Abstract
Our recent studies implicated key and distinct roles for the highly related RalA and RalB small GTPases (82% sequence identity) in pancreatic ductal adenocarcinoma (PDAC) tumorigenesis and invasive and metastatic growth, respectively. How RalB may promote PDAC invasion and metastasis has not been determined. In light of known Ral effector functions in regulation of actin organization and secretion, we addressed a possible role for RalB in formation of invadopodia, actin-rich membrane protrusions that contribute to tissue invasion and matrix remodeling. We determined that a majority of KRAS mutant PDAC cell lines exhibited invadopodia and that expression of activated K-Ras is both necessary and sufficient for invadopodium formation. Invadopodium formation was not dependent on the canonical Raf-MEK-ERK effector pathway and was instead dependent on the Ral effector pathway. However, this process was more dependent on RalB than on RalA. Surprisingly, RalB-mediated invadopodium formation was dependent on RalBP1/RLIP76 but not Sec5 and Exo84 exocyst effector function. Unexpectedly, the requirement for RalBP1 was independent of its best known function as a GTPase-activating protein for Rho small GTPases. Instead, disruption of the ATPase function of RalBP1 impaired invadopodium formation. Our results identify a novel RalB-mediated biochemical and signaling mechanism for invadopodium formation.
INTRODUCTION
The ability of cancer cells to degrade the extracellular matrix and invade through the basement membrane is essential for metastatic disease (23, 53). Pancreatic cancer is a highly aggressive and invasive disease, though the mechanisms by which pancreatic ductal adenocarcinoma (PDAC) cells mediate invasion and metastasis are largely unknown (65). Mutational activation of K-Ras is an early initiating event that occurs in essentially 100% of human pancreatic tumors (28). However, K-Ras activity may also contribute to invasion and metastasis of PDAC (12), suggesting that K-Ras plays a role in multiple steps of tumor progression.
The formation of dynamic, actin-rich, extracellular matrix-degrading protrusions known as invadopodia is linked to the invasive phenotype of cancer cells (10, 11, 21, 66). Invadopodia have been identified in multiple malignancies, including melanoma, glioblastoma, breast, and head and neck squamous cell carcinoma, and whether they are seen in PDAC has not been determined. While members of the Rho family of small GTPases (Rac1, RhoA, and Cdc42) have been implicated in invadopodium formation (19, 37, 52), whether aberrant Ras activation can promote invadopodium formation has not been addressed.
KRAS encodes a small GTPase that serves as a signaling node activating multiple downstream pathways in response to extracellular stimuli (18, 31). Activating mutations in KRAS encode a constitutively activated K-Ras protein which stimulates persistent, deregulated activation of downstream signaling pathways. The canonical effectors of Ras are the Raf serine/threonine kinases, which phosphorylate and activate the MEK1 and MEK2 dual specificity kinases, and MEK1/MEK2 phosphorylate and activate the ERK1/ERK2 mitogen-activated protein kinases (MAPKs) (18, 31). A second important group of effectors is the class IA catalytic subunits of phosphoinositide 3-kinases (PI3Ks). The third-best validated class of effectors required for Ras-mediated oncogenesis is comprised of guanine nucleotide exchange factors (RalGEFs) for the highly related RalA and RalB small GTPases (7, 51). In PDAC, our studies suggest that the RalGEF-Ral effector pathway may be more essential than Raf or PI3K signaling for mutant KRAS-dependent tumor growth (34). Interestingly, we determined that RalA and RalB have distinct roles in PDAC, with RalA required for anchorage-independent growth in vitro and tumorigenic growth in vivo, whereas Matrigel invasion and lung colonization metastasis were more dependent on RalB (36). The distinct functions of RalA and RalB are unexpected in that they can interact with the same set of downstream effectors, including subunits of the exocyst complex and the RalBP1/RLIP76 Rho GTPase-activating protein (RhoGAP) (7, 51). Whether RalA and RalB utilize distinct effectors to mediate their distinct functions in PDAC has not been determined.
How RalB promotes PDAC invasion and metastasis has not been determined. Since the effector functions of RalB can regulate actin organization and exocytosis, activities critical for invadopodium formation, we investigated whether invadopodia are associated with pancreatic cancer invasiveness and regulated by K-Ras and Ral signaling. We determined that invadopodia are seen in a majority of PDAC cell lines, that mutant K-Ras is necessary and sufficient for invadopodium induction, and that the Ral effector pathway, but not the Raf effector pathway, is required for this activity. RalB, and to a lesser extent RalA, was required and, unexpectedly, did not require the exocyst and instead required RalBP1 through a RhoGAP-independent mechanism. Instead, we have found that the ATPase function of RalBP1 is critical for invadopodia in PDAC. Our studies establish the first association for K-Ras and RalB for invadopodium formation and establish a novel effector function in this process.
MATERIALS AND METHODS
Human pancreatic tumor cell lines.
The MIA PaCa-2, CFPAC-1, PANC-1, Capan-1, Capan-2, Panc 10.05, and T3M4 PDAC cell lines were obtained from the ATCC (Manassas, VA), and the HuPT3 cell line was obtained from Dan Billadeau (Mayo Clinic, Rochester, MN). The immortalized human pancreatic duct-derived (HPNE) cells and the matched K-Ras-transformed cell line (HPNE-KRAS) were described previously (13, 33).
Plasmids and viral infection.
pSUPER.retro.blast plasmids encoding short hairpin RNA (shRNA) directed against sequences encoding green fluorescent protein (GFP; nonspecific control) (4) or human RalA and RalB have been validated and characterized previously (41). pBabe-puro plasmids expressing RNA interference (RNAi)-resistant sequences encoding wild-type (WT) RalB or the D49N and D49E effector mutants were used to infect PDAC cells (as described previously) (41). Blasticidin-resistant (Invitrogen) and puromycin-resistant (Invitrogen) stable cell lines were generated (for blasticidin, MIA PaCa-2 and Capan-2 [5 μg/ml], CFPAC-1 [10 μg/ml], and PANC-1 [20 μg/ml]; for puromycin, CFPAC-1 and PANC-1 [2 μg/ml]). pSUPER.retro.puro plasmids encoding shRNA directed against the coding sequence for human KRAS G12V (5′-GTTGGAGCTGGTGGCGTAG-3′) were described previously (9). The pLKO.1 plasmid encoding nonspecific (NS) RNA (5′-CCTCTTGATGAACCATCTATT-3′) or shRNA directed against the 3′ untranslated region (UTR) of human KRAS (5′-CAGTTGAGACCTTCTAATTGG-3′) were obtained from Jeffrey Settleman (Charlestown, MA) and were previously described (61). Lentiviral particles were generated using the pLKO.1 plasmid encoding nonspecific (NS) RNAs and shRNAs directed against RalBP1 obtained from Open Biosystems. The sequences for the clones are as follows: for clone 47920, 5′-GCACAAGAGATAGCCAGTCTT-3′, and for clone 47922, 5′-GCCAGTTTGCTGAAGCAGTAT-3′. The pSUPER.retro.blast plasmid encoding shRNA against human RalBP1 has been previously described (26). The pBabe-HAII-puro expression plasmid encoding an RNAi-resistant sequence for WT RalBP1 was generated by introducing silent mutations as described previously (41). The R232A/K268A (RK/AA) RalBP1 mutant was generated by introducing missense mutations in two consecutive mutagenesis reactions using the following mutations (mutated base pairs are italicized): R232A-5′-GAAGTGTGAAGGCATCTACGCAGTATCAGGAATTAAATC-3′ and K268A-5′-GTAGCCAGTTTGCTGGCGCAGTATTTGCGAG-3′. The cDNA sequence encoding the R208L/K244R (RK/LR) RalBP1 mutant was obtained from Christopher Counter (Duke University) and subcloned into the pBabe-HA expression construct. The K74M and K425M RalBP1 mutants were generated by introducing missense mutations using the following primers (mutated base pairs are italicized): K74M-5′-GAAGAAAAAAGGGATGTTTAAGAAAAAGG-3′ and K425M-5′-GATAAAGGATTTGTCTATGGAAGAAAGATTATGGG-3′. The K74M/K425M RalBP1 double mutant was generated by introducing missense mutations in two consecutive reactions using the above-described primers.
Immunoblot analyses.
Whole-cell lysates were isolated in NP-40 lysis buffer (50 mM Tris-HCl [pH 7.4], 10 mM MgCl2, 150 mM NaCl, 1% NP-40, 10% glycerol, 0.25% sodium deoxycholate). Proteins were separated by SDS-PAGE and transferred to Immobilon-P polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA). Membranes were blocked with 5% bovine serum albumin (Sigma) in Tris-buffered saline-Tween 20 (TBST) (0.1% Tween 20) buffer, incubated with primary antibodies, including mouse anti-K-Ras (clone OP24) (Millipore), mouse anti-RalB (Millipore), mouse anti-β-tubulin (Sigma), mouse antivinculin (Sigma), mouse anti-β-actin (Sigma), rabbit anti-phospho-ERK1/2 (Cell Signaling Technology), rabbit anti-ERK1/2 (Cell Signaling Technology), rabbit anti-phospho-Ser473 Akt (Cell Signaling Technology), rabbit anti-Akt (Cell Signaling Technology), mouse anti-RalA (BD Biosciences), mouse antihemagglutinin (anti-HA) (clone 16B12) (Covance), or mouse anti-RalBP1 (Abnova), overnight at 4°C, washed, and incubated with species-appropriate horseradish peroxidase-conjugated secondary antibodies (Pierce). Membranes probing with anti-RalBP1 were blocked in 5% (wt/vol) nonfat dry milk in phosphate-buffered saline–Tween 20 (PBST) (0.1% Tween 20) as suggested by the manufacturer. Membranes were subjected to detection using enhanced chemiluminescence.
Fluorescent matrix degradation assay.
The fluorescent matrix degradation assay was utilized to evaluate invadopodia and was described previously (16). Briefly, human fibronectin (BD Biosciences) was coupled to Alexa Fluor 488 using a protein labeling kit (Invitrogen). A 50-μg/ml solution was prepared in phosphate-buffered saline (PBS) and incubated on cross-linked gelatin (300 Bloom) (Sigma) on glass coverslips. Coverslips were sterilized with 70% ethanol and equilibrated with growth medium for 30 min prior to adding cells. Cells were seeded on coverslips, incubated for 24 h, fixed in 4% paraformaldehyde, permeabilized in 0.5% Triton X-100–PBS, and blocked in 3% bovine serum albumin–PBS. Coverslips were then incubated with mouse anticortactin (clone 4F11) antibody (Millipore) or fluorescent phalloidin. For inhibitor experiments, cells were seeded on the coverslips in the presence of U0126 (Sigma) or LY294002 (Cell Signaling Technology). Confocal images were acquired using an LSM 510 Meta laser scanning microscope (Carl Zeiss) with a 63×, 1.3-numerical-aperture oil immersion lens, and images were processed by Photoshop software (Adobe Systems).
Gelatin zymography.
Cells expressing GFP (control) or RalB-specific shRNA were seeded at 50% confluence and allowed to attach overnight. Serum-free medium was added to the cells for 24 h, conditioned media were collected, and cells were lysed in NP-40 lysis buffer. Protein concentrations for conditioned media were normalized to total cell numbers (total protein concentration from each cell lysate) and determined using the Bio-Rad Bradford protein assay. Nonreducing 6× Laemmli sample buffer was added to normalized conditioned medium, and samples were loaded onto 10% SDS-PAGE containing 0.2% gelatin (175 Bloom) (Sigma) and run under nonreducing conditions. Gels were washed with 2.5% Triton 100-X twice for 15 min and incubated at 37°C in substrate buffer (50 mM Tris-HCl [pH 7.5], 10 mM CaCl2) overnight, stained with 0.5% Coomassie blue, 50% methanol, and 10% acetic acid, and destained in 50% methanol and 10% acetic acid.
Surface biotinylation and avidin pulldown.
Surface biotinylation and avidin pulldown experiments were performed using the Pierce cell surface protein isolation kit (Pierce). Briefly, CFPAC-1 cells were seeded on tissue culture flasks coated with 2.5% gelatin–2.5% sucrose–PBS, labeled with biotin, harvested, and lysed. NeutrAvidin agarose was used to isolate biotinylated surface proteins from lysates. Eluates were loaded onto 10% SDS-PAGE and subjected to immunoblot analysis using rabbit anti-MT1-MMP antibody (Calbiochem).
GTP hydrolysis assay.
The human RalBP1 cDNA (residues 124 to 468) was subcloned into the NcoI and XhoI sites of the bacterial expression vector pProEX-HTb to express an N-terminal His6-tagged recombinant RalBP1 protein. The RalBP1 protein was expressed in the Escherichia coli strain Rosetta 2 (DE3) for 5 h at 25°C and purified on a nickel-charged metal chelating column (GE Healthcare), followed by separation on an S200 size exclusion column (GE Healthcare) equilibrated in a buffer containing 50 mM Tris (pH 8.0), 200 mM NaCl, 1 mM EDTA, 2 mM dithiothreitol (DTT), and 10% glycerol. The purified protein was concentrated to 10 mg/ml and snap-frozen in liquid N2. Site-directed mutagenesis of pProEx-HTb RalBP1 was carried out using the QuikChange kit (Stratagene) to replace arginine 232 and lysine 268 with alanines. RalBP1 (residues 124 to 468) R232A/K268A was expressed and purified exactly as described above for the wild-type protein. Recombinant human Cdc42 (C188S) was bacterially expressed and purified as previously described (45).
The GAP activity of RalBP1 was measured biochemically using the N-[2-(1-maleimidyl)ethyl]-7-(diethylamino)coumarin-3-carboxamide (MDCC) fluorophore-tagged phosphate-binding protein (PBP) fluorescent-based GAP assay as described previously (60). Briefly, Cdc42 was preloaded with GTP in nucleotide exchange buffer (10 mM HEPES [pH 7.5], 50 mM NaCl, 5 mM EDTA, 2 mM DTT and 1 mM GTP) for 20 min at 22°C before being stabilized with 20 mM MgCl2. GTP-loaded Cdc42 was then rapidly desalted on a PD10 column (GE Healthcare) equilibrated in 10 mM HEPES (pH 7.5), 50 mM NaCl, 1 mM MgCl2, 5% glycerol, and 2 mM dithiothreitol to remove excess GTP. Following desalting, single-turnover hydrolysis assays were performed with 3 μM MDCC-PBP and 2 μM Cdc42 in a reaction buffer containing 20 mM Tris (pH 7.5), 50 mM NaCl, and 2 mM MgCl2. Assays were initiated with addition of 1 μM wild-type or R232A/K268A RalBP1 to the reaction buffer. The real-time fluorescent signal (λex = 425 nm, λem = 465 nm) of the GAP reaction was monitored using a FluorMax 4 (Horiba) spectrofluorimeter with constant stirring at 15°C. An increase in fluorescence corresponds to a proportional increase in Pi production from GTP hydrolysis. The data were graphed in GraphPad Prism.
RESULTS
KRAS mutant pancreatic carcinoma cells form invadopodia.
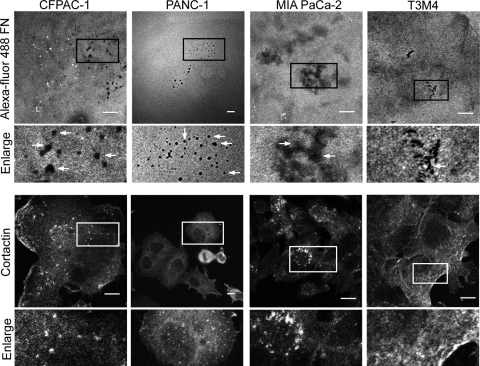
The signaling mechanisms that drive PDAC invasion and metastasis remain poorly characterized. Since the formation of invadopodia is associated with the invasive and metastatic potential of breast and other cancers, we first addressed the possibility that these structures are also found in PDAC cells. A panel of eight PDAC cell lines was analyzed for invadopodium activity using a fluorescent matrix degradation assay. Cells were seeded on fluorescently labeled fibronectin, and areas of matrix degradation that overlapped with anti-cortactin staining were identified as active invadopodia and scored. The majority of the cell lines analyzed exhibited moderate (more than 1 invadopodium per cell, percentage of invadopodium-positive cells per field of greater than 30%) to high (more than 3 invadopodia per cell, percentage of invadopodium-positive cells per field of greater than 40%) invadopodium activity (Fig. 1).
Fig 1.
PDAC cells form invadopodia. Eight PDAC cell lines (seven KRAS mutant and one wild-type [WT] KRAS) were analyzed for invadopodium activity using the in vitro matrix degradation assay. Cells were cultured for 24 h on cross-linked gelatin overlaid with Alexa Fluor 488-coupled fibronectin, fixed, and stained with antibody against cortactin. Quantitation was conducted to determine high (more than 3 invadopodia and a percentage of invadopodium-positive cells/field of greater than 40%), moderate (more than 1 invadopodium and a percentage of invadopodium-positive cells/field of greater than 30%), and low (0 to 1 invadopodia and a percentage of invadopodium-positive cells/field of 0 to 30%) invadopodium activity. Representative confocal images from a matrix degradation assay using CFPAC-1, PANC-1, MIA PaCa-2, and T3M4 cells. Boxed areas of interest (illustrating staining of cortactin in degraded areas of the matrix) are enlarged (2×). Scale bars, 10 μm. Data shown are representative of three independent experiments.
Activated K-Ras growth transformation of human pancreatic cells is associated with increased invadopodium activity.
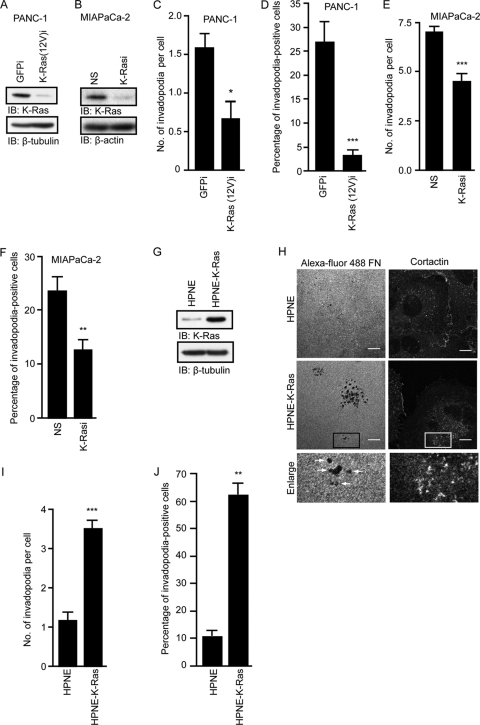
There appeared to be a correlation between high levels of invadopodium formation and the presence of a mutant KRAS allele. To determine if K-Ras expression is essential for invadopodium formation, we knocked down expression of the activated KRAS allele in PANC-1 cells (heterozygous for mutant KRAS) by stable expression of shRNA specific for this mutant sequence (9). We also knocked down total K-Ras protein expression in MIA PaCa-2 cells (homozygous for the mutant KRAS allele) using KRAS-specific shRNA sequences described previously (61). Stable expression of both shRNA constructs resulted in substantial reductions in the K-Ras protein (Fig. 2A and B). We observed that stable suppression of endogenous K-Ras(12V) expression in PANC-1 cells and total K-Ras expression in MIA PaCa-2 cells was associated with a significant decrease in invadopodia (Fig. 2C to F). These results suggest that continued expression of activated K-Ras is essential for invadopodia in PDAC cells.
Fig 2.
Activated K-Ras growth transformation of human pancreatic cells is associated with increased invadopodium activity. Expression of K-Ras(12V)-specific shRNA [K-Ras(12V)i] in PANC-1 cells and total K-Ras-specific shRNA (K-Rasi) in MIA PaCa-2 cells decreases invadopodium activity. (A) Western blot analysis of lysates from PANC-1 cells expressing GFP control shRNA or K-Ras(12V)-specific shRNA. (B) Western blot analysis of lysates from MIA PaCa-2 cells expressing nonspecific (NS) shRNA or total K-Ras-specific shRNA. (C and D) Expression of K-Ras(12V)-specific shRNA in PANC-1 cells decreases the number of invadopodia per cell (C) and the percentage of invadopodium-positive cells (D). Statistically significant differences between GFP shRNA- and K-Ras(12V)-specific shRNA-expressing cells, as determined by a Mann-Whitney U test, are indicated by a single asterisk (P value < 0.05) or three asterisks (P value < 0.005). (E and F) Expression of K-Ras-specific shRNA in MIA Paca-2 cells decreases the number of invadopodia per cell (E) and the percentage of invadopodium-positive cells (F). Statistically significant differences between NS shRNA- and K-Ras-specific shRNA-expressing cells, as determined by a Mann-Whitney U test, are indicated by two asterisks (P value < 0.01) or three asterisks (P value < 0.005). (G) Western blot analysis to verify K-Ras(12D) expression in HPNE-K-Ras cells. (H) Confocal images of HPNE and HPNE-K-Ras cells. Cells were seeded on coverslips coated with Alexa Fluor 488-coupled fibronectin and stained with antibody against cortactin. Boxed areas of interest (illustrating staining of cortactin in degraded areas of the matrix) are enlarged (2×). Scale bars, 10 μm. (I and J) Expression of K-Ras(12D) increases the number of invadopodia per cell (I) and the percentage of invadopodium-positive cells (J). Statistically significant differences between HPNE and HPNE-KRAS cells, as determined by a Mann-Whitney U test, are indicated by two asterisks (P value < 0.01) or three asterisks (P value < 0.0005). Ten fields (containing 15 to 40 cells each) per condition were used for all quantitation. Data are represented in graphs as means ± standard errors of the mean (SEM). Data shown are representative of two or more independent experiments.
Next, we determined whether expression of activated K-Ras in normal human pancreatic duct-derived epithelial cells (HPNE) is sufficient to induce invadopodium formation (13, 33). We showed previously that mutant KRAS induced the tumorigenic growth and invasion of HPNE cells (13, 43). Activated K-Ras(12D) was stably expressed in HPNE cells (HPNE-K-Ras), and the matrix degradation assay was performed. HPNE-K-Ras cells exhibited a significant increase in invadopodia compared to control HPNE cells (Fig. 2G to J). We conclude that mutant K-Ras is both necessary and sufficient for invadopodium formation in PDAC cells.
Activation of RalB is critical for invadopodium formation in KRAS mutant PDAC cell lines.
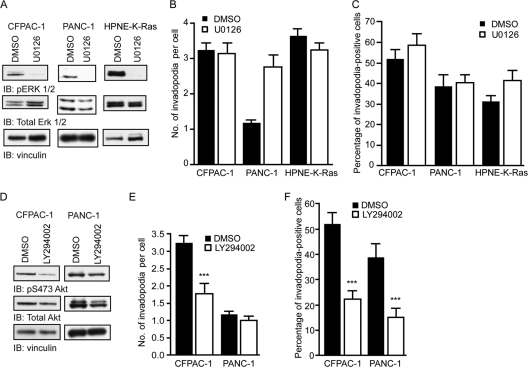
K-Ras activates multiple downstream signaling pathways, including the Raf-MEK-ERK cascade, the phosphatidylinositol 3-kinase (PI3K)-Akt, and the Ral small GTPase guanine nucleotide exchange factor (RalGEF)-Ral effector pathways (56). We next sought to investigate which downstream pathways were important for K-Ras-mediated invadopodium activity in PDAC cell lines. It was shown previously that ARF6 small GTPase- and transforming growth factor β-mediated invadopodium formation in melanoma and human breast cancer cells, respectively, is dependent on ERK signaling (40, 63). We therefore investigated whether this pathway was important for invadopodia in PDAC cell lines and HPNE cells expressing the activating KRAS(12D) mutant (HPNE-K-Ras). Invadopodium activity was assessed in PANC-1, CFPAC-1, and HPNE-K-Ras cells treated with the U0126 MEK1/MEK2-selective inhibitor. ERK1/ERK2 phosphorylation was decreased in cells treated with U0126 (Fig. 3A), but neither the number of invadopodia per cell (Fig. 3B) nor the percentage of cells with invadopodia (Fig. 3C) was decreased by inhibitor treatment. These results indicate that the Raf effector signaling pathway does not play a significant role in invadopodium formation in PDAC cells.
Fig 3.
The Raf-MEK-ERK signaling pathway downstream of activated K-Ras is not required for invadopodia, but pharmacological inhibition of PI3K decreases invadopodium activity. (A) Western blot analysis for phospho-ERK 1/2 (pERK1/2), total ERK, and vinculin of CFPAC-1, PANC-1, and HPNE-K-Ras cell lysates following treatment with 10 μM U0126 for 24 h. (B and C) Number of invadopodia per cell (B) and percentage of invadopodium-positive cells (C). (D) Western blot analysis for phosphoserine 473 Akt (pS473 Akt), total Akt, and vinculin of CFPAC-1 and PANC-1 cell lysates following treatment with 10 μM LY294002 for 24 h. (E and F) Number of invadopodia per cell (E) and percentage of invadopodium-positive cells (F). Statistically significant differences between dimethyl sulfoxide (DMSO)- and LY294002-treated cells, as determined by a Mann-Whitney U test, are indicated by three asterisks (P value < 0.005). Ten fields (containing 15 to 40 cells each) per condition were used for all quantitation. Data are means ± SEM. Data shown are representative of at least two independent experiments.
The phosphatidylinositol 3-kinase (PI3K) signaling pathway has an established role in invadopodium formation in melanoma (50) and breast cancer (70) cell lines. Therefore, we sought to investigate whether treatment of PDAC cells with an inhibitor of the PI3K pathway affected invadopodium formation. Treatment of CFPAC-1 and PANC-1 cells with LY294002 significantly reduced the levels of phosphorylation of Akt on serine 473 (Fig. 3D). This reduction in phosphorylation was accompanied by a significant, but not complete, reduction in the number of invadopodia per cell in CFPAC-1 cells (Fig. 3E) and in the percentage of invadopodium-positive cells in both CFPAC-1 and PANC-1 cells (Fig. 3F). These data suggest that elevated basal PI3K activity is necessary for invadopodium formation in PDAC cells. However, since our previous analyses did find that pAkt levels are not associated with mutant KRAS in PDAC cell lines (34) or patient tumors (36) and that shRNA suppression of mutant KRAS expression did not reduce pAkt levels (data not shown), PI3K activation may not be through its role as a K-Ras effector.
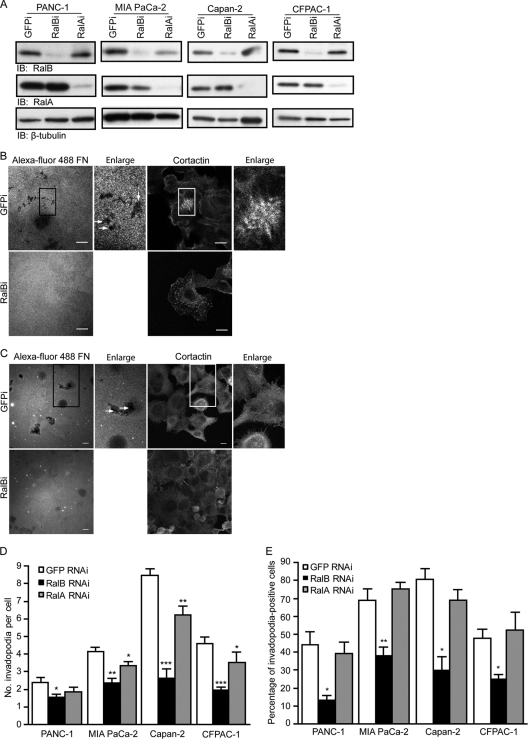
Our previous studies established a critical role for the RalGEF-RalB pathway in PDAC Matrigel invasion in vitro and/or metastasis as determined in the tail vein lung colonization assay (36, 64). Based on these observations, we speculated that Ral GTPases may be involved in PDAC invadopodium formation. To determine if expression of RalA or RalB is required for invadopodia in PDACs, we measured invadopodium formation in cells depleted of endogenous RalA or RalB by using RalA- and RalB-specific shRNAs. All four cell lines stably expressing RalB-specific shRNA showed significant decreases in numbers of invadopodia and in the percentages of invadopodium-positive cells (Fig. 4A to E). Decreases in numbers of invadopodia were also seen in three of the four cell lines expressing RalA-specific shRNA that were tested (Fig. 4D), although these effects were not as significant as those seen with a RalB knockdown. No changes were observed in the percentage of invadopodium-positive cells in cells expressing RalA-specific shRNA (Fig. 4E). These results suggest an essential role for RalB, and to a lesser extent RalA, in the establishment of an invasive phenotype.
Fig 4.
RalB plays a more predominant role than RalA in invadopodium formation in human KRAS mutant PDAC cell lines. (A) Western blot analysis of lysates from indicated cell types stably expressing RalB-specific (RalBi), RalA-specific (RalAi), or control GFP (GFPi) shRNA. Confocal images of CFPAC-1 (B) and PANC-1 (C) cells stably expressing RalB-specific (RalBi) or GFP (GFPi) shRNA seeded on coverslips coated with Alexa Fluor 488-coupled fibronectin and stained with anticortactin antibody. Boxed areas of interest (illustrating staining of cortactin in degraded areas of the matrix) are enlarged (2×). Scale bars, 10 μm. (D) The number of invadopodia per cell is decreased in cells expressing RalB-specific shRNA. (E) The percentage of invadopodium-positive cells is decreased more significantly in cells expressing RalB-specific shRNA. Ten fields (containing 15 to 40 cells each) per condition were used for all quantitation. Statistically significant differences between RalB shRNA or RalA shRNA and GFP shRNA cells, as determined by a Mann-Whitney U test, are indicated by one (P value < 0.05), two (P value < 0.01), or three (P value < 0.005) asterisks. Data are means ± SEM. Data shown are representative of three independent experiments.
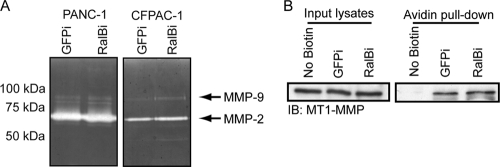
One of the major characteristics of productive invadopodia is the secretion and plasma membrane localization of matrix metalloproteinases (MMPs), including MMP-2, MMP-9, and MT1-MMP (19, 37, 52). The exocyst plays a central role in the transport and secretion of MMPs critical for invadopodium activity (38, 58). Because two of the major downstream effectors of activated RalB are Sec5 and Exo84, components of the exocyst, we examined whether the secretion and activity of MMP-2 and MMP-9 were impaired in PDAC cells expressing RalB-specific shRNA. Gelatin zymography was performed using conditioned media from control GFP shRNA- and RalB-specific shRNA-expressing cells. Unexpectedly, we observed no significant changes in the secretion or activity of MMP-2 or MMP-9 (Fig. 5A). We also examined whether plasma membrane localization of the transmembrane protease MT1-MMP was altered in cells with RalB suppression by using surface biotinylation followed by an avidin pulldown. Cells expressing RalB-specific shRNA showed no impairment in MT1-MMP plasma membrane localization (Fig. 5B). These data suggest that RalB does not mediate the secretion, localization, and activity of three of the major proteases involved in invadopodium activity in tumor cells.
Fig 5.
RalB suppression and impaired invadopodium formation are not associated with reduced MMP-2/MMP-9 gelatinase activity or MT1-MMP plasma membrane localization. (A) Representative zymograms from 24-h-conditioned medium showing secreted MMP-2 and MMP-9. (B) Representative Western blot analysis of MT1-MMP in input lysates and avidin pulldown following surface biotinylation in CFPAC-1 cells stably expressing control GFP (GFPi) or RalB-specific shRNA (RalBi). Data shown are representative of two independent experiments.
RalBP1 is essential for RalB-mediated invadopodium formation.
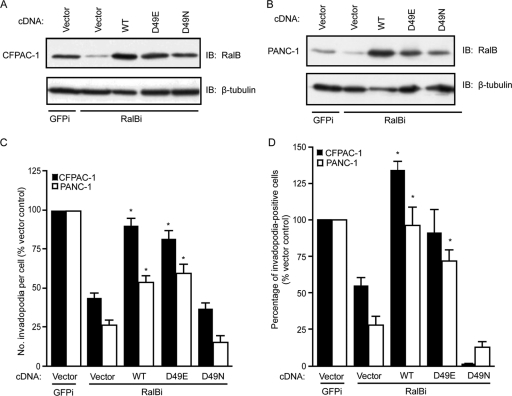
To date, the best-validated effectors of Ral function are two subunits (Sec5 and Exo84) of the octomeric exocyst complex. In particular, Sec5-mediated activation of the TBK1 protein kinase has been implicated in mutant KRAS-dependent promotion of tumor cell survival (5, 17). Furthermore, both Sec5 and Exo84 have been implicated in mutant KRAS-dependent, anchorage-independent growth transformation of human cells (26). Finally, our studies linked a third Ral effector, RalBP1/RLIP76, to RalA-dependent PDAC anchorage-independent and tumorigenic growth (35). That RalBP1 can function as a Rho-selective GTPase-activating protein (RhoGAP) and as a negative regulator of Rac1 and Cdc42, both implicated previously in invadopodium activity (32, 50, 58, 69), suggests that it may be an important effector of RalB-mediated invadopodia. Therefore, to address possible roles for Sec5, Exo84, and RalBP1 in RalB-dependent invadopodium formation, we utilized two well-characterized effector binding mutants of RalB (47, 48, 55). Introduction of a D49N point mutant into the Ral core effector binding domain impairs Ral binding to RalBP1 while retaining the ability of Ral to bind both Sec5 and Exo84. In contrast, a D49E point mutation retains RalBP1 binding but is impaired for both Sec5 and Exo84 binding. Both mutants have been utilized widely to dissect Ral effector function. We found that expression of WT RalB and RalB(D49E) in cells depleted of endogenous RalB restored the ability of these cells to form invadopodia. These results verify the specificity of the RalB shRNA and show that the two exocyst subunits are dispensable for RalB-induced invadopodia. In contrast, RalB(D49N) failed to rescue the deficiency in invadopodia caused by the suppression of endogenous RalB expression (Fig. 6A to D). These results suggest that RalBP1 may be a critical downstream effector necessary for invadopodium activity in PDAC cells.
Fig 6.
RalB D49N effector mutant with impaired RalBP1 binding fails to rescue invadopodium formation in PDAC cells expressing RalB-specific shRNA. Western blot analysis of lysates from CFPAC-1 (A) and PANC-1 (B) cells expressing GFP shRNA (GFPi), RalB-specific shRNA (RalBi), and either an empty vector or wild-type (WT), D49E, or D49N RalB. (C) Number of invadopodia per cell. (D) Percentage of invadopodium-positive cells per field. Ten fields (containing 15 to 40 cells each) per condition were used for all quantitation. Statistically significant differences between RalB-specific shRNA/vector and RalB-specific shRNA/WT or D49E RalB cells, as determined by a Mann-Whitney U test, are indicated by an asterisk (P value < 0.005). Data are means ± SEM. Data shown are representative of three independent experiments.
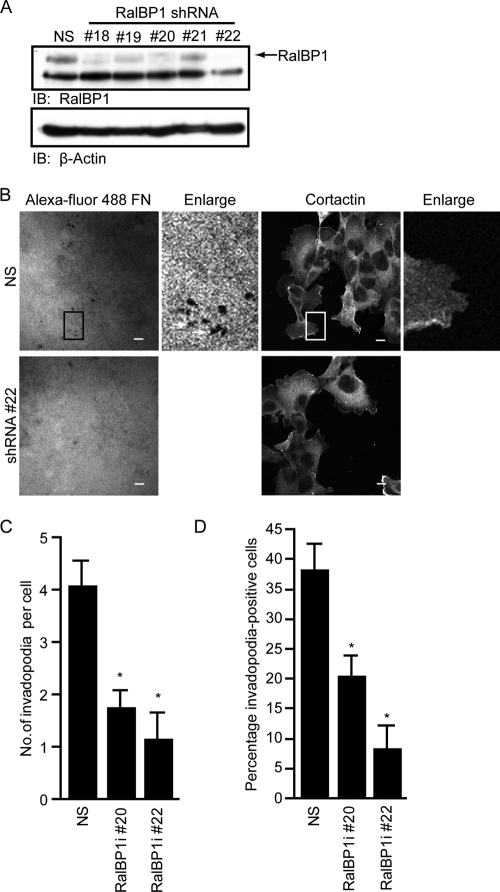
To directly determine the requirement for RalBP1 for invadopodium formation in PDAC cells, we utilized RalBP1-specific shRNA to suppress endogenous RalBP1 expression and analyzed the consequences on invadopodium formation. Suppression of endogenous RalBP1 expression using two shRNAs that target different RalBP1 sequences resulted in severely impaired invadopodium activity in CFPAC-1 cells (Fig. 7A to D). When taken together with the results from rescue experiments with RalB effector binding mutants, our analyses support a key role for RalBP1 as the critical RalB effector in this process.
Fig 7.
RalBP1 is required for PDAC cell invadopodium formation. (A) Western blot analysis of lysates from CFPAC-1 cells expressing nonspecific (NS) shRNA or shRNA clones directed against RalBP1 (RalBP1i). Clones 20 and 22 exhibited the most efficient knockdown of protein by Western blot and were chosen for further analyses. (B) Representative confocal images of CFPAC-1 cells expressing NS shRNA or RalBP1-specific shRNA clone 22. Cells were seeded on coverslips coated with Alexa Fluor 488-coupled fibronectin and stained for cortactin. Boxed areas of interest (illustrating staining of cortactin in degraded areas of the matrix) are enlarged (2×). Scale bars, 10 μm. (C) Number of invadopodia per cell. (D) Percentage of invadopodium-positive cells per field. Ten fields (containing 15 to 40 cells each) per condition were used for all quantitation. Statistically significant differences between NS shRNA- and RalBP1-specific shRNA-expressing cells, as determined by a Mann-Whitney U test, are indicated by an asterisk (P value < 0.005). Data are means ± SEM. Data shown are shown are representative of two independent experiments.
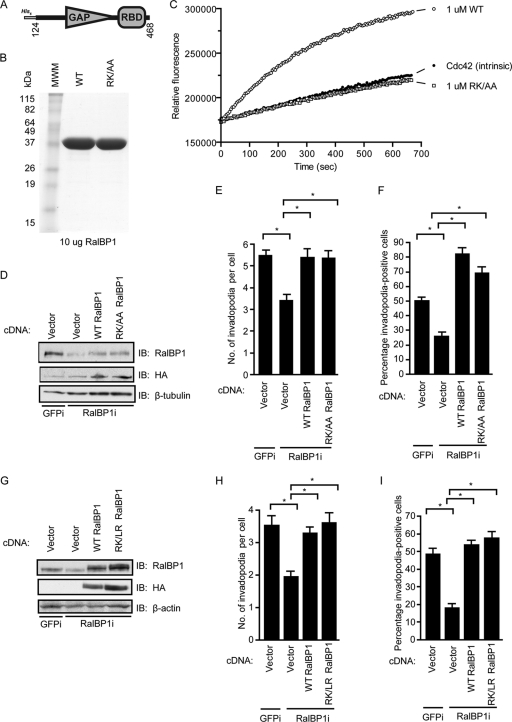
Although RalBP1 is a multidomain protein with diverse biochemical functions, its best known function is as a Rho-selective GTPase-activating protein (RhoGAP) and negative regulator of Cdc42 and Rac1 (14, 29, 54). Because Cdc42 is known to play an important role in invadopodium formation (50, 58), we investigated whether the GAP activity of RalBP1 is required for its role in invadopodium activity in PDAC cells. We generated a GAP-deficient mutant by introducing missense mutations in the arginine finger (R232) and the conserved lysine residue (K268) in the GAP catalytic domain of human RalBP1. We tested the ability of bacterially expressed wild-type and R232A/K268A RalBP1 GAP domains (amino acids [aa] 124 to 468) (Fig. 8A and B) to stimulate the intrinsic GTP hydrolysis activity of Cdc42 using a fluorescent-conjugated phosphate-binding protein assay (60). The presence of purified wild-type RalBP1 stimulated the intrinsic GTP hydrolysis of Cdc42, while the presence of an equivalent concentration of R232A/K268A RalBP1 failed to enhance hydrolysis (Fig. 8C).
Fig 8.
RalBP1 supports PDAC invadopodium formation through a GAP-independent mechanism. (A) Schematic of the domains of bacterially purified RalBP1 used in the GAP activity assay. (B) Representative Coomassie brilliant blue-stained gel of purified wild-type (WT) and R232A/K268A (RK/AA) RalBP1. (C) Real-time GTP hydrolysis rate of 2 μM purified GTP-loaded Cdc42 alone (no GAP) or in the presence of 1 μM WT or RK/AA RalBP1. (D) Western blot analysis of CFPAC-1 cells stably expressing GFP (GFPi) or RalBP1-specific shRNA (RalBP1i) and transiently expressing either empty vector or HA-tagged WT or RK/AA (GAP-deficient) RalBP1. (E) Number of invadopodia per cell. (F) Percentage of invadopodium-positive cells. Twenty fields (containing 15 to 40 cells each) per condition were used for quantitation, and only HA-positive cells were counted. Statistically significant differences between cells stably expressing RalBP1 shRNA and empty vector and either GFP shRNA and empty vector, RalBP1 shRNA and WT RalBP1, or RalBP1 shRNA and RK/AA RalBP1, as determined by a Mann-Whitney U test, are indicated by an asterisk (P < 0.005). (G) Western blot analysis of CFPAC-1 cells stably expressing GFP or RalBP1-specific shRNA and stably expressing either empty vector or HA-tagged WT or R208L/K244R (RK/LR) (GAP-deficient) RalBP1. (H) Number of invadopodia per cell. (I) Percentage of invadopodium-positive cells. Ten fields (containing 15 to 40 cells each) per condition were used for quantitation. Statistically significant differences between cells stably expressing RalBP1 shRNA and empty vector and either GFP shRNA and empty vector, RalBP1 shRNA and WT RalBP1, or RalBP1 shRNA and RK/LR RalBP1, as determined by a Mann-Whitney U test, are indicated by an asterisk (P < 0.005). Data are means ± SEM. Data shown are representative of three independent experiments.
We then generated HA-tagged, full-length wild-type and R232A/K268A RalBP1 RNAi-insensitive mammalian expression constructs to determine whether the role of RalBP1 in invadopodium formation was dependent on its catalytic GAP activity. We were unable to generate CFPAC-1 RalBP1 interference (RalBP1i) cells stably expressing the RK/AA RalBP1 mutant due to lethality, which was perhaps due to a dominant negative phenotype. Therefore, invadopodium activity was quantitated in transiently infected cells. HA epitope-tagged wild-type or R232A/K268A RalBP1-infected cells were identified by immunofluorescence staining with an anti-HA antibody. Only cells staining positive for HA were used for quantitation. We found that restoration of wild-type RalBP1 expression rescued impairment in invadopodium activity in cells expressing RalBP1-specific shRNA, demonstrating the specificity of the RalBP1 shRNA (Fig. 8D to F). Surprisingly, expression of GAP-deficient RalBP1(R232A/K268A) also rescued the loss of endogenous RalBP1. To verify that the role of RalBP1 in invadopodium activity was indeed GAP independent, we examined whether a previously validated GAP-deficient RalBP1 mutant (8) would also rescue the invadopodium deficiency in cells expressing RalBP1-specific shRNA. We stably expressed an R208L/K244R (RK/LR) RalBP1 RNAi-insensitive expression construct in RalBP1 shRNA-expressing CFPAC-1 cells. Expression of the RK/LR mutant protein also rescued the invadopodium-deficient phenotype in RalBP1i cells (Fig. 8G to I). These results demonstrate a GAP-independent role for RalBP1 in invadopodium formation in PDAC cells.
RalBP1 ATP-binding mutants fail to rescue invadopodium defects in cells expressing RalBP1 shRNA.
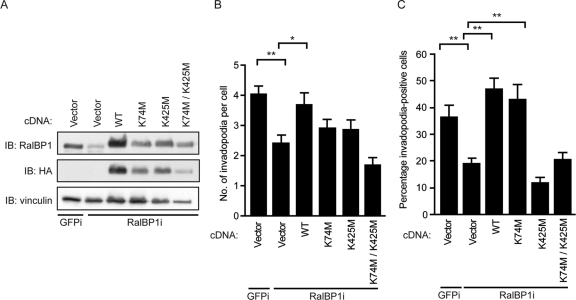
Another well-established role of RalBP1 is its function as an ATP-binding plasma membrane transporter pump (2, 3, 62). RalBP1 contains two validated ATP-binding sites at residues 69 to 74 and 418 to 425 (2). Mutation of lysines 74 and 425 to methionine in recombinant RalBP1 N and C terminus peptides impaired RalBP1 binding to ATP and ATPase activity (2). We sought to investigate whether the ATPase function of RalBP1 was important for its role in supporting invadopodium formation. RalBP1 variants containing the K74M, K425M, or combined K74M/K425M mutations were generated, stably expressed in RalBP1 shRNA-expressing cells, and evaluated for their ability to rescue the loss of endogenous RalBP1. The K425M mutant, either alone or in combination with the K74M mutation, failed to rescue the invadopodium defects in cells expressing RalBP1 shRNA (Fig. 9A to C). These results suggest that ATP binding to RalBP1, and perhaps its plasma membrane transport role, is essential for invadopodia in PDAC cells.
Fig 9.
RalBP1 ATP-binding mutants fail to rescue invadopodium defects in cells expressing RalBP1 shRNA. (A) Western blot analysis of CFPAC-1 cells stably expressing GFP (GFPi) or RalBP1-specific shRNA (RalBP1i) and stably expressing either empty vector or HA-tagged WT, K74M, K425M, or K74M/K425M RalBP1. (B) Number of invadopodia per cell. (C) Percentage of invadopodium-positive cells. Ten fields (containing 15 to 40 cells each) per condition were used for quantitation. Statistically significant differences between the groups within the brackets, as determined by a Mann-Whitney U test, are indicated by one (P value < 0.01) or two (P value < 0.005) asterisks. Data are means ± SEM. Data shown are representative of three independent experiments.
DISCUSSION
Although the best-studied effector pathways of Ras are those involving Raf or PI3K, there is greater appreciation that the dependency of human tumor cells on mutant KRAS must involve additional effectors (39, 59, 61). In support of this possibility, our recent studies suggest that the RalGEF effector pathway, rather than the Raf or PI3K effector pathway, may be a major mediator of KRAS-dependent PDAC growth (34). Although RalGEF leads to activation of the highly related RalA and RalB proteins, there is now considerable evidence that they each serve distinct roles in oncogenesis (7, 51). For PDAC, we found that RalB was dispensable for anchorage-independent and tumorigenic growth and instead was important for invasion and metastasis. To further explore the signaling mechanisms that drive PDAC invasion and metastasis, we first determined that PDAC tumor cells form invadopodia, plasma membrane protrusions that promote matrix degradation, and that their formation was dependent on mutant KRAS function. We then found that this activity was dependent on RalB, and to a lesser extent RalA, and not the Raf effector pathway. Surprisingly, RalB-mediated invadopodium formation was not dependent on exocyst regulation and instead was dependent on a RhoGAP-independent function of the RalBP1 effector.
Previous studies have described invadopodium formation for Src-transformed cells and a variety of human cancers, in particular, breast carcinoma, melanoma, and glioblastoma. Our studies provide the first description of invadopodia in PDAC and its association with K-Ras. The six cell lines that showed moderate to high invadopodium activity possessed activating KRAS mutations, suggesting a correlation between KRAS mutation status and invadopodium activity. We conclude that invadopodia are a common feature of PDAC cells, thus expanding the spectrum of human cancers for which they may contribute to invasive and malignant growth. The essential roles of mutationally activated K-Ras in initiation and progression in the early stages of pancreatic cancer are well established. However, our results suggest that oncogenic K-Ras can additionally promote the invasive phenotype that occurs later in tumor progression. One mechanism by which this may be achieved is a switch in K-Ras effector utilization at different stages of tumor progression. Recently, we described such a mechanism where Ras can switch from use of Raf to use of RalGEF, leading to changes in Ras-driven cell fate (71). Finally, whether a K-Ras-dependent induction of invadopodia will be seen in other human cancers (lung and colorectal) with frequent KRAS mutations is important to determine.
Our data strongly implicate the RalB-mediated signaling pathway downstream of K-Ras and suggest the absence of a role for the MAPK pathway. In contrast, there appears to be a clear role for the PI3K signaling pathway in PDAC invadopodium activity based on our inhibitor studies. However, there is no definitive association between KRAS mutation status and activation of the PI3K pathway in PDAC cells. This is illustrated by our previous data showing that pS473 Akt levels are not elevated in human PDAC cell lines (34) or patient tumors (36). In addition, a recent study identified the frequent coexistence of KRAS and PIK3CA mutations in multiple advanced-stage cancers (27), suggesting that the PI3 kinase pathway may function independently of K-Ras activation.
Our implication of K-Ras and RalB in PDAC invadopodium formation extends the repertoire of small GTPases associated with this process. Previous studies have implicated Rho, Rac, Cdc42, and Wrch-1 Rho family small GTPases in cytoskeleton-associated processes and Arf6 in transport processes that are associated with invadopodia (22, 32, 49, 50, 58, 63, 69). It is essential to determine whether RalB is also important for invadopodium activity in breast, melanoma, and other cancers to assess whether RalB-dependent invadopodium formation is unique for PDAC or shared with other cancers.
Our finding that RalB, to a greater extent than RalA, is required for invadopodium formation is consistent with our previous determination of a more critical role of RalB in PDAC invasion and metastasis. However, what was unexpected was our determination of the effectors that mediate RalB function. Since secretion of proteases is a key property of invadopodia, we anticipated that the Sec5 and Exo84 subunits of the exocyst would be required. Previously, the Sec3, Exo70, and Sec8 exocyst subunits were shown to be required for invadopodium formation in KRAS mutant MDA-MB-231 breast carcinoma cells, primarily for MMP regulation and matrix degradation (38, 58). However, our finding that exocyst binding was not required for RalB function is consistent with our result that RalB suppression did not reduce MMP protease activity in PDAC. Perhaps non-Ral-dependent mechanisms that regulate exocyst function and MMP secretion are involved in PDAC invadopodia. Finally, our exclusion of the importance of Sec5 and Exo84 from RalB function contrasts with the majority of other reported functions of RalB, including regulation of cytokinesis (15), innate immunity (17), or the cellular starvation response (6), in which exocyst subunit function was found to be essential.
In light of the established involvement of Rac and Cdc42 in invadopodia, our finding that the RalBP1 requirement for RalB-mediated invadopodium formation is largely RhoGAP independent was unexpected. This result differs from our finding that RalA-dependent PDAC growth was dependent on RalBP1 and involved negative regulation of Rac and Cdc42 function (35). However, RalBP1 is a large multidomain protein with a variety of other functions mediated through interactions with a spectrum of functionally distinct proteins. Instead, we found that an ATPase-deficient mutant of RalBP1 was drastically impaired in its ability to rescue the loss of endogenous RalBP1 and restore invadopodium activity. The ATP binding and ATPase activity of RalBP1 have been reported to be required for RalBP1 function as a non-ABC transporter of structurally divergent molecules, including chemotherapeutic drugs, out of cells (1, 2). Interestingly, while the K425 residue of the C-terminal ATP-binding motif does not directly overlap the Ral-GTP binding domain of RalBP1, it is speculated that the RalB-GTP association may regulate ATP binding (20). Whether it is this function as a transporter that is essential for RalBP1 function in invadopodium formation and whether this function is regulated by RalB binding are important future issues to resolve.
RalBP1 is also known to associate with several proteins involved in endocytosis, including the μ2 subunit of the clathrin adaptor complex AP-2 (30), activin receptor-interacting protein 2 (ARIP2) (44), partner of RalBP1 (POB1) (46), and Reps1 (68). RalBP1 is involved in stress responses and mitosis through its interactions with heat shock factor 1 (HSF-1) (24) and Cdk1 (57), respectively. The multidomain structure of RalBP1 links it to a number of physiological roles within the cell that may be important for invadopodia. Putative membrane association domains were previously identified for RalBP1 (67). In addition, active Ral can recruit RalBP1 to the membrane (42), and it is likely that this localization is important for its function in invadopodia. RalBP1 serves as an endocytic adaptor protein (25, 30, 68), and it is likely to play a scaffolding role in other cellular processes, including perhaps invadopodia. Future detailed mutagenesis analyses of RalBP1 will be required to identify the function(s) essential for invadopodium formation in PDAC cells.
In summary, our studies implicate for the first time the importance of the aberrant activation of K-Ras and the downstream Ral effector signaling pathway for invadopodium activity in pancreatic cancer cells. Specifically, we demonstrate that RalB is necessary for invadopodium function through its effector RalBP1. Interestingly, we show that RalBP1 regulates invadopodium activity in a GAP-independent manner. Further elucidation of the mechanism by which RalBP1 promotes invadopodia may define novel directions for therapeutic strategies for metastatic PDAC. In summary, while the Src oncoprotein and Rho GTPases are well-characterized regulators of invadopodia, our delineation of a K-Ras- and RalB-mediated mechanism defines a novel signaling mechanism for invadopodium formation.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants CA042978 (C.J.D.) and CA140424 (J.J.Y.) from the National Cancer Institute. N.F.N. was supported by a fellowship from the American Cancer Society (18777-PF-10-023-01-CSM).
We thank Alissa Weaver (Vanderbilt University School of Medicine) for providing technical advice. We thank the laboratories of Jeffrey Settleman (MGH-Harvard) and Christopher Counter (Duke University) for providing reagents. We also acknowledge the Michael Hooker Microscope facility at UNC for providing the Zeiss microscope on which the images in the article were acquired.
Footnotes
Published ahead of print 13 February 2012
REFERENCES
- 1. Awasthi S, et al. 2000. Novel function of human RLIP76: ATP-dependent transport of glutathione conjugates and doxorubicin. Biochemistry 39: 9327–9334 [DOI] [PubMed] [Google Scholar]
- 2. Awasthi S, et al. 2001. Functional reassembly of ATP-dependent xenobiotic transport by the N- and C-terminal domains of RLIP76 and identification of ATP binding sequences. Biochemistry 40: 4159–4168 [DOI] [PubMed] [Google Scholar]
- 3. Awasthi S, Singhal SS, Sharma R, Zimniak P, Awasthi YC. 2003. Transport of glutathione conjugates and chemotherapeutic drugs by RLIP76 (RALBP1): a novel link between G-protein and tyrosine kinase signaling and drug resistance. Int. J. Cancer 106: 635–646 [DOI] [PubMed] [Google Scholar]
- 4. Baines AT, et al. 2006. Use of retrovirus expression of interfering RNA to determine the contribution of activated K-Ras and ras effector expression to human tumor cell growth. Methods Enzymol. 407: 556–574 [DOI] [PubMed] [Google Scholar]
- 5. Barbie DA, et al. 2009. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bodemann BO, et al. 2011. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 144: 253–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bodemann BO, White MA. 2008. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat. Rev. Cancer 8: 133–140 [DOI] [PubMed] [Google Scholar]
- 8. Boissel L, et al. 2007. Recruitment of Cdc42 through the GAP domain of RLIP participates in remodeling of the actin cytoskeleton and is involved in Xenopus gastrulation. Dev. Biol. 312: 331–343 [DOI] [PubMed] [Google Scholar]
- 9. Brummelkamp TR, Bernards R, Agami R. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2: 243–247 [DOI] [PubMed] [Google Scholar]
- 10. Buccione R, Caldieri G, Ayala I. 2009. Invadopodia: specialized tumor cell structures for the focal degradation of the extracellular matrix. Cancer Metastasis Rev. 28: 137–149 [DOI] [PubMed] [Google Scholar]
- 11. Buccione R, Orth JD, McNiven MA. 2004. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat. Rev. Mol. Cell Biol. 5: 647–657 [DOI] [PubMed] [Google Scholar]
- 12. Campbell PM, Der CJ. 2004. Oncogenic Ras and its role in tumor cell invasion and metastasis. Semin. Cancer Biol. 14: 105–114 [DOI] [PubMed] [Google Scholar]
- 13. Campbell PM, et al. 2007. K-Ras promotes growth transformation and invasion of immortalized human pancreatic cells by Raf and phosphatidylinositol 3-kinase signaling. Cancer Res. 67: 2098–2106 [DOI] [PubMed] [Google Scholar]
- 14. Cantor SB, Urano T, Feig LA. 1995. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol. Cell. Biol. 15: 4578–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cascone I, et al. 2008. Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. EMBO J. 27: 2375–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen W-T, Yunyon Y, Nakahara H. 1994. An in vitro cell invasion assay: determination of cell surface proteolytic activity that degrades extracellular matrix. J. Tissue Cult. Methods 16: 177–181 [Google Scholar]
- 17. Chien Y, et al. 2006. RalB GTPase-mediated activation of the IκB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 127: 157–170 [DOI] [PubMed] [Google Scholar]
- 18. Cox AD, Der CJ. 2010. Ras history: The saga continues. Small Gtpases 1: 2–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Destaing O, Block MR, Planus E, Albiges-Rizo C. 2011. Invadosome regulation by adhesion signaling. Curr. Opin. Cell Biol. 23: 597–606 [DOI] [PubMed] [Google Scholar]
- 20. Fenwick RB, et al. 2010. The RalB-RLIP76 complex reveals a novel mode of ral-effector interaction. Structure 18: 985–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gimona M, Buccione R, Courtneidge SA, Linder S. 2008. Assembly and biological role of podosomes and invadopodia. Curr. Opin. Cell Biol. 20: 235–241 [DOI] [PubMed] [Google Scholar]
- 22. Hashimoto S, et al. 2004. Requirement for Arf6 in breast cancer invasive activities. Proc. Natl. Acad. Sci. U. S. A. 101: 6647–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoon DS, et al. 2006. Molecular mechanisms of metastasis. Cancer Metastasis Rev. 25: 203–220 [DOI] [PubMed] [Google Scholar]
- 24. Hu Y, Mivechi NF. 2003. HSF-1 interacts with Ral-binding protein 1 in a stress-responsive, multiprotein complex with HSP90 in vivo. J. Biol. Chem. 278: 17299–17306 [DOI] [PubMed] [Google Scholar]
- 25. Ikeda M, Ishida O, Hinoi T, Kishida S, Kikuchi A. 1998. Identification and characterization of a novel protein interacting with Ral-binding protein 1, a putative effector protein of Ral. J. Biol. Chem. 273: 814–821 [DOI] [PubMed] [Google Scholar]
- 26. Issaq SH, Lim KH, Counter CM. 2010. Sec5 and Exo84 foster oncogenic ras-mediated tumorigenesis. Mol. Cancer Res. 8: 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Janku F, et al. 2011. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One 6: e22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jones S, et al. 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jullien-Flores V, et al. 1995. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J. Biol. Chem. 270: 22473–22477 [DOI] [PubMed] [Google Scholar]
- 30. Jullien-Flores V, et al. 2000. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. J. Cell Sci. 113(Pt 16): 2837–2844 [DOI] [PubMed] [Google Scholar]
- 31. Karnoub AE, Weinberg RA. 2008. Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 9: 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuroiwa M, Oneyama C, Nada S, Okada M. 2011. The guanine nucleotide exchange factor Arhgef5 plays crucial roles in Src-induced podosome formation. J. Cell Sci. 124: 1726–1738 [DOI] [PubMed] [Google Scholar]
- 33. Lee KM, Nguyen C, Ulrich AB, Pour PM, Ouellette MM. 2003. Immortalization with telomerase of the Nestin-positive cells of the human pancreas. Biochem. Biophys. Res. Commun. 301: 1038–1044 [DOI] [PubMed] [Google Scholar]
- 34. Lim KH, et al. 2005. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell 7: 533–545 [DOI] [PubMed] [Google Scholar]
- 35. Lim KH, et al. 2010. Aurora-A phosphorylates, activates, and relocalizes the small GTPase RalA. Mol. Cell. Biol. 30: 508–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lim KH, et al. 2006. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr. Biol. 16: 2385–2394 [DOI] [PubMed] [Google Scholar]
- 37. Linder S, Wiesner C, Himmel M. 2011. Degrading devices: invadosomes in proteolytic cell invasion. Annu. Rev. Cell Dev. Biol. 27: 185–211 [DOI] [PubMed] [Google Scholar]
- 38. Liu J, Yue P, Artym VV, Mueller SC, Guo W. 2009. The role of the exocyst in matrix metalloproteinase secretion and actin dynamics during tumor cell invadopodia formation. Mol. Biol. Cell 20: 3763–3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Luo J, et al. 2009. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mandal S, Johnson KR, Wheelock MJ. 2008. TGF-beta induces formation of F-actin cores and matrix degradation in human breast cancer cells via distinct signaling pathways. Exp. Cell Res. 314: 3478–3493 [DOI] [PubMed] [Google Scholar]
- 41. Martin TD, Samuel JC, Routh ED, Der CJ, Yeh JJ. 2011. Activation and involvement of Ral GTPases in colorectal cancer. Cancer Res. 71: 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matsubara K, Hinoi T, Koyama S, Kikuchi A. 1997. The post-translational modifications of Ral and Rac1 are important for the action of Ral-binding protein 1, a putative effector protein of Ral. FEBS Lett. 410: 169–174 [DOI] [PubMed] [Google Scholar]
- 43. Matsuo Y, et al. 2009. K-Ras promotes angiogenesis mediated by immortalized human pancreatic epithelial cells through mitogen-activated protein kinase signaling pathways. Mol. Cancer Res. 7: 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matsuzaki T, et al. 2002. Regulation of endocytosis of activin type II receptors by a novel PDZ protein through Ral/Ral-binding protein 1-dependent pathway. J. Biol. Chem. 277: 19008–19018 [DOI] [PubMed] [Google Scholar]
- 45. Mitin N, et al. 2007. Release of autoinhibition of ASEF by APC leads to CDC42 activation and tumor suppression. Nat. Struct. Mol. Biol. 14: 814–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morinaka K, et al. 1999. Epsin binds to the EH domain of POB1 and regulates receptor-mediated endocytosis. Oncogene 18: 5915–5922 [DOI] [PubMed] [Google Scholar]
- 47. Moskalenko S, et al. 2002. The exocyst is a Ral effector complex. Nat. Cell Biol. 4: 66–72 [DOI] [PubMed] [Google Scholar]
- 48. Moskalenko S, et al. 2003. Ral GTPases regulate exocyst assembly through dual subunit interactions. J. Biol. Chem. 278: 51743–51748 [DOI] [PubMed] [Google Scholar]
- 49. Muralidharan-Chari V, et al. 2009. ADP-ribosylation factor 6 regulates tumorigenic and invasive properties in vivo. Cancer Res. 69: 2201–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nakahara H, et al. 2003. Involvement of Cdc42 and Rac small G proteins in invadopodia formation of RPMI7951 cells. Genes Cells 8: 1019–1027 [DOI] [PubMed] [Google Scholar]
- 51. Neel NF, et al. 2011. The RalGEF-Ral effector signaling network: the road less traveled for anti-Ras drug discovery. Genes Cancer 2: 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nurnberg A, Kitzing T, Grosse R. 2011. Nucleating actin for invasion. Nat. Rev. Cancer. 11: 177–187 [DOI] [PubMed] [Google Scholar]
- 53. Pantel K, Brakenhoff RH. 2004. Dissecting the metastatic cascade. Nat. Rev. Cancer 4: 448–456 [DOI] [PubMed] [Google Scholar]
- 54. Park SH, Weinberg RA. 1995. A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene 11: 2349–2355 [PubMed] [Google Scholar]
- 55. Polzin A, Shipitsin M, Goi T, Feig LA, Turner TJ. 2002. Ral-GTPase influences the regulation of the readily releasable pool of synaptic vesicles. Mol. Cell. Biol. 22: 1714–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Repasky GA, Chenette EJ, Der CJ. 2004. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 14: 639–647 [DOI] [PubMed] [Google Scholar]
- 57. Rosse C, L'Hoste S, Offner N, Picard A, Camonis J. 2003. RLIP, an effector of the Ral GTPases, is a platform for Cdk1 to phosphorylate epsin during the switch off of endocytosis in mitosis. J. Biol. Chem. 278: 30597–30604 [DOI] [PubMed] [Google Scholar]
- 58. Sakurai-Yageta M, et al. 2008. The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J. Cell Biol. 181: 985–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Scholl C, et al. 2009. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 137: 821–834 [DOI] [PubMed] [Google Scholar]
- 60. Shutes A, Der CJ. 2006. Real-time in vitro measurement of intrinsic and Ras GAP-mediated GTP hydrolysis. Methods Enzymol. 407: 9–22 [DOI] [PubMed] [Google Scholar]
- 61. Singh A, et al. 2009. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 15: 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Singhal SS, Yadav S, Roth C, Singhal J. 2009. RLIP76: a novel glutathione-conjugate and multi-drug transporter. Biochem. Pharmacol. 77: 761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tague SE, Muralidharan V, D'Souza-Schorey C. 2004. ADP-ribosylation factor 6 regulates tumor cell invasion through the activation of the MEK/ERK signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 101: 9671–9676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vigil D, et al. 2010. Aberrant overexpression of the Rgl2 Ral small GTPase-specific guanine nucleotide exchange factor promotes pancreatic cancer growth through Ral-dependent and Ral-independent mechanisms. J. Biol. Chem. 285: 34729–34740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. 2011. Pancreatic cancer. Lancet 378: 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wolf K, Friedl P. 2009. Mapping proteolytic cancer cell-extracellular matrix interfaces. Clin. Exp. Metastasis 26: 289–298 [DOI] [PubMed] [Google Scholar]
- 67. Yadav S, et al. 2004. Identification of membrane-anchoring domains of RLIP76 using deletion mutant analyses. Biochemistry 43: 16243–16253 [DOI] [PubMed] [Google Scholar]
- 68. Yamaguchi A, Urano T, Goi T, Feig LA. 1997. An Eps homology (EH) domain protein that binds to the Ral-GTPase target, RalBP1. J. Biol. Chem. 272: 31230–31234 [DOI] [PubMed] [Google Scholar]
- 69. Yamaguchi H, et al. 2005. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J. Cell Biol. 168: 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yamaguchi H, et al. 2011. Phosphoinositide 3-kinase signaling pathway mediated by p110alpha regulates invadopodia formation. J. Cell Biol. 193: 1275–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zand TP, Reiner DJ, Der CJ. 2011. Ras effector switching promotes divergent cell fates in C. elegans vulval patterning. Dev. Cell 20: 84–96 [DOI] [PMC free article] [PubMed] [Google Scholar]