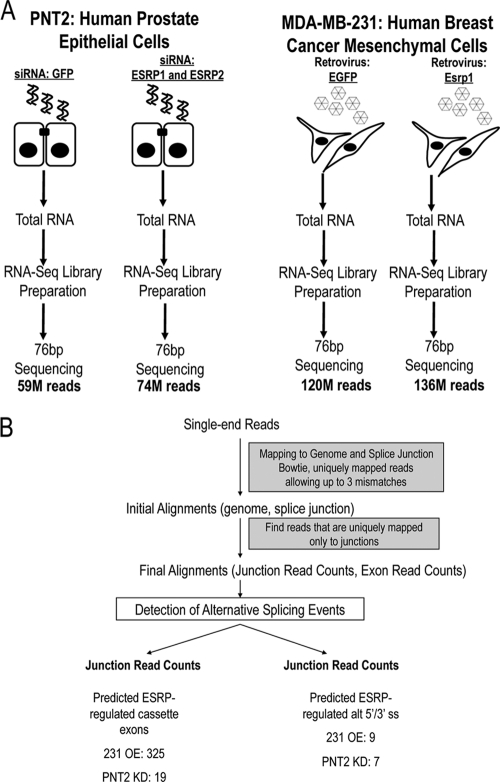
We carried out whole-transcriptome sequencing by RNA-Seq to identify changes in splicing in the presence and absence of ESRP. We prepared RNA-Seq libraries from epithelial cells (PNT2) in which ESRP1 and ESRP2 are knocked down by small interfering RNAs (siRNAs) and mesenchymal cells (MDA-MB-231) in which ESRP1 is ectopically expressed, as well as their respective controls (Fig. 1A). The libraries were sequenced using 76-bp single-end reads, and we mapped the first 50 bases of each read using Bowtie (18) (Fig. 1B). Allowing three mismatches per read, these sequences were mapped to the human genome assembly (hg19), with an overall mapping rate of ∼80% and a unique mapping rate of ∼65% (see Table S1 in the supplemental material). Reads were also mapped to a custom splice junction database of all possible exon-exon junctions within the same gene. In total, ∼8.5% of the sequenced reads from all four samples were mapped uniquely (i.e., to only one junction and not directly to the genome) to either known or hypothetical exon-exon junctions (see Table S1).
Fig 1.
RNA-Seq analysis detects ESRP-regulated alternative splicing events. (A) Outline of the experimental systems and RNA-Seq protocol used to identify ESRP-regulated exons. (B) Flowchart summarizing the bioinformatics analytical pipeline applied to detect ESRP-regulated alternative splicing events.
Identification of ESRP-regulated cassette exons.
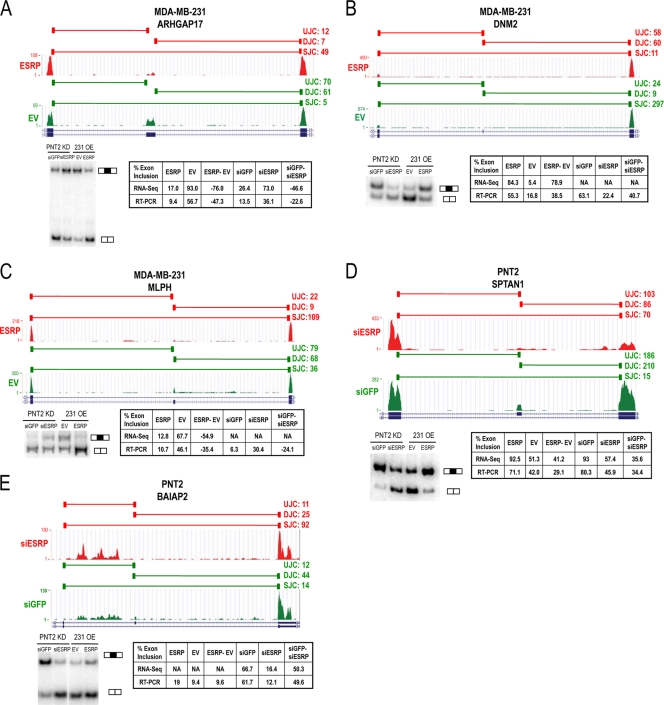
To uncover novel ESRP-regulated cassette exons, we calculated an exon inclusion level (ψ) for each alternative exon using exon junction counts uniquely mapped to the upstream (UJC), downstream (DJC), and skipping (SJC) junctions. To detect differences in exon inclusion between samples, we used a Bayesian statistics method to calculate a P value and false discovery rate (FDR) that the change in inclusion level was at least 5% (see Materials and Methods). We thereby identified 325 cassette exons predicted to undergo changes in splicing upon ectopic ESRP overexpression in MDA-MB-231 cells (FDR < 0.05) (see Table S2 in the supplemental material). Of these targets, 281 had not been identified previously in microarray analyses and included some exons without previous annotated evidence of skipping. Of 100 newly identified events (92%) tested by RT-PCR, 92 were validated with changes of ≥5% in the predicted direction, or 83% using a cutoff of at least a 10% change in splicing (see Table S3 in the supplemental material). In the PNT2 ESRP knockdown samples, only 19 cassette exons were identified using the same stringent thresholds (see Table S2). While a number of explanations might account for this discrepancy, we suspect that the reduced number of reads as well as the less complete change in ESRP expression between samples in the knockdown system compared to the ectopic expression system plays a role. In the PNT2 cells, knockdown of ESRP1 reduced the RNA expression level from 100.4 to 20.6 reads per kilobase of exon model per million mapped reads (RPKM), or an approximately 5-fold reduction. Knockdown of ESRP2 expression reduces its level from 16.0 to 7.5 RPKM. In contrast, the expression of ESRP1 in MDA-MB-231 control cells was a negligible 0.015 RPKM and increased to 297.9 RPKM with ectopic expression. Therefore, deeper RNA-Seq coverage may be needed to comprehensively capture ESRP-regulated exons in the knockdown system. Of note, we tested 11 cassette exons identified in the MDA-MB-231 system by RT-PCR, most of which showed the expected change upon ESRP1 and ESRP2 knockdown in PNT2 cells (data not shown). We thus suspect that many events in the PNT2 system were false negatives at the current sequencing depth using the strict cutoffs selected here. Three of the 19 exons from the PNT2 analysis had previously been validated, and we therefore tested the remaining 16 exons by RT-PCR, of which 8 yielded the expected products. Of these, 6 were validated with a >5% change in splicing (75%; 4/8 [50%] using a 10% cutoff) (see Table S3). Several examples of novel ESRP-regulated cassette exons uncovered by RNA-Seq that were validated in both experimental systems are presented schematically in Fig. 2. In ARHGAP17 and MLPH, ESRP-induced silencing is indicated by a switch from predominantly exon inclusion junction counts (UJC and DJC) to exon skipping counts (SJC) upon ectopic ESRP expression in MDA-MB-231 cells (Fig. 2A and C). In DNM2, ectopic expression of ESRP induced exon inclusion with a switch from exon skipping toward increased exon inclusion counts (Fig. 2B). Two ESRP-enhanced exons in SPTAN1 and BAIAP2 transcripts were identified in the PNT2 knockdown system (Fig. 2D and E). Among the validated ESRP-silenced exons from the MDA-MB-231 analysis were 11 novel alternatively spliced known exons for which there was no previous mRNA or EST evidence of exon skipping (see Table S3).
Fig 2.
Examples of validated ESRP-regulated enhancement or silencing of RNA-Seq predicted target cassette exons. The University of California, Santa Cruz (UCSC), genome browser view of the transcripts that contain or skip the exon is shown with tracks representing the junction reads (horizontal bars on top) and exon body read counts (vertical bars below) in either MDA-MB-231 cells with ESRP overexpression (A to C) or PNT2 cells with ESRP knockdown (D and E). The green tracks represent control cells and the red tracks represent ESRP knockdown or overexpression. The upstream junction read count (UJC), downstream junction read count (DJC), and skipping junction read count (SJC) are on the right in each panel, and at the bottom are RT-PCR validation gels with bands corresponding to exon inclusion and skipping indicated. Tables present the exon inclusion levels from RNA-Seq and RT-PCR. For the percent change in exon inclusion (ESRP-EV or siGFP-siESRP), negative values indicate ESRP-silenced exons and positive values indicate ESRP-enhanced exons. (A) ESRP-silenced exon in ARHGAP17; (B) ESRP-enhanced exon in DNM2; (C) ESRP-silenced exon in MLPH; (D) ESRP-enhanced exon in SPTAN1; (E) ESRP-enhanced exon in BAIAP2.
SELEX-Seq reveals a high-confidence ESRP binding motif.
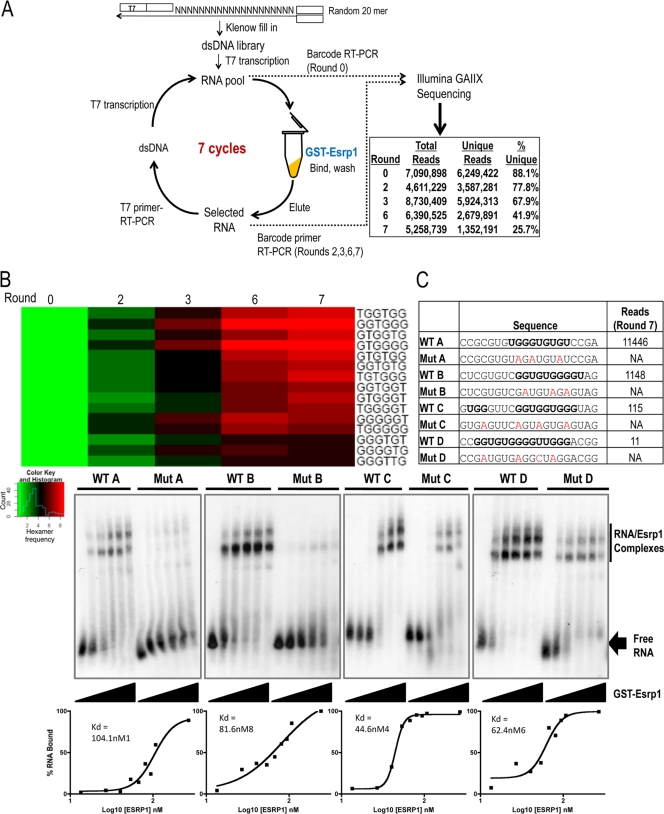
A previous bioinformatics-based analysis noted that UG-rich motifs were enriched downstream of ESRP-enhanced exons and within the body of ESRP-silenced exons (47). These observations were consistent with a model, or RNA map, wherein ESRP binding to such motifs downstream of an exon promoted its inclusion, whereas binding within the exon promoted skipping. To further define the ESRP binding motif experimentally in an unbiased manner, we performed systematic evolution of ligands by exponential enrichment (SELEX) (40). When coupled with high-throughput sequencing (SELEX-Seq), this approach can identify substantially more binding sequences and thus more accurately define optimal high-affinity, sequence-specific interactions than the standard approach using Sanger sequencing. We used recombinant GST-ESRP1 to screen a library of random 20-mers using seven cycles of SELEX. Using a barcoding approach, we sequenced RT-PCR products from RNAs in the input library prior to selection for ESRP1 binding (round 0), as well as after 2, 3, 5, and 7 cycles of binding in a single lane of single-end Illumina sequencing. As expected, the number of unique 20-mers decreased with increasing cycle numbers, most likely due to the PCR amplification steps at each step (Fig. 3A). Three of the hexamers previously identified as being enriched downstream of ESRP-enhanced exons were among the top 12 hexamers selected after 7 rounds of SELEX, further validating their identification as bona fide direct ESRP binding motifs (see Fig. S1B in the supplemental material). It was also apparent that the top n-mers obtained after 7 rounds were already enriched after 2 to 3 rounds of SELEX (Fig. 3B; also, see Table S4 in the supplemental material). Position weight matrix (PWM) models for both 6-mers and 8-mers similarly showed that common motifs were evident after the first two or three rounds (see Fig. S1 in the supplemental material). These findings suggested that by coupling high-throughput sequencing with SELEX, as few as two or three rounds of selection may be sufficient to identify a high-confidence RNA binding motif.
Fig 3.
SELEX-Seq defines a UG-rich ESRP binding motif (A) Schematic for SELEX-Seq protocol. The total and unique number of reads obtained by Illumina sequencing in each sequencing round are shown. (B) SELEX-Seq-identified 6-mer motifs after seven rounds of selection and their enrichment in each round. (C) EMSA analysis of ESRP1 binding affinity to selected 20-mer sequences using increasing amounts of GST-Esrp1 (0 to 250 ng), shown from left to right. Dissociation constants (Kds) were calculated from EMSAs using additional protein concentrations. Potential ESRP binding sites within the wild-type 20-mer sequences are in bold, and mutations that are expected to abolish ESRP binding are in red. The number of reads obtained for each selected sequence from round 7 is also shown.
To further validate the sequence binding specificities, we carried out EMSA analysis of four selected 20-mers as well as corresponding 20-mer sequences with G-to-A mutations introduced into the putative ESRP binding motifs. All four sequences displayed sequence-specific binding to recombinant ESRP1, with reduced or abolished binding to the mutated sequences (Fig. 3C). In principle, cases in which larger numbers of the same sequence are observed with increased SELEX cycle numbers might represent higher-affinity binding sites (15). However, sequences present in greater numbers by round 7 did not display evident differences in binding affinity, suggesting that the increased frequency with which some sequences were amplified by PCR was largely stochastic.
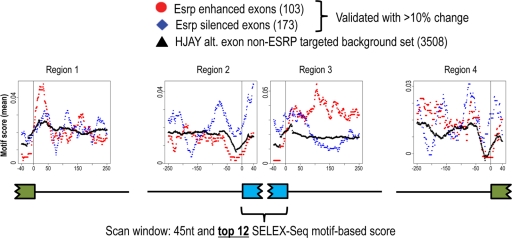
High-resolution functional RNA map for ESRP splicing regulation of cassette exons.
The expanded set of ESRP-regulated cassette exons uncovered by RNA-Seq coupled with an improved definition of ESRP binding preferences allowed us to define an RNA map of position-dependent ESRP functions. We used the top 12 6-mer motifs to define an ESRP binding score using an approach similar to that previously used for Nova binding sites (41). We evaluated the ESRP score at each position across the set of ESRP-regulated cassette exons and the 250 nt of intronic sequences flanking these exons as well as the upstream and downstream exons (see Materials and Methods). The complete set of ESRP-regulated exons with validated switches in splicing of at least 10% in either experimental system comprised a total of 276 exons (see Table S3 in the supplemental material). A control data set consisted of alternatively spliced exons that are expressed in these cells but for which there was no evidence of ESRP-mediated regulation. As shown in Fig. 4, there was enrichment of ESRP binding sites in the intronic region 75 to 250 nt downstream of ESRP-enhanced exons The peak of this enriched region was in the region from +90 to +135, where 37 nucleotide positions were enriched above the background by a P value of <0.001 (see Table S5 in the supplemental material). We also noted that the same motifs were underrepresented in the same region downstream of ESRP-silenced exons. In contrast, the ESRP binding motifs were enriched within the silenced exons as well as in the 125 nt of intronic sequence upstream of these exons, including the polypyrimidine tract, as well as positions −250 to −230, with numerous positions in these regions having P values for enrichment of <0.001 (see Table S5). These same motifs were underrepresented in these positions in the enhanced exon data set. These findings thus provide a high-resolution map consistent with the position-dependent binding functions of the ESRPs to either enhance exon inclusion or promote skipping. ESRP binding motifs also appeared to be enriched in the region ∼10 to 75 nt downstream of all ESRP-regulated exons and ∼10 to 50 nt downstream of the flanking upstream exons, but neither of these peaks was determined to be statistically significant (see Table S5). We also add that while several ESRP targets have previously been confirmed as direct targets, the list of ESRP-regulated spicing events used for this analysis may also include indirect targets. Nonetheless, given the unbiased determination of the ESRP binding site and the consistent experimental data, we suggest that while such indirect targets may introduce some background noise, they should not significantly affect the general model of the map.
Fig 4.
A functional map for ESRP position-dependent regulation of alternative splicing. The top 12 6-mer ESRP binding motifs from SELEX-Seq were used to derive an ESRP binding score, which is mapped across the set of 276 validated ESRP-regulated cassette exons with at least 10% change and the 250-nt intronic sequences flanking these exons, with enhanced exons in red and silenced exons in blue. This was also mapped across a set of alternative exons present on the HJAY arrays but not ESRP regulated (black). Each position in the RNA map represents the ESRP score within a 45-nt window, centered at the current position and averaged over all exons in a given group.
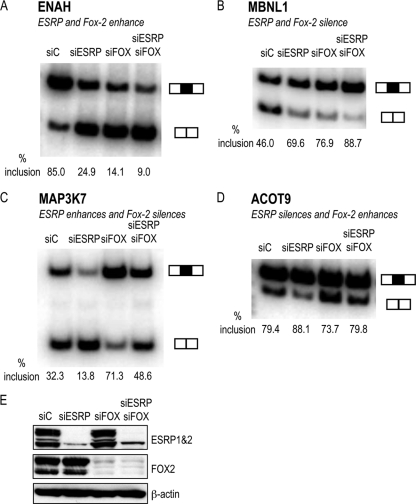
The ESRPs and FOX2 splicing regulators combinatorially influence alternative splicing of overlapping target exons.
We previously noted that the well-defined UGCAUG binding site for the Fox family of splicing regulators was enriched downstream of both ESRP-enhanced and ESRP-silenced exons (42, 47, 54, 56). Because FOX2, but neither FOX1 nor FOX3, was expressed in both of our cell lines, we further investigated the role of this splicing regulator in the combinatorial regulation of shared target exons. Using the complete set of ESRP-regulated cassettes described above, we compared the intersection of the largest set of FOX2-validated target exons (44). Among the 276 ESRP-regulated exons, 27 (9.8%) are also regulated by FOX2, whereas in a background set of 3,508 non-ESRP target exons, only 15 (0.4%) are known FOX2 targets, a >20-fold enrichment (P value < 2.2e-16), indicating a significant overlap of coregulated targets of these regulators. Because Fox proteins similarly silence exons from upstream binding sites and enhance from downstream sites, these observations suggested that the ESRPs and Fox proteins could have either additive or antagonistic effects on splicing of common regulated exons. Consistent with this observation, of the complete set of ESRP and FOX2 targets that have each been validated by RT-PCR, 18 have the opposite effect on exon splicing and 9 promote the same change (see Table S6 in the supplemental material). To determine whether the functions of ESRP and FOX2 on these common targets are redundant or additive, we tested the response of several of the exons to the knockdown of the ESRPs (ESRP1 and ESRP2) or FOX2 alone, as well as upon combined knockdown in PNT2 epithelial cells (Fig. 5E). ESRP and FOX2 knockdown resulted in similar decreases in splicing of an ENAH exon, whereas the combined knockdown resulted in an additive decrease in exon splicing (Fig. 5A). In an MBNL1 exon that is silenced by both proteins, combined knockdown of ESRP and FOX2 caused a greater increase in exon inclusion than knockdown of either alone (Fig. 5B). In exons in ACOT9, which is ESRP silenced and FOX2 enhanced, and MAP3K7, which is ESRP enhanced and FOX2 silenced, knockdown of either factor alone in PNT2 cells promoted the expected changes in exon splicing (Fig. 5C and D). However, the combined knockdown of ESRP and FOX2 caused exon inclusion levels to approximate those in the control knockdown cells. In MDA-MB-231 cells, for exons where they promoted opposite changes, a combination of ESRP overexpression and FOX2 knockdown induced a larger change than either treatment alone (see Fig. S2 in the supplemental material). These collective observations indicate how the integrated maps of ESRP and FOX2 binding sites can account for their combinatorial effects to promote the same or opposite effects on splicing. We also noted that 38 (13.8%) ESRP validated exons had a Fox binding site within the 250-nt upstream intron and 55 (19.9%) had a Fox binding site within the 250-nt downstream intron, excluding the splice site sequence (versus 7.0% and 8.8% in the control exon data set, respectively), suggesting that the overlap extends beyond known Fox targets (see Table S6). We therefore tested 8 exons that had not previously been experimentally validated as Fox targets, of which 5 showed the predicted changes in splicing upon knockdown of FOX2 in MDA-MB-231 cells (see Fig. S3 in the supplemental material). These findings further indicate that the ESRPs and FOX2 exhibit widespread combinatorial regulation of an overlapping set of coregulated target transcripts. Of note, similar observations were recently reported for Fox and Nova (55). While the biological significance of these overlapping splicing networks is not clear, these results illustrate the role of combinatorial regulation on a global scale. These findings also highlight how the overlapping RNA maps of several splicing factors, coupled with expression data for each, might be predictive of splicing outcome in different cell types, a feature central to the concept of the global splicing code (1).
Fig 5.
The ESRPs and FOX2 combinatorially regulate the alternative splicing of common cassette exons. RT-PCR analysis of ESRP and FOX2 coregulated exons was performed in PNT2 cells with knockdown of ESRP1 and ESRP2 (siESRP), FOX2 (siFOX), and ESRP1 and ESRP2 and FOX2 combined (siESRP siFOX) and in control cells (siC). Data are shown for alternative exons in ENAH, which is enhanced by ESRP and FOX2 (A), MBNL1, which is silenced by ESRP and FOX2 (B), MAP3K7, which is enhanced by ESRP and silenced by FOX2 (C), and ACOT9, which is silenced by ESRP and enhanced by FOX2 (D). (E) A Western blot demonstrates the knockdown of ESRP and FOX2.
ESRP-regulated exons substantially overlap exons that exhibit cell-type-specific differences in a large set of epithelial versus mesenchymal cell types and that switch during EMT.
We previously observed that a subset of ESRP-regulated target exons had cell-type-specific splicing that could serve as a splicing signature to distinguish breast cancer cell lines defined as basal B (or claudin low) from those defined as luminal (47). Basal B/claudin-low cell lines display increased invasive properties and a mesenchymal gene expression signature relative to an epithelial phenotype of luminal cells that express the ESRPs (2, 26, 47). A recent study using HJAY arrays revealed a set of alternatively spliced exons whose inclusion or skipping was strongly associated with these cell subtypes across a large spectrum of cell lines (19). Because these cell line characteristics parallel those associated with the EMT, these findings thus describe a set of alternative splicing events that are broadly associated with the EMT and can distinguish epithelial from mesenchymal cells (2, 32). We used the MADS+ HJAY analysis pipeline (35) to identify exons with splicing patterns that displayed the highest statistical evidence to distinguish basal B/mesenchymal from luminal/epithelial cells. Among the top exons from this list, we noted extensive overlap of ESRP-regulated exons (see Table S7 in the supplemental material). For example, 18 of the top 20 basal B versus luminal differentially spliced exons were validated ESRP targets, and in each case the ESRPs promote the luminal splicing pattern. These observations thus suggest that a majority of splicing differences that distinguish basal B from luminal cells are predominantly regulated by the ESRPs, which promote the global splicing pattern seen in less aggressive luminal cancer cells.
Further evidence for a central role for the ESRPs in the EMT is provided by a more recent study that used RNA-Seq to identify changes in alternative splicing in a Twist-induced model of the EMT in mammary epithelial cells (34). This study verified our previous observation that both ESRP1 and ESRP2 are significantly downregulated during EMT and showed that these changes in ESRP expression levels exceeded those of any other splicing factors or RNA binding proteins. This analysis also revealed a significant overlap in predicted changes in splicing during EMT with our published set of ESRP-regulated exons from array analysis. Among the 25 cassette exons with a Twist-induced predicted change of >30% exon inclusion and an FDR of <0.05 that were validated by those authors, 20 of the 24 successful RT-PCRs showed the expected changes upon both ESRP expression in MDA-MB-231 cells and ESRP knockdown in PNT2 cells. These observations suggest that the ESRPs are central regulators of splicing changes associated with the EMT, although these events are also influenced by the combinatorial effects of additional splicing regulators, including FOX2.
Identification of ESRP-regulated alternative 3′ and 5′ splice sites.
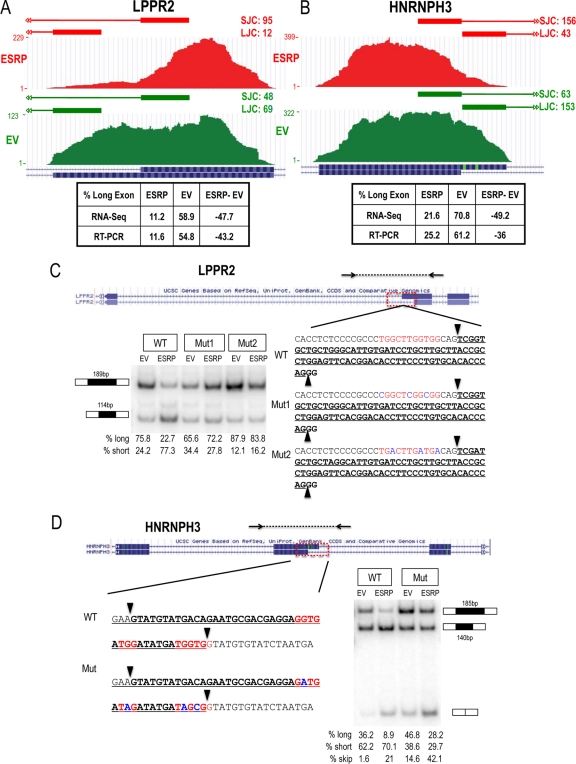
We used RNA-Seq junction reads to identify nine candidate ESRP-regulated alternative 3′ or 5′ splice sites in the MDA-MB-231 data set and seven in the PNT2 system (see Table S8 in the supplemental material). Two of six events from MDA-MB-231 were validated at the 5% cutoff level. To verify that these events also included direct ESRP targets, we used minigene models to investigate the functions of ESRP to regulate splicing of LPPR2 and HNRNPH3 via binding to ESRP binding motifs near their alternative 3′ and 5′ splice sites (Fig. 6). Minigenes were constructed using conserved regions, including the target exon and flanking introns. Candidate ESRP binding sites were mutated to test ESRP's role in alternative splice site use. In the case of alternative 3′ splice sites in LPPR2, we noted UGG elements within the polypyrimidine tract (PPT) associated with the upstream 3′ splice site that is repressed by the ESRPs. Cotransfection of the wild-type LPPR2 minigene with an ESRP1 cDNA in 293T cells reproduced ESRP-mediated silencing of splicing at this upstream splice site. Because mutations in the region of the PPT can also affect splicing by influencing recruitment of the general splicing factor U2AF, we tested the effects of two sets of mutations that were predicted to be relatively neutral with respect to changing PPT “strength.” While each mutated minigene had modest and opposite effects on splicing in the control transfections, each set of mutations nearly abolished the ability of ESRP1 to repress this alternative 3′ splice site. In HNRNPH3, a wild-type minigene recapitulated the function of ESRP to induce a switch from a downstream to an upstream 5′ splice site. We identified ESRP binding motifs overlapping the downstream 5′ splice site. Mutations predicted to abolish ESRP binding decreased the ability of ESRP to block splicing at this 5′ splice site. In addition to increased splicing at the upstream 5′ splice site in response to ESRP1 cotransfection, we also noted that the mutation caused a small increase in ESRP-mediated skipping of the complete exon. These examples are both consistent with a molecular mechanism whereby ESRP bound within or near the splice sites sterically blocks binding by components of the constitutive splicing apparatus, thereby favoring the other splice site. However, we suspect that there are other mechanisms that can account for ESRP regulation of alternative 3′ and 5′ splice sites.
Fig 6.
The ESRPs regulate alternative 3′ and 5′ splice sites. The UCSC browser view of the alternative 3′ splice sites of LPPR2 exon 4 (A) and alternative 5′ splice sites of HNRNPH3 exon 4 (B) are shown with tracks representing the junction reads (horizontal bars on top) and exon body read counts (vertical bars below) in MDA-MB-231 control cells (EV, green) or ESRP-overexpressing cells (ESRP, red). Tables present the predicted levels of the long exon from RNA-Seq and the experimentally determined levels from RT-PCR. Minigene splicing reporters were constructed containing LPPR2 exon 4 (bold) (C) or HNRNPH3 exon 4 (bold) (D) and conserved flanking intronic sequences. The long form of each exon is underlined, and alternative splice sites are indicated by arrowheads. Conserved intronic and exonic UGG-rich elements near the alternative splice sites are in red, and point mutations within UGG motifs are in blue. RT-PCR analysis for the minigenes is shown.
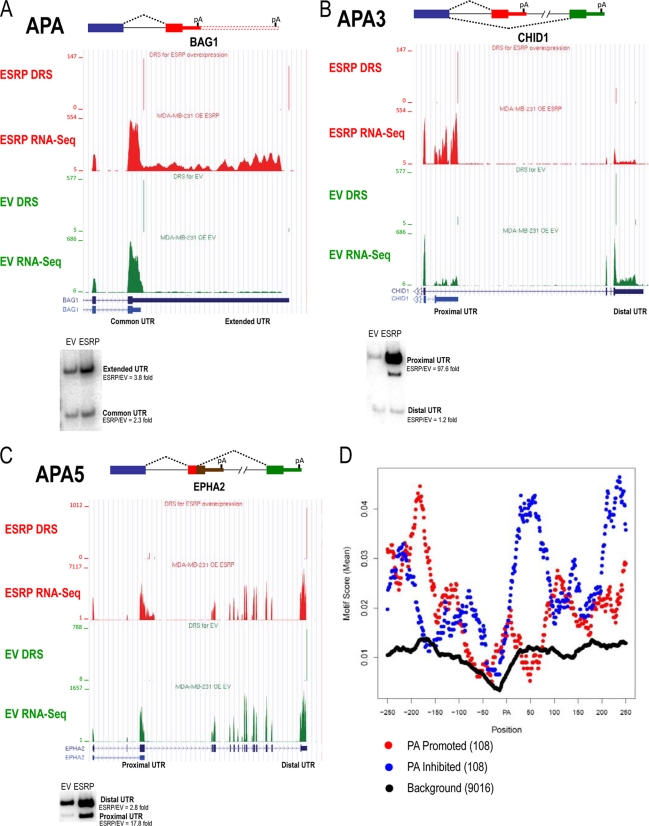
Identification of ESRP-regulated changes in alternative 3′-end formation by coupling RNA-Seq with direct RNA sequencing (DRS) of poly(A) sites.
The past few years have seen a growing appreciation of the widespread prevalence of alternative polyadenylation and coupling of alternative splicing with formation of alternative poly(A) sites (reviewed in references 8 and 22). Several recent studies have described new methods for high-throughput direct sequencing of the 3′ ends of mRNAs (10, 11, 28, 37). We previously used exon arrays to identify a few examples of ESRP-regulated alternative poly(A) sites (49). To identify a broader set of ESRP-regulated poly(A) events we developed a pipeline that couples direct sequencing of the 3′ cleavage region with the whole transcriptome RNA-Seq data set described previously. While different definitions and nomenclatures have been used, we considered alternative polyadenylation to consist of three distinct subtypes. The most basic form of alternative polyadenylation occurs through use of alternative poly(A) sites within the same 3′ UTR (APA) (Fig. 7A). Two mechanisms of alternative splice site use associated with alternative poly(A) sites can give rise to completely different 3′ terminal exons and 3′ UTRs. One type occurs when an upstream exon is either spliced to the 3′ splice site in an exon with a functional polyadenylation site or skips the exon entirely and splices to a 3′ splice site in a downstream exon (APA3; nomenclature is according to reference 55) (Fig. 7B). The second type is when alternative use of a 5′ splice site in an otherwise 3′ terminal exon can lead to splicing to a downstream exon (APA5) (Fig. 7C). In either case, the downstream exon can itself be an alternative 3′ terminal exon, or multiple additional cassette exons can be included before the downstream terminal exon is reached. Thus, these types of events have the potential to more significantly affect protein size and function than most simple alternative cassette exons. All three types of alternative 3′ end events share the potential to render the resulting transcripts susceptible to differential regulation by microRNAs or proteins that regulate RNA stability. For example, a general decrease in microRNA-mediated repression occurs in transcripts expressed in proliferating cells, including cancer cells, that switch to isoforms with shorter 3′ UTRs (23, 31).
Fig 7.
The ESRPs regulate alternative 3′-end formation. Examples of three types of alternative 3′-end formation regulated by the ESRPs are shown with UCSC browser views of RNA-Seq and direct RNA sequencing (DRS) read counts from MDA-MB-231 control (EV, green) versus ESRP-overexpressing cells (ESRP, red) and RT-PCR validations. (A) Alternative polyadenylation within the same 3′ UTR (APA); (B) alternative polyadenylation associated with alternative 3′ splice site usage (APA3); (C) alternative polyadenylation associated with alternative 5′ splice site usage (APA5). (D) A functional map for ESRP position-dependent regulation of alternative polyadenylation. The top 12 6-mer ESRP binding motifs from SELEX-Seq were used to derive an ESRP binding score, which is shown mapped across the set of 108 DRS-identified and RNA-Seq cross-validated ESRP-regulated poly(A) sites and the 250 nt upstream and downstream with promoted sites in red and silenced sites in blue. These motifs were also mapped across a background set of annotated poly(A) sites (black).
Sequencing of mRNA 3′ ends was performed using the Helicos direct RNA sequencing (DRS) platform through capture of poly(A) RNA on poly(dT)-coated flow cells as previously described (28). One channel each was used to sequence RNA from control MDA-MB-231 cells and cells with ectopic expression of ESRP1. Reads obtained through the pipeline were filtered for a minimum length of 25 and to remove reads derived from internal genomically encoded poly(A) stretches. Clustered reads were then used to identify 335 candidate pairs of poly(A) sites with significant differential use between control and ESRP-expressing cells (see Table S9 in the supplemental material; also, see Materials and Methods). To validate these events, we used mapped reads from the RNA-Seq analysis within the 300-nt region upstream of the poly(A) sites. Overall, 71.8% of the DRS-predicted changes in poly(A) site use were supported by RNA-Seq, and many of the nonvalidated cases lacked sufficient RNA-Seq coverage in the relevant genomic location. These data suggested that the DRS pipeline was robust and accurate. Nonetheless, to obtain a more confident set of ESRP-regulated poly(A) sites, we filtered the DRS-predicted poly(A) sites to include only those with statistical RNA-Seq validation. This resulted in a total of 160 high-confidence changes in poly(A) site use in response to ESRP1 expression, of which 32 were categorized as APA, 76 as APA3, and 52 as APA5 (see Table S9).
In six cases, we also used competitive RT-PCR using a common forward primer and specific reverse primers that would recognize each alternative form. Although these competitive PCRs are less quantitative than those using common primer sets, these validations supported each of the events tested (Fig. 7; also, see Fig. S4A to C). One example of alternative poly(A) use in the same UTR (APA) was BAG1, where ESRP1 promoted expression of the isoform with an extended 3′ UTR (Fig. 7A). We also validated an APA3-type event in the CHID1 transcript, an example where ESRP promotes use of a proximal terminal exon (Fig. 7B). In the EPHA2 transcript, ESRP promotes the use of a 5′ splice site in the proximal APA5-type terminal exon.
We noted several examples where ESRP expression induced proximal APA3- or APA5-type 3′ terminal exons that were very close to the 5′ end of transcripts and associated with a significant decrease in expression of the downstream exons (see Fig. S4D to F in the supplemental material). For example, in COL5A1, we identified a novel APA3 in the fourth intron and an APA5 event in the first exon (Fig S4D). ESRP expression promoted both of these events, leading to short truncated products and a nearly 35-fold reduction in expression of full-length transcripts (see Table S9 in the supplemental material). While it is not known whether the truncated transcripts encode polypeptides, these examples illustrated how early induction of polyadenylation might downregulate gene expression, although this mechanism may also involve contributions by microRNAs. These examples are reminiscent of a previous observation in cleavage stimulation factor 77 (CstF-77) transcripts, where the use of a conserved alternative poly(A) site in the third intron was proposed to serve as a means of utilizing alternative polyadenylation to directly modulate expression of the full-length functional isoform (30).
Similar to our observations on the ESRPs, a limited number of other examples have been described wherein splicing factors can also regulate polyadenylation (8). Previous studies of the Nova proteins (and their Drosophila melanogaster ortholog Pasilla) have shown that the binding sites or known motifs support a position dependent function to regulate these types of regulation (3, 21). We therefore similarly sought to explore whether the ESRP binding motif was enriched in the set of alternatively polyadenylated transcripts in a position-dependent manner. Using a more refined set of 108 such events (see Materials and Methods) regulated by ESRP, we evaluated the positions of these motifs relative to a background set of alternative poly(A) sites. As shown in Fig. 7D, there was a highly significant level of enrichment for ESRP binding sites both upstream and downstream of the ESRP-regulated poly(A) sites, suggesting that they can directly impact polyadenylation. While there were regions relative to the poly(A) site in which ESRP binding motifs were enriched in both ESRP-enhanced and -repressed sites, there were also regions in which binding sites were more statistically associated with ESRP-enhanced or repressed sites. For example, in the region from ∼−220 to −160 upstream of ESRP-regulated poly(A) sites, there was greater enrichment in enhanced than repressed poly(A) sites (Fig. 7D; also, see Table S10 in the supplemental material). However, for ESRP-repressed sites there was greater enrichment of the motifs +7 to +86 and +200 to +250 nt downstream of the poly(A) site. These observations are in agreement with previous studies showing enrichment for binding motifs of other splicing factors, such as Nova and hnRNP H, that also regulate polyadenylation (17, 21). While these observations implicate the ESRPs in the regulation of polyadenylation, it is worth noting that the regulation of APA3 and APA5 events may also occur through the regulation of splicing via binding to regions near the regulated splice sites. Such events may further involve coupled recruitment or inhibition of the splicing and polyadenylation machineries. However, given the limited number of each subtype (APA3 and APA5), we were unable to derive a separate confident map for ESRP binding sites in these events. While this motif analysis supports a role for ESRPs in direct regulation of polyadenylation, we cannot be sure that all of the events we identified are direct targets. Nonetheless, the current set of over 100 high-confidence changes in polyadenylation provides a useful data set for downstream analysis and illustrates the potential of integrating high-throughput sequencing of mRNA 3′ ends with transcriptome-wide sequencing to uncover larger sets of regulatory networks.
DISCUSSION
We used RNA-Seq to provide a more comprehensive view of the genome-wide ESRP-regulated splicing regulatory network. In addition to identifying and validating a larger set of cassette exon targets and alternative 3′ and 5′ splice sites, the RNA-Seq data, together with the DRS analysis of mRNA 3′ ends, further implicates the ESRPs in the genome-wide regulation of alternative polyadenylation. This work highlights the emerging potential of high-throughput sequencing methods to comprehensively identify differential patterns of alternative splicing and polyadenylation. The pipeline using RNA-Seq and DRS to investigate regulation of polyadenylation demonstrates how coupling these different high-throughput technologies can increase the power of detection while also providing intrinsic cross-validation. Our studies also add the ESRPs to the list of known splicing regulators, such as Nova, hnRNP F, hnRNP H, PTB, and U1A, that also regulate polyadenylation (7, 8, 25). While the proposed map of ESRP binding sites suggests that the ESRPs can promote or inhibit polyadenylation in a position-dependent manner, further experiments are needed to validate this. In addition, future investigations are needed to understand the mechanisms by which the ESRPs regulate polyadenylation. hnRNP F, a homolog of ESRP, was previously suggested to regulate alternative polyadenylation of IgM heavy chain through inhibition of Cstf-64 binding to the downstream poly(A) (45). A similar role for PTB-mediated inhibition via reduced Cstf-64 binding was also shown, whereas binding of PTB upstream of the poly(A) site can promote polyadenylation (5). Based upon the pattern of enrichment of ESRP binding motifs relative to regulated poly(A) sites, we envision that they might operate through similar mechanisms to regulate polyadenylation. We also noted numerous examples of high-confidence ESRP-regulated poly(A) sites that represent novel, previously unannotated sites of polyadenylation. In fact, 72 of the 271 (26.6%) of the regulated poly(A) sites supported by both DRS and RNA-Seq were novel, similar to findings in other recent 3′-end sequencing studies that discovered large numbers of novel poly(A) sites (10, 11, 28). Thus, as more studies using similar technologies are performed for additional regulators and in different cell contexts, the percentage of human genes that are known to undergo regulated alternative polyadenylation will almost surely continue to rise.
A common feature of most RNA binding proteins is that the sequences that they bind exhibit various degrees of degeneracy, which can complicate the prediction of in vivo binding sites. SELEX-Seq offers considerable advantages over previous SELEX-based approaches. While we are not aware of previous reports on this approach for RNA binding proteins, a study using a similar SELEX-Seq approach characterized the DNA binding sites for several transcription factors (15). Similar to results in that report, we also noted that while the high-affinity binding sites were enriched by cycle number, the consensus motifs that mediate binding emerged after just 2 to 3 cycles, highlighting an additional advantage of coupling SELEX with high-throughput sequencing. One potential limitation of the SELEX-Seq approach is that in vitro binding specificity may not always recapitulate in vivo binding preferences. However, there are now several examples in which the in vitro SELEX-determined motif has been shown to be the same as that determined in vivo using cross-linking and immunoprecipitation (CLIP) (e.g., FOX2, PTB, and Nova) (21, 52, 54). Furthermore, in the case of ESRP the SELEX motif matches one that was both bioinformatically predicted and functionally validated.
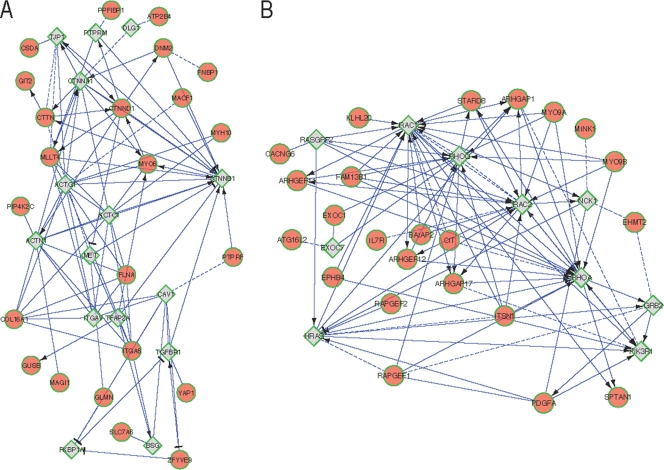
Gene ontology (GO) analysis previously performed for HJAY-predicted cassette exons demonstrated enrichment for processes and functions that are functionally relevant for the EMT, including cell-cell adhesion, cell motility, and regulation of the actin cytoskeleton (47). A comparable analysis using the broader set of validated ESRP cassette targets identified from either HJAY or RNA-Seq with at least 10% change in exon inclusion further supported a role for ESRP target transcripts in EMT relevant categories, most notably for genes that function in GTPase regulator activity (data not shown). We also projected the same validated list onto a functional interaction (FI) network derived by combining curated interactions from well annotated pathway databases with predicted protein functional interactions based on heterogeneous sources, including physical protein-protein interactions, coexpression data, protein domain interactions, and GO annotations (51). We were able to map 250 of the 276 (90.6%) validated ESRP target transcripts with regulated cassette exons to this extended FI network and used an edge betweenness algorithm to identify 19 network modules (see Table S11 in the supplemental material). Module 1 was enriched for pathways related to cell-cell adhesion, including those for E-cadherin-based adherens junctions, tight junctions, and nectin-based adhesion components (Fig. 8A). Module 2 was enriched for the Rho GTPase signaling pathway (Fig. 8B). This analysis provides further evidence that the ESRP splicing network encodes proteins that are biologically coherent and can also be used to generate hypotheses for future experimental investigations into the roles of ESRP target splicing events. The roles of most ESRP-regulated genes in the EMT and the functional consequences of the splicing changes they direct remain largely unexplored. In a very few select cases (e.g., CTNND1 and CD44), differential activities of the epithelial and mesenchymal isoforms that directly impact the EMT have been demonstrated (4, 53). A noteworthy observation from these examples is that the different isoforms can have opposing functions, implying that many seemingly contradictory roles of many other genes can also be accounted for by alternative splicing or polyadenylation. However, for most of the ESRP target genes, isoform-specific functions have not been investigated, and the relevance of these differences for developmental or pathophysiological EMT remains unclear. The genome-wide determination of this splicing program opens the way for more detailed investigations of the functional consequences of these splicing changes during the EMT at the level of individual gene transcripts as well as for systems-level analyses to determine how the global changes in splicing alter cellular pathways on a larger scale to influence cell phenotype and function. A challenge for future work will be to develop methods by which the isoform-specific functions of these transcripts can be systematically determined and at the same time to reveal how alternative splicing alters relevant protein interaction networks that influence distinct cell behaviors during development and disease.
Fig 8.
The subnetwork of functional interactions of ESRP-target networks. Circles represent genes of ESRP targets, while diamonds represent linker genes. FIs extracted from pathways are shown as solid lines, while predicted FIs based on the naïve Bayes classifier are shown as dashed lines. FIs involved in activation, expression regulation, or catalysis are shown with an arrowhead, while T bars indicate inhibition. (A) Detailed view of module 1, enriched for pathways related to cell-cell adhesion, including those for E-cadherin-based adherens junctions, tight junctions, and nectin-based adhesion components. (B) Detailed view of module 2, enriched for the Rho GTPase signaling pathway.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jonathan Schug and members of the Functional Genomics Core Facility for assistance with the RNA-Seq analysis and design of the SELEX-Seq experiments. We thank the University of Iowa Institute for Clinical and Translational Science (NIH grant UL1 RR024979) for computer support. We thank Claude Warzecha for careful review of the manuscript and members of the Carstens and Xing labs for thoughtful suggestions and advice. We also thank Steve Liebhaber and Xinjun Ji for helpful suggestions and discussion.
This work was supported by NIH grants R01 GM088809 (R.P.C.) and R01 GM088342 and P30 DK054759 (Y.X.), and a junior faculty grant from the Edward Mallinckrodt Jr. Foundation (Y.X.). J.W.P. is supported by an NIH T32 postdoctoral fellow training grant (T32HL007638).
Footnotes
Published ahead of print 21 February 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Barash Y, et al. 2010. Deciphering the splicing code. Nature 465: 53–59 [DOI] [PubMed] [Google Scholar]
- 2. Blick T, et al. 2008. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin. Exp. Metastasis 25: 629–642 [DOI] [PubMed] [Google Scholar]
- 3. Brooks AN, et al. 2011. Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res. 21: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown RL, et al. 2011. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Invest. 121: 1064–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Castelo-Branco P, et al. 2004. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol. Cell. Biol. 24: 4174–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen M, Manley JL. 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Danckwardt S, Hentze MW, Kulozik AE. 2008. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 27: 482–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Di Giammartino DC, Nishida K, Manley JL. 2011. Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43: 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flicek P, et al. 2011. Ensembl 2011. Nucleic Acids Res. 39: D800–D806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fox-Walsh K, Davis-Turak J, Zhou Y, Li H, Fu XD. 2011. A multiplex RNA-seq strategy to profile poly(A+) RNA: application to analysis of transcription response and 3′ end formation. Genomics 98: 266–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fu Y, et al. 2011. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 21: 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hallegger M, Llorian M, Smith CW. 2010. Alternative splicing: global insights. FEBS J. 277: 856–866 [DOI] [PubMed] [Google Scholar]
- 13. Hovhannisyan RH, Carstens RP. 2007. Heterogeneous ribonucleoprotein m is a splicing regulatory protein that can enhance or silence splicing of alternatively spliced exons. J. Biol. Chem. 282: 36265–36274 [DOI] [PubMed] [Google Scholar]
- 14. Hovhannisyan RH, Carstens RP. 2005. A novel intronic cis element, ISE/ISS-3, regulates rat fibroblast growth factor receptor 2 splicing through activation of an upstream exon and repression of a downstream exon containing a noncanonical branch point sequence. Mol. Cell. Biol. 25: 250–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jolma A, et al. 2010. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 20: 861–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32: D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7: 1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lapuk A, et al. 2010. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol. Cancer Res. 8: 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Licatalosi DD, Darnell RB. 2010. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 11: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Licatalosi DD, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lutz CS, Moreira A. 2011. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip. Rev. RNA 2: 22–31 [DOI] [PubMed] [Google Scholar]
- 23. Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mi H, Guo N, Kejariwal A, Thomas PD. 2007. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 35: D247–D252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Millevoi S, Vagner S. 2010. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 38: 2757–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Neve RM, et al. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10: 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nilsen TW, Graveley BR. 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ozsolak F, et al. 2010. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143: 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- 30. Pan Z, et al. 2006. An intronic polyadenylation site in human and mouse CstF-77 genes suggests an evolutionarily conserved regulatory mechanism. Gene 366: 325–334 [DOI] [PubMed] [Google Scholar]
- 31. Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sarrio D, et al. 2008. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 68: 989–997 [DOI] [PubMed] [Google Scholar]
- 33. Shannon P, et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shapiro IM, et al. 2011. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 7: e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shen S, Warzecha CC, Carstens RP, Xing Y. 2010. MADS+: discovery of differential splicing events from Affymetrix exon junction array data. Bioinformatics 26: 268–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shen S, et al. 2012. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res. [Epub ahead of print.] doi:10.1093/nar/gkr1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shepard PJ, et al. 2011. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thiery JP, Acloque H, Huang RY, Nieto MA. 2009. Epithelial-mesenchymal transitions in development and disease. Cell 139: 871–890 [DOI] [PubMed] [Google Scholar]
- 39. Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tuerk C, Gold L. 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510 [DOI] [PubMed] [Google Scholar]
- 41. Ule J, et al. 2006. An RNA map predicting Nova-dependent splicing regulation. Nature 444: 580–586 [DOI] [PubMed] [Google Scholar]
- 42. Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. 2005. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 25: 10005–10016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vastrik I, et al. 2007. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 8: R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Venables JP, et al. 2009. Cancer-associated regulation of alternative splicing. Nat. Struct. Mol. Biol. 16: 670–676 [DOI] [PubMed] [Google Scholar]
- 45. Veraldi KL, et al. 2001. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 21: 1228–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang ET, et al. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Warzecha CC, et al. 2010. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 29: 3286–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. 2009. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol. Cell 33: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Warzecha CC, Shen S, Xing Y, Carstens RP. 2009. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 6: 546–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu G, Feng X, Stein L. 2010. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 11: R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu G, Feng X, Stein L. 2010. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 11: R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xue Y, et al. 2009. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell 36: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yanagisawa M, et al. 2008. A p120 catenin isoform switch affects Rho activity, induces tumor cell invasion, and predicts metastatic disease. J. Biol. Chem. 283: 18344–18354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yeo GW, et al. 2009. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 16: 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang C, et al. 2010. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science 329: 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang C, et al. 2008. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 22: 2550–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.