Abstract
We have used the RAW 264.7 murine macrophage-like cell line as a platform to characterize the recognition and early signaling response to recombinant adenoviral vectors (rAdV). Infection of RAW 264.7 cells triggers an early response (2 to 6 h postinfection) that includes phosphorylation of the interferon (IFN) response factor 3 (IRF3) transcription factor, upregulation of IRF3 primary response genes (interferon-stimulated gene 56 [ISG56], beta IFN [IFN-β]), and subsequent type I IFN secondary signaling (STAT1/2 phosphorylation). Using short hairpin RNA (shRNA) lentiviral vectors, we show an essential role for Tank binding kinase 1 (TBK1) in this pathway. Data also support a role for STING (MITA) as an adaptor functioning in response to rAdV infection. Using UV/psoralen (Ps)-inactivated virus to block viral transcription, Ps-inactivated virus stimulated primary (IRF3) and secondary (STAT1/2) activation events to the same degree as untreated virus. IRF3 phosphorylation was not blocked in RAW 264.7 cells pretreated with the RNA polymerase III inhibitor ML60218. However, they were compromised in the type I IFN-dependent secondary response (phosphorylation of STAT1/STAT2). At 24 h postinfection, ML60218-treated cells were compromised in the overall antiviral response. Therefore, initial sensing of rAdV or viral DNA (vDNA) does not depend on viral template transcription, but ML60218 treatment influences cellular cascades required for an antiviral response to rAdV. Using overexpression or knockdown assays, we examined how four DNA sensors influence the antiviral response. Knockdown of DNA Activator of Interferon (DAI) and p204, the murine ortholog to IFI16, had minimal influence on IRF3 phosphorylation. However, knockdown of absent in melanoma 2 (AIM2) and the helicase DDX41 resulted in diminished levels of pser388IRF3 following rAdV infection. Based on these data, multiple DNA sensors contribute to an antiviral DNA recognition response, leading to TBK1-dependent IRF3 phosphorylation in RAW 264.7 cells.
INTRODUCTION
Early recognition of viral infection by sentinel immune cells is key to induction of the innate and adaptive arms of antiviral immunity. In the case of adenovirus (Ad) and recombinant adenoviral vectors (rAdV), early recognition by antigen-presenting cells (APCs) (macrophages and dendritic cells) generates an antiviral response that is biased toward a type I interferon (IFN) pathway (8, 32, 49). Multiple viral components contribute to anti-Ad recognition by APCs, including viral capsid proteins (6, 34, 38), virus-dependent transcription (46), and the viral genome (8, 29, 31, 32, 49). In the murine model, recognition of the double-stranded DNA (dsDNA) viral genome occurs in a cell type-specific manner, where the Toll-like receptor 9 (TLR9) endosomal receptor responds to rAd DNA in plasmacytoid dendritic cells (DCs) (49), while a cytosolic DNA sensor is implicated in primary murine macrophages and classical dendritic cells (29, 32, 49).
Depending on the cellular environment, the murine APC response to rAdV can trigger distinct antiviral cascades. The classic antiviral interferon response is initiated by activation of interferon response factor 3 (IRF3) and contributes to type I IFN gene expression. Activated IRF3 in combination with NF-κB and ATF-2/cJUN binds to the beta interferon (IFN-β) promoter, leading to early expression of IFN-β mRNA as well as a number of other IRF3-dependent transcription units (31). Secretion of type I IFNs (and other chemokines and cytokines) by the activated cell leads to paracrine-autocrine signaling, which amplifies the antiviral response. Type I interferon binding to the IFN-α/β receptor triggers Janus kinase phosphorylation of Stat1 and STAT2, which combine with IRF9 to form the heterotrimeric ISGF3 transcription factor. In the case of macrophage and conventional dendritic cells, the culmination of the primary and secondary antiviral cascades is APC maturation from a naïve to a mature phenotype (8, 31, 32).
A second antiviral DNA response leads to inflammasome formation (29) and is characterized by caspase-1 cleavage of pro-interleukin-1 beta (pro-IL-1β) to IL-1β (as well as processing of pro-IL-18 to IL-18) (reviewed in references 22 and 33). In the case of rAdV, initial studies characterizing inflammasome activation were carried out using primary murine macrophage primed with lipopolysaccharide (LPS) (29). rAdV stimulation of the Caspase-1/IL-1β inflammasome pathway was dependent on both NOD-like receptor 3 (NLRP3) and apoptosis-associated speck-like protein (ASC). Macrophages derived from either ASC or NLRP3 knockout (KO) mice were compromised in inflammasome activation by rAdV, but IRF3 activation remained intact. In contrast to viral infection, when viral DNA (vDNA) was introduced through chemical transduction, ASC but not NLRP3 was required for Caspase-1 cleavage and secretion of IL-1β. Neither ASC nor NLRP3 contains known DNA binding domains. For both inflammasome and interferon antiviral response cascades, a cytosolic DNA sensor has been proposed, but the nature of the Ad DNA sensor has not been elucidated.
Studies using direct DNA transfection in an array of cell types under various sets of conditions have established a role for DNA sensor proteins in the type I IFN response (15, 41) as well as the inflammasome pathway (29). The first interferon-activating cytosolic DNA sensor identified was the response factor DNA Activator of Interferon (DAI; previously named DLM1 and ZBP1) (42). RNA interference (RNAi) knockdown studies in L929 cells revealed reduced antiviral response at the level of IFN-β mRNA and IRF3 dimer formation following DNA transfection. Recent studies have also shown that, independently of DNA activation, DAI overexpression results in NF-κB induction through RIP1/3 interactions (19). In vivo studies previously showed that mice deficient for DAI were not compromised in the innate or adaptive response to DNA vaccination (16). Primary mouse embryonic fibroblasts from the DAI KO mice were not impaired in IFN stimulation following DNA or herpes simplex virus (HSV) stimulation (16). Therefore, additional DNA sensors have been proposed to contribute to the type I interferon response. One such sensor is the HIN-200 family member IFI16 (mouse homolog, IFI/P204), which stimulates type I IFN expression through a STING/TBK1 cascade following DNA presentation or infection by HSV-1 (43), and another candidate DNA sensor leading to type I IFN induction is the DDX41 helicase (48), which also stimulates the type I IFN response through the STING/TBK1 cascade.
A second class of cytosolic DNA sensors is represented by the absent in melanoma 2 protein (AIM2). Several laboratories identified Aim2 as a DNA sensor that anchors formation of an AIM2-dependent inflammasome complex following DNA transfection (3, 9, 14, 36) (reviewed in reference 21). Aim2 binds DNA through the HIN-200 domain. It contains a C-terminal pyrin domain through which it binds ASC and demonstrates a cytosolic presence, unlike other HIN-200 family members. Binding to ASC results in caspase-1 activation, leading to conversion of pro-IL-1β to IL-1β. IL-1β activation contributes to autocrine-paracrine activation through the IL-1 receptor. IL-1 receptor ligation triggers an MyD88-dependent TRAF6 cascade leading to activation of NF-κB and cJUN transcription factors, stimulating proinflammatory genes (IL-6, tumor necrosis factor [TNF], IFN, and transforming growth factor β [TGFβ]). Potent stimulation of inflammasome pathways leads to cell death through pyroptosis (reviewed in reference 23). A recent report identified IFI16 as a nuclear DNA sensor for Kaposi's sarcoma-associated herpesvirus (KSHV), leading to formation of an ASC-dependent inflammasome complex (20).
Foreign DNA can also be detected through an indirect mechanism via RNA polymerase III (Pol III) (1, 4). This pathway operates through transcription of dsDNA rich in dA-dT domains. The RNA polymerase III sensor appears to function predominantly on short, duplex dA-dT templates and generates an RNA transcript that engages the RNA RIG-I/IPS pathway, leading to IRF3 activation and type I IFN stimulation. RNA polymerase III is responsible for transcription of adenoviral VAI and VAII transcripts, which may be involved in triggering an IFN antiviral response (46).
In the present study, we used the RAW 264.7 murine macrophage-like cell line as a model to determine involvement of DNA sensors in rAdV IRF3 activation. The data reveal that viral transcription is not required for antiviral recognition by these cells. When we characterized the effects of short hairpin RNA (shRNA) knockdowns targeting 4 previously identified DNA sensors, we found that DAI and p204 are not required for the antiviral response, whereas AIM2 and DDX41 knockdowns influence the levels of IRF3 activation following rAdV infection.
MATERIALS AND METHODS
Viruses and AdV DNA.
Ad5CiG (cytomegalovirus-chloramphenicol acetyltransferase internal ribosome entry site green fluorescent protein [CMV-CATiresGFP] reporter in E1 region) (38) and ts1-Ad2 (13) were previously described. Viruses were grown on a large scale in 293 cells, followed by 2 rounds of CsCl banding and dialysis against 4% sucrose–50 mM Tris (pH 8.0)–2 mM MgCl2, and stored at −80°C. Viral particle numbers were quantified by spectrophotometric detection of intact virions and determination of optical density at 260 nm (OD260) (1012 particles [p]/OD260 unit). Stocks of psoralen (Ps; Sigma) (100×; 33 mg/ml) were prepared in dimethyl sulfoxide (DMSO) (at concentrations which did not cause psoralen precipitation during treatment). Psoralen-UV inactivation was performed by first mixing 0.01 volumes of psoralen (Sigma) with 400 μl of purified viral vector (∼109 particles/μl) in a sterile 12-well tissue culture (TC) dish. The mixture was allowed to rest at room temperature for 20 min and then placed on ice and exposed to a 365-nm UV light source (2 cm from the light filter) for 60 min. For each treatment, a matching control virus was carried through a mock protocol. Residual psoralen was removed by dialysis. Virus particle numbers at OD260 were determined for both treated and mock-treated samples. AdV DNA was purified by standard procedures (38).
Cells.
RAW 264.7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Cellgro) with 5% fetal bovine serum (characterized; HyClone) and 1% penicillin-streptomycin (Gibco).
Overexpressing cell lines.
DAI and AIM2 were cloned by reverse transcription (RT) of total RNA isolated from primary murine bone marrow-derived macrophages by the use of an AccuScript High Fidelity RT-PCR system (Stratagene catalog no. 600180). DAI was cloned into a pcDNA3 vector in frame with an N-terminal hemagglutinin (HA) tag. AIM2 was cloned into a pcDNA4 vector in frame with a C-terminal HA tag. All clones were confirmed by DNA sequencing. RAW 264.7 cells were transfected with the expression plasmids by the use of Fugene 6 (Roche catalog no. 11815091001) and selected for plasmid-expressing cells with G418 (Gemini Bioproducts catalog no. 400-111P) (400 μg/ml) for HA-DAI or Zeocin (InvivoGen catalog no. ant-zn-1p) (300 μg/ml) for AIM2-HA. Single-cell clones were isolated, tested for expression, maintained under drug selection, and used in experiments as indicated.
Cell treatment.
Cells (5 × 105) per well were plated in 2 ml of media in 6-well plates 48 h before treatment. At time point 0, cells were either infected with virus of the indicated amounts or transfected with AdV DNA, poly(I·C) (Invivogen; catalog no. tlrc-picw) (low molecular weight [LMW]), or poly(dA-dT) (Sigma catalog no. P0883) by the use of Lipofectamine2000 (Invitrogen), according to the manufacturers' instructions, at a ratio of 1 μg of nucleic acid/2.5 μl of LF2000/125 μl of Opti-MEM (Invitrogen). LPS (LPS-EB Ultrapure; Invivogen) was added to reach a final concentration of 100 ng/ml from a stock solution of 100 μg/ml. Equal volumes of Opti-MEM were also used for mock treatment and as the dilution medium for the virus. For the polymerase III inhibition experiments, the medium was removed from the cells 10 h before time point 0 and mixed with either DMSO (Hybri-Max; Sigma) or ML60218 (Calbiochem catalog no. 557403) at a final concentration of 20 μM (10 mM stock dissolved in DMSO) (45) and readded to the cells at 2 ml per well.
Western blot analysis.
Whole-cell extracts were prepared by washing cells twice with ice-cold phosphate-buffered saline (PBS) and incubating them in lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40) with the addition of Phosphatase Inhibitor Cocktail 1 and 2 (Sigma catalog no. P2850 and P5726) and protease inhibitors (30 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mM benzamidine) for 30 min at 4°C on a rocking platform before scraping and transferring to tubes. The lysates were cleared by centrifugation at 13,000 × g for 20 min at 4°C, and protein quantification was performed with a DC protein assay kit (Bio-Rad Laboratories).
For Western blot analysis, 20 μg of total protein was separated using standard 10% sodium dodecyl sulfate (SDS) polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon P from Millipore). All blots were blocked in 5% skim milk–Tris-buffered saline (TBS)–Tween (0.1%) at room temperature for 1 h. Phospho-IRF3 (Ser396; catalog no. 4961), phospho-STAT1 (58D6) (Tyr701; catalog no. 9167), TBK1 (catalog no. 3013), IκB kinase ε (Ikkε) (catalog no. 2690), beta-actin (catalog no. 4967), HA tag (6E2) (catalog no. 2367), DYKDDDDK tag (catalog no. 2368) (binds to the same epitope as Sigma's anti-FLAG M2 antibody), and horseradish peroxidase (HRP)-linked anti-rabbit IgG (catalog no. 7074) antibodies were from Cell Signaling. Total IRF3 antibody was from Santa Cruz Laboratories (catalog no. SC-9082). pSTAT2 (Tyr689; catalog no. 07-224) antibody was from Millipore. HRP-linked donkey anti-goat IgG (catalog no. A50-201P) was from Bethyl Laboratories. HRP-linked anti-mouse IgG (catalog no. NA931V) was from Amersham. All primary antibodies were used at a dilution of 1:1,000 in 5% bovine serum albumin (BSA)–TBS except beta-actin, HA tag, and pSTAT2 antibody, which were used at a dilution of 1:2,000. Signals were visualized by the use of an ECL Plus kit (Amersham Biosciences).
sh knockdown.
sh constructs targeting AIM2 were obtained from Sigma (TRCN0000096104 to TRCN0000096108). sh constructs targeting DDX41 were purchased from OpenBiosystems through Thermo Scientific (TRCN0000104010 to TRCN0000104014). sh constructs for knockdown of DAI and TBK1 were generated by cloning published target sequences (42) into pLKO.1 (28) (Addgene plasmid 10878) by the use of primer extension with phi29 to generate the double-stranded hairpin insert (26). Target sequences for P204 were identified with the small interfering RNA (siRNA) at the Whitehead website (47) and cloned as previously described. Additionally, a scrambled nucleotide sequence determined to not target any murine genes was cloned into pLKO.1. The constructs were used to generate lentiviral particles in 293T cells. Cells targeted for knockdown were infected with sh lentivirus and, beginning at 48 h postinfection, selected with puromycin for approximately 7 days, at which point there was no survival of control cells lacking the puromycin cassette. The puromycin-selected sh cells were used in experiments as described above.
SYBR green I qRT-PCR.
For quantitative RT-PCR (qRT-PCR), total cellular RNA was isolated using Tri reagent, Tri reagent RT, or RNAzol RT (Molecular Research Center) as instructed by the manufacturer. Purified RNA was treated using an Ambion Turbo DNA-free kit (Applied Biosystems) to remove residual genomic DNA. Concentrations and purity were quantified by NanoDrop spectrophotometry. A two-step qRT-PCR protocol was employed as follows: first, cDNA was synthesized from 1 μg of total RNA in a volume of 20 μl by the use of a Maxima First Strand cDNA synthesis kit (Fermentas); second, amplifications were carried out in a total volume of 15 μl by using a Maxima SYBR green/ROX qPCR Master Mix (Fermentas) in an Applied Biosystems Prism 7900H sequence detection system with SDS 2.1 software. Cycles consisted of an initial incubation at 95°C for 10 min, 40 cycles at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s, and a melting curve analysis cycle. Data acquisition was performed during the extension step. All determinations were performed in technical triplicate. Nontemplate and non-RT controls were run with every assay and had cycle threshold (CT) values that were significantly higher than those of experimental samples or were undetermined. The relative abundance of each mRNA was calculated by the ΔΔCT method (25, 37) by normalizing to actin or HPRT expression and comparing to one reference sample as indicated on the graph. ΔΔCT values were set in the exponent to the base 2, and the resulting values were multiplied by 1 (see Fig. 3C), 100 (see Fig. 2C and D and 4C and D), or 10,000 (see Fig. 3A) to facilitate plotting of the bar graph. Sequences of primers are available on request.
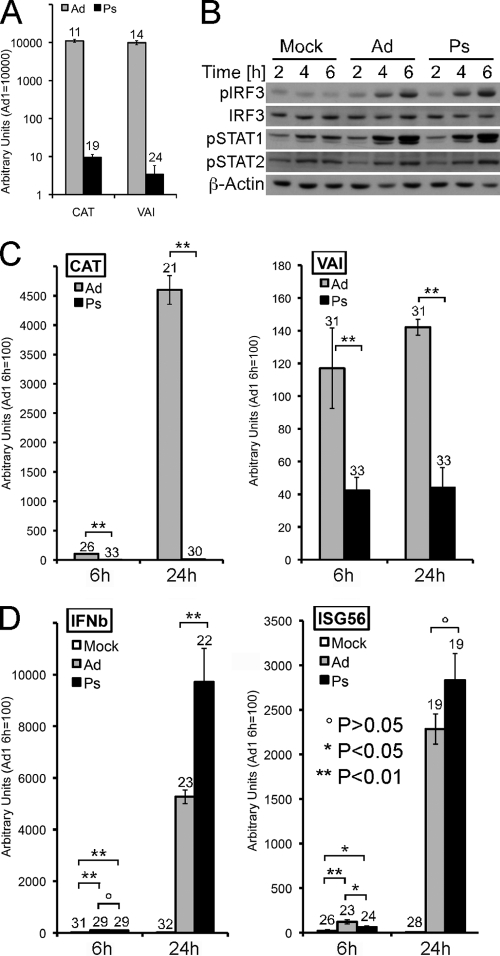
Fig 3.
UV/psoralen-inactivated Ad5CiG virus induces RAW 264.7 cells. (A) RNA harvested from 293 cells 24 h postinfection with mock-treated (Ad) or psoralen/UV-treated (Ps) Ad5CiG virus (1,000 p/cell) (as described in Materials and Methods) was analyzed in a two-step qRT-PCR assay for expression of the CAT reporter gene or the RNA polymerase III viral transcript VAI and normalized to cellular actin. Numbers above each bar represent the unnormalized average CT value for each transcript. (B) RAW 264.7 cells were infected with the indicated viruses at 20,000 p/cell and harvested at the indicated times postinfection. Samples were analyzed by Western blotting as previously described. (C) RNA from RAW 264.7 cells infected with the indicated viruses harvested at 6 or 24 h postinfection was used in a two-step qRT-PCR assay screening for virus-generated CAT or VAI transcripts. (D) RNA from RAW 264.7 cells infected with the indicated viruses harvested at 6 or 24 h postinfection was used in a two-step qRT-PCR assay screening for the IRF3-inducible transcripts IFN-β and interferon-stimulated gene 56 (ISG56). All samples were normalized to actin. Numbers above each bar represent unnormalized average CT values from biological triplicates for each transcript.
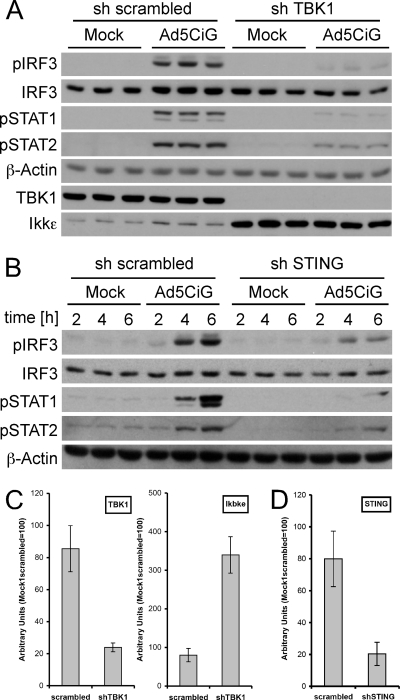
Fig 2.
shRNA knockdown of TBK1 or STING proteins compromises the antiviral response in RAW 264.7 cells. (A) RAW 264.7 cells infected with either scrambled or anti-TBK1 shRNA lentiviral vectors (puromycin selected for 7 days) were used in a standard AdV infection assay (20,000p/cell) and harvested at 6 h postinfection for Western blot analysis as previously described. (B) Infection was performed as described for panel A, except the STING shRNA lentiviral vector was used in place of sh-TBK1. (C) RNA isolated from scrambled- or shTBK1-uninfected cells was characterized for knockdown of TBK1 mRNA or changes in IKKε mRNA by a two-step qRT-PCR assay. RNAs were normalized to total HPRT mRNA by the use of the ΔΔCT method as described in Materials and Methods. (D) Two-step qRT-PCR assay of mRNA harvested from scrambled- or sh-STING-infected cells, measuring relative levels of STING mRNA normalized to HPRT mRNA.
Statistical analysis.
Data were expressed as means ± the standard errors of the means. Statistical analysis was performed with Student's t test. A P value of less than 0.05 was considered to represent statistical significance.
RESULTS
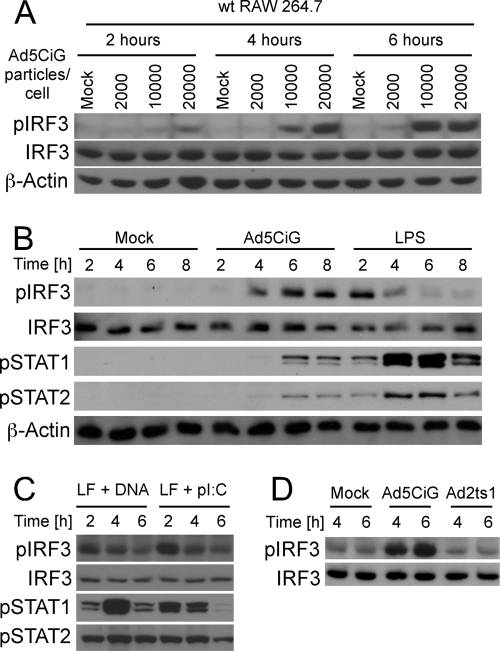
RAW 264.7 cells are a murine leukemic monocyte/macrophage cell line that retains many of the characteristics of APCs (44) while offering the advantage of stable, long-term growth in culture. To characterize the early activation response to rAdV infection, RAW 264.7 cells were exposed to increasing concentrations of the E1-deleted Ad5CiG vector (38) and harvested over a time course of 6 h. Cell lysates were characterized by Western blot analysis using a phosphoserine 388-specific anti-IRF3 antibody (αpser388IRF3) (corresponding to p-ser 396 in human IRF3) (Fig. 1). Phosphorylation at this site is a key indicator of IRF3 activation (40) and provides a robust, reliable marker for the early response to virus infection. Data reveal a dose-dependent stimulation of IRF3 C-terminal phosphorylation (Fig. 1A); similar to observations made in primary APCs, accumulation of pser388IRF3 was modest at 2 h and increased at 4 and 6 h postinfection in RAW 264.7 cells. This gradual increase in pser388IRF3 contrasts the rapid activation found with LPS treatment (TRIF-dependent activation of IRF3) (Fig. 1B) or direct liposome-mediated transfection of rAdV DNA or poly(I·C) (cytosolic RNA-sensing response through RIG-I/MAVS) (Fig. 1C). The activation of IRF3 is associated with an antiviral response characterized by expression of type I IFN (32). Virally induced type I IFN stimulates autocrine-paracrine secondary signaling, leading to phosphorylation of STAT1 and STAT2 (31, 32). Levels of pSTAT1/2 (Stat1 pTyr 701 and Stat 2-pTyr247) provide a sensitive and practical assessment of secreted type I IFN secondary signaling in response to virus or nucleic acid treatments (Fig. 1B and C). The Ad2ts1 mutant (mt) binds and is internalized but is unable to escape the endosomal compartment (11, 13). However, endosomal escape of rAdV is a prerequisite for IRF3 activation (8, 31, 32). When RAW 264.7 cells were infected with the Ad2ts1 mt virus, IRF3 phosphorylation was at background levels (Fig. 1D). Therefore, the early antiviral recognition response detected in RAW 264.7 cells is consistent with what has been found in primary murine APC.
Fig 1.
Adenovirus activation of RAW 264.7 cells. (A) RAW 264.7 cells infected with the indicated amount of virus (particles/cell) were harvested at 2, 4, and 6 h postinfection as described in Materials and Methods and analyzed by Western blotting (probed with pser388IRF3-, IRF3-, or actin-specific antibody). (B) The IRF3 activation assay was performed as described for panel A, comparing cells exposed to 20,000 p/cell of Ad5CiG or bacterial endotoxin LPS (100 ng/ml). A Western blot was probed with phosphospecific antibodies pser388IRF3, ptyr701STAT1, and ptyr689STAT2 as well as IRF3- and actin-specific antibodies as total protein controls. (C) RAW 264.7 cells were transfected with Ad5CiG DNA or poly(I·C), and cell lysates were harvested and analyzed by Western blotting as previously described. (D) IRF3 activation assay comparing cells exposed to 20,000 p/cell of Ad5CiG or the endosomal escape mutant Ad2ts1. Samples were harvested at 4 and 6 h postinfection and analyzed by Western blotting as previously described. All experiments have been done a minimum of three times.
TBK1/STING signaling cascade and IRF3 activation by rAdV.
Tank binding kinase 1 (TBK1) is one of two kinases associated with IRF3 phosphorylation (10, 27) and undergoes early activation in primary macrophages following rAdV infection (31). To assess TBK1 involvement in rAdV activation of IRF3 in RAW 264.7 cells, an anti-TBK1 shRNA vector was constructed using the pLKO.1 lentiviral system (see Materials and Methods). Lentiviral infection of RAW 264.7 cells, followed by puromycin selection, was used to establish stable cell line pools derived from each shRNA vector. Each pool was used in a rAdV infection assay to assess shRNA influence on IRF3 activation. Cell lysates harvested 6 h postinfection were examined by Western blot analysis for pser388IRF3 (Fig. 2A). TBK1 knockdown severely limited responses to rAdV infection at the level of IRF3 phosphorylation and generated correspondingly low levels of STAT1/2 phosphorylation. Interestingly, while steady-state levels of TBK1 were greatly reduced by the shRNA treatment, we observed an enhancement of steady-state IKKε levels (Fig. 2A and C). These data indicate that TBK1 is critical to IRF3 phosphorylation following rAdV infection. IKKε does not compensate for loss of TBK1 in this model system.
A similar approach was used to assess the contribution of the adaptor molecule STING to rAdV induction of pser388IRF3. STING, a mitochondrial/endoplasmic reticulum-associated protein, plays a critical role in several DNA sensing cascades, acting as a bridge between DNA recognition complexes and TBK1 (reviewed in reference 17). Anti-STING shRNA lentiviral treatment reduced STING mRNA levels by approximately 75% (Fig. 2D). Following rAdV infection, levels of both the primary (pser388IRF3) and secondary (pSTAT1/2) activation markers were correspondingly reduced (Fig. 2B). Similarly, sh-STING cells were compromised in their response to vDNA transfection (data not shown). Under these conditions, the data are consistent with STING acting as an adaptor leading to TBK1 activation in response to rAdV infection. The data support the idea of an antiviral sensing pathway in which early recognition by a putative DNA sensor complex triggers a STING/TBK1 adapter kinase cascade phosphorylating IRF3.
Viral transcription is not required for antiviral stimulation.
Adenoviral DNA is transcribed by RNA polymerase II and III, and early viral transcripts may activate an antiviral response, including IRF3 activation. To determine if viral transcripts contribute to IRF3 activation during the early phase of virus infection, we used psoralen-UV-inactivated adenovirus (see Materials and Methods) to reduce viral transcript without compromising entry. This treatment diminishes both transcription and DNA replication by cross-linking viral duplex DNA (5). The efficiency of inactivation was determined by infecting permissive 293 cells with 1,000 particles/cell (approximate wild-type [wt] multiplicity of infection [MOI], 20 to 30) of unmodified or Ps-treated virus. At 24 h postinfection, the unmodified Ad5CiG virus generates uniform, strong GFP fluorescence. In contrast, the Ps-treated virus produced no discernible GFP fluorescence by microscopy (data not shown). A two-step qRT-PCR assay (see Materials and Methods) was established to characterize levels of viral transcript (the RNA polymerase II CAT reporter mRNA and a viral RNA polymerase III transcript, VAI) from infected 293 cells (Fig. 3A). Consistent with the lack of GFP fluorescence, CAT and VAI RNA levels were greatly diminished by UV/Ps treatment. VAI RNAs were reduced by 3 logs, while CAT mRNA levels were down by 2 to 3 orders of magnitude in late-stage replication-competent viral infections.
These vectors were used to infect RAW 264.7 cells (20,000 particles/cell). Western analysis of proteins harvested over a 6-h time course demonstrated a strong induction of pIRF3 and pSTAT1/pSTAT2 in cells infected with each virus (Fig. 3B). Template inactivation did not compromise the antiviral response at the level of primary or secondary signals. To assess the impact of inactivation on RNA levels, qRT-PCR was used to characterize levels of viral (Fig. 3C) and IRF3-responsive (Fig. 3D) transcripts from RAW 264.7 cells harvested 6 and 24 h postinfection. All qRT-PCR assays were normalized to the polymerase II transcript actin, and in each primer-pair grouping, the AdV 1 DMSO sample was given an arbitrary value of 100, allowing direct comparison to all other samples. To provide a reference indicator of transcript levels, the unnormalized average cycle threshold (CT) number from 3 distinct RNA isolates has been provided at the top of each bar (where a level of 3.3 CT units represents an approximately 1 log difference in yield). CT values of 30 or greater represent extremely low levels of steady-state transcript.
Consistent with the established inactivation profile for these viruses, Ps-inactivated virus generated CAT mRNA levels that were between 2 and 3 logs lower than those seen with untreated virus (Fig. 3C). In comparison, levels of VAI from untreated Ad5CiG vector were extremely low in both the 6- and 24-h samples (CT only slightly above background), and levels from treated vector were significantly reduced. (In this assay, 32 to 33 CT is baseline for mock treatment due to primer-generated noise.) Both IFN-β and ISG56 mRNA levels were induced from inactivated vector harvested at either the 6 or 24 h time point. At the level of pser388IRF3 activation, induction of IRF3-dependent transcript, and secondary signaling responses, the data indicate that rAdV activation of RAW 264.7 cells can be uncoupled from transcription of the viral genome.
Treatment with the RNA polymerase III inhibitor ML60218 does not prevent AdV-induced IRF3 phosphorylation.
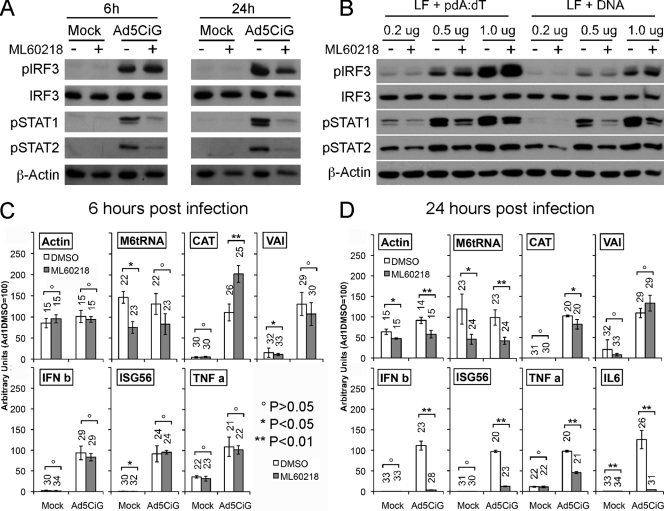
Two studies have indicated that rAdV induction of IFN-β was diminished in RAW 264.7 cells treated with the RNA polymerase III inhibitor ML60218 at 20 μM (4, 46). In the latter study, the adenovirus polymerase III transcript VAI was proposed to induce expression of type I interferon in a manner that is influenced by ML60218 treatment (46). Since IRF3 activation is an important step in β-IFN induction and since UV/PS inactivation experiments indicate that transcription of viral DNA is not essential to the antiviral response, we asked if exposure of RAW 264.7 cells to the RNA polymerase III inhibitor ML60218 influenced IRF3 activation following treatment with virus.
Cells were pretreated for 10 h with 20 μM ML60218 or DMSO (4, 46) and exposed to rAd5CiG or mock treatment. At 6 h postinfection, we found similar levels of pser388IRF3 in ML62018- and DMSO-treated cells with or without rAd5CiG (Fig. 4A). Therefore, initial activation of IRF3 was not compromised by drug treatment. This observation is consistent with viral RNA not acting as the primary ligand leading to IRF3 activation. However, at 6 h postinfection, levels of pSTAT1/2 were diminished in ML60218-treated cells. Therefore, treatment with the inhibitor uncoupled the induction of pIRF3 from stimulation of type I IFN-dependent downstream signaling pathways. To determine if IRF3 activation under these conditions was unique to virus infection, an experiment was carried out using liposome transfection of vDNA or poly(dA-dT). IRF3 activation by transfected vDNA or poly(dA-dT) was dose dependent and was not influenced by pretreatment with ML60218 (Fig. 4B). However, in all ML60218 pretreatment assays, levels of pSTAT1/2 were reduced compared to untreated samples. The antiviral response to rAdV undergoes amplification over time through autocrine-paracrine stimulation. To assess how an extended infection may be compromised by ML60218 treatment, Ad5CiG-infected lysates were harvested at 24 h postinfection. Consistent with diminished autocrine-paracrine signaling, the extended treatment resulted in diminished levels of pSTAT1/2 and pser388IRF3 (Fig. 4A).
Fig 4.
Activation of RAW 264.7 cells treated with RNA polymerase III inhibitor ML60218. (A) RAW 264.7 cells pretreated with 20 μm of ML60218 (+) or DMSO (− [solvent control]) for 10 h were mock infected or infected with Ad5CiG virus (20,000 p/cell) and lysates harvested at 6 and 24 h postinfection. Protein lysates were analyzed by Western blotting as previously described. (B) RAW 264.7 cells pretreated with 20 μm of ML60218 (+) or DMSO (− [solvent control]) for 10 h were Lipofectamine 2000 transfected with increasing amounts of poly(dA-dT) or Ad5CiG DNA; lysates harvested at 5 h were analyzed by Western blotting as previously described. (C and D) RAW 264.7 cells treated as described for panel A were harvested for RNA at 6 h postinfection (C) or 24 h postinfection (D). qRT-PCR values for cellular control RNAs (Actin or M6tRNA), for virus-specific transcript (CAT or VAI), or for antiviral response-inducible transcripts (IFNB, ISG56, or TNF-α) were normalized to cellular HPRT as described in the text and Materials and Methods. The number above each bar indicates the unnormalized average CT value determined using biological triplicates for each transcript (see text).
Using qRT-PCR, we determined the influence of ML60218 on host and viral transcripts at 6 and 24 h postinfection. All qRT-PCR assays were normalized to the polymerase II transcript hypoxanthine phosphoribosyltransferase (HPRT). We first determined how ML60218 treatment influenced cellular transcripts. The M6tRNA RNA polymerase III transcript level was reduced at 6 and 24 h postinfection (4C and D, respectively). In comparison, the RNA polymerase II transcript for actin was unaffected by ML60218 treatment at 6 h postinfection, but at 24 h a modest reduction was observed, possibly associated with the stress induced by extended exposure to ML60218. At 6 h postinfection, Ad5CiG induced similar levels of IFN-β, ISG56, and TNF-α in the presence or absence of the inhibitor. However, at 24 h, there was a clear impact of drug treatment on expression of IRF3-responsive transcripts (IFN-β and ISG56) as well as on expression of TNF-α and IL-6. Using the same normalization, we characterized transcripts originating from Ad5CiG. Treatment with ML60218 did not reduce CAT expression levels at 6 or 24 h (Fig. 4C and D). Although overall levels of VAI transcript were extremely low, the qRT-PCR assay showed no significant impact of ML60218 treatment on levels of VAI RNA at either 6 or 24 h. Under these conditions, we do not find gene expression from Ad5CiG to be influenced by drug treatment in a statistically significant manner. However, treatment with ML60218 did impair a complete antiviral response, presumably through perturbation of cellular transcription-translation machineries.
DNA sensors: redundant, complementary, or antagonistic functions?
A number of proteins have been identified as DNA binding pattern recognition receptors (PRRs), DNA sensors that contribute to an innate inflammatory response following DNA transfection or virus infection. Using the stable RAW 264.7 cell line, we have used knockdown or overexpression assays to determine the contribution of known sensors to rAdV induction of pser388IRF3.
To assess the involvement of the DNA sensor DAI in the antiadenoviral response, we employed stable overexpression/shRNA knockdown strategies. An HA-tagged DAI expression vector was constructed using cDNA from primary murine macrophage (see Materials and Methods). Five cell lines expressing various levels of HA-DAI were selected for an rAdV activation assay (Fig. 5A). Cell lysates from mock- or rAdV-infected cells were screened for HA-DAI, pIRF3, pSTAT1, or total TBK1 by Western blotting. There was no apparent enhancement of pser388IRF3 or pSTAT1 levels in HA-DAI-expressing cells exposed to rAdV.
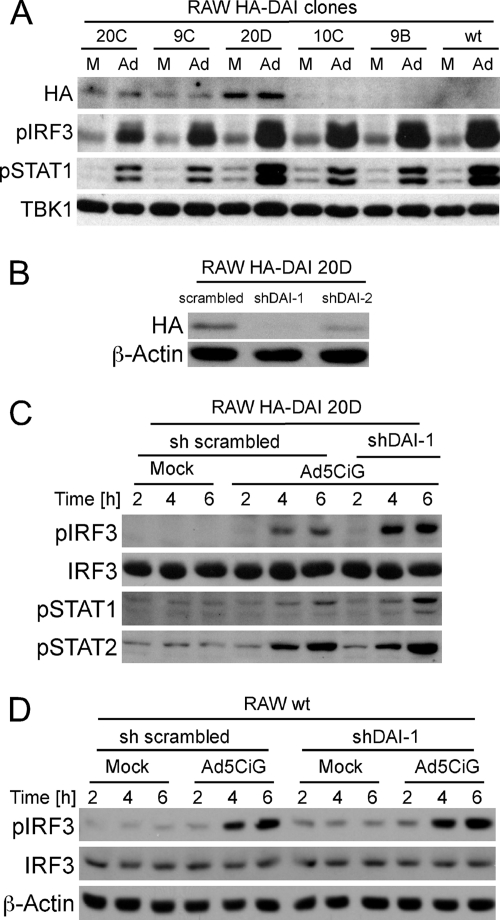
Fig 5.
Impact of DAI overexpression or knockdown on IRF3 activation. (A) Independent G418-selected clones expressing HA-tagged DAI (see Materials and Methods) were mock infected (M) or infected with 10,000 particles of Ad5CiG per cell (Ad). Lysates harvested at 5 h postinfection were analyzed by Western blotting as previously described. Numbers correspond to individual clonal isolates. (B) The 20D cell line was infected with sh-SC (scrambled), sh-DAI-1, or sh-DAI-2 lentiviral vector and screened for knockdown of HA-DAI by Western analysis after 7 days of puromycin selection. (C) RAW 20D cells expressing a scrambled or DAI-targeting hairpin were mock infected or infected with 20,000 particles per cell. Cells were harvested at the indicated time points and analyzed by Western blotting. (D) RAW wt cells infected with sh-SC- or sh-DAI-1-targeting hairpin lentivirus were mock infected or infected with 20,000 particles per cell. Cells lysates were harvested at the indicated time points and analyzed by Western blotting.
Two anti-DAI shRNAs (42) and a scrambled shRNA were cloned into pLKO.1, which was used to generate lentiviral vectors. Infection of the 20D HA-DAI cell line, followed by puromycin selection, was used to establish stable cell line pools derived from each shRNA vector. Anti-HA Western blot analyses indicated that sh-DAI-1 most effectively reduced HA-DAI protein levels (Fig. 5B). rAdV-infected shSc (Scrambled) and sh-DAI-1 knockdown pools were harvested over a 6-h time course and screened by Western blotting (Fig. 5C). Knockdown of DAI did not compromise rAdV-induced phospho-IRF3, STAT1, or STAT2. To characterize the influence of a DAI knockdown in the absence of overexpression, the sh-DAI-1 lentiviral vector was used to infect wt RAW cells. qRT-PCR demonstrated knockdown of DAI mRNA to levels below 30% of those of Sc-treated samples (data not shown). Levels of rAdV-induced pIRF3 were similar to those found in control RAW cells treated with the scrambled shRNA vector (Fig. 5D). Based on these observations, we conclude that DAI is not essential to rAdV activation of IRF3.
The Aim2 DNA sensor contributes to IRF3 activation.
We next applied the overexpression/knockdown strategy to the DNA sensor absent in melanoma 2. An AIM2-HA expression vector was generated and verified by sequence analysis. AIM2-HA RAW 264.7 cell lines were established following transfection and G418 selection. Cell lines expressing AIM2-HA showed no obvious enhancement of pIRF3 induction over a 6-h time course following rAdV infection (Fig. 6A).
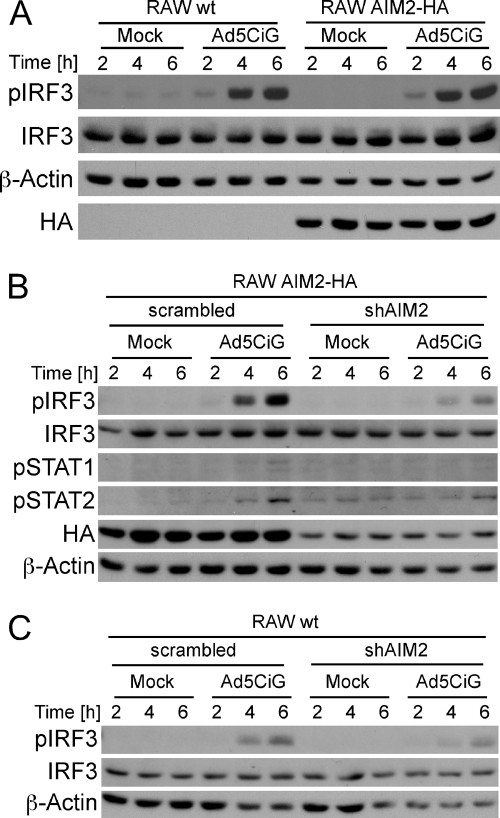
Fig 6.
Impact of AIM2 overexpression or knockdown on IRF3 activation. (A) wt RAW 264.7 cells or an AIM2-HA-expressing RAW 264.7 cell clone (see Materials and Methods) were either mock or Ad5CiG infected (20,000 p/cell). Cell lysates were harvested at the indicated time points and analyzed by Western blotting as previously described. (B) AIM2-HA RAW cells infected with sh-SC (scrambled nontargeting) or sh-AIM2 (hairpin targeting AIM2) were mock or Ad5CiG infected (20,000 p/cell). Cell lysates were harvested at the indicated time points and analyzed by Western blotting. (C) wt RAW 264.7 cells infected with sh-SC (scrambled nontargeting) or sh-AIM2 (hairpin targeting AIM2) were mock or Ad5CiG infected (20,000 p/cell). Cell lysates were harvested at the indicated time points and analyzed by Western blotting.
A series of shAim2 lentiviral vectors were screened using the AIM2-HA cell line. The sh-AIM2-HA cell line pool most depleted in HA-tagged Aim2 (data not shown) was used to carry out an rAdV IRF3 activation assay (Fig. 6B). In sh-AIM2 knockdown cells, levels of induced pser388IRF3 were greatly reduced compared to the parent AIM2-HA-RAW 264.7 cell line. The type I IFN secondary signaling response as measured by pSTAT1 and pSTAT2 analysis was diminished in the sh-AIM2-treated cells. We extended use of the sh-AIM2 lentiviral construct to wt RAW 264.7 cells (Fig. 6C) (65% knockdown by qRT-PCR; data not shown) and found reduced levels of rAdV-induced pIRF3 in cells treated with the sh-AIM2 vector. Assessing secondary signaling (pSTAT1/2) in the RAW wt cells treated with sh-AIM2 vector was complicated by increased background levels of pSTAT1 and pSTAT2, but the results are consistent with those from the AIM2-HA cell line. Based on the data presented, we found that AIM2 contributed to the cascades involved in rAdV induction of pIRF3.
DDX41, but not p204, participates in rAdV activation of IRF3.
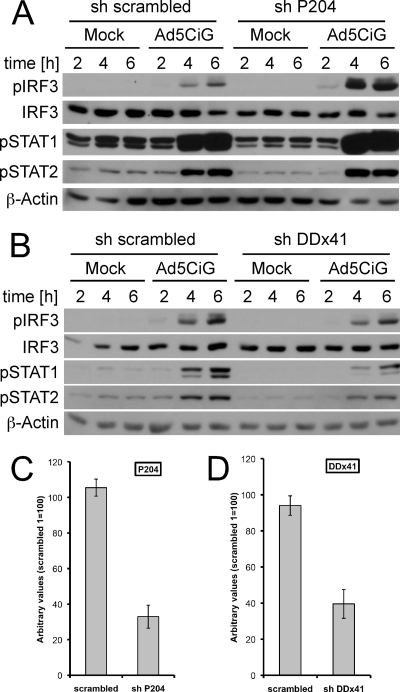
AIM2 is a member of the Hin-200 family of proteins and is associated with inflammasome activation through DNA sensing but not type I interferon induction (9, 14, 21). Another Hin-200 family member, p204, has been identified as a DNA sensor that contributes to type I interferon induction (43), and, more recently, the human ortholog IFI16 has also been found to form an IFI16 inflammasome (20). In contrast to the Hin-200 proteins, DDX41 is not an interferon-inducible gene and functions as a DNA sensor in response to viral and intracellular bacterial infections (48). To test the involvement of p204 and DDX41 in rAdV induction of IRF3, shRNA vectors targeting p204 and DDX41 were constructed (see Materials and Methods). Selected cell pools corresponding to shScrambled, shP204, or shDDX41 were used in IRF3 activation assays following Ad5CiG infection (Fig. 7A and B). qRT-PCR indicated knockdown of p204 by approximately 70% and shDDX41 knockdown by roughly 60% (Fig. 7C and D, respectively). Knockdown of p204 resulted in increased pser388IRF3 following virus infection, and levels of phospho-STAT1 and -2 were not diminished. In contrast, the knockdown of DDX41 resulted in diminished levels of pser388IRF3 and of phospho-STAT1 and -2. Since the shDDX41 knockdown was not complete, the DDX41 that remained may account for residual IRF3 activation. Alternatively, there may be redundant or overlapping functions served by AIM2 and DDX41 with respect to rAdV recognition and activation of the TBK/IRF3 cascade.
Fig 7.
DDX41 but not p204 contributes to the rAdV recognition response. (A) wt RAW 264.7 cells infected with sh-SC (scrambled nontargeting) or shP204 (hairpin targeting p204) were mock or Ad5CiG infected (20,000 p/cell). Cell lysates were harvested at the indicated time points and analyzed by Western blotting. (B) wt RAW 264.7 cells infected with sh-SC (scrambled nontargeting) or shDDX41 (hairpin targeting DDX41) were mock or Ad5CiG infected (20,000 p/cell). Cell lysates were harvested at the indicated time points and analyzed by Western blotting. (C) RNA isolated from scrambled- or shP204-uninfected cells was characterized for knockdown of P204 mRNA by a two-step qRT-PCR assay. RNAs were normalized to total HPRT mRNA by the ΔΔCT method as described in Materials and Methods. (D) Two-step qRT-PCR assay of mRNA harvested from scrambled- or sh-DDX41-infected cells, measuring relative levels of DDX41 mRNA normalized to HPRT mRNA.
DISCUSSION
The adenoviral genome is a critical ligand for induction of the type I IFN response in antigen-presenting cells. To sort out the array of DNA sensor complexes involved in recognition of rAdV in the murine model, we have established a simplified assay focusing on early signaling events (phosphorylation of IRF3) in RAW 264.7 cells. At the level of IRF3 activation, the overall response pattern was consistent with previous studies in primary bone marrow-derived murine macrophage (BMMO) and conventional dendritic cells (32). TBK1 activation in BMMO cells is an early response to rAdV infection (31). We have now shown through shRNA knockdown that TBK1 is required for rAdV-induced IRF3 phosphorylation in RAW 264.7 cells. Furthermore, shRNA knockdown of the adaptor protein STING compromised IRF3 activation by rAdV. Several studies have indicated that STING operates as a critical adaptor bridging DNA sensing cascades to a TBK1-dependent type I IFN response (17, 18, 43, 48). This holds for DNA transfection and infection with DNA viruses; however, STING is not required for type I IFN induction following poly(IC) transfection or infection by influenza virus (48). A dominant role for the STING adaptor emphasizes participation of a DNA sensor in recognition of rAdV (see Fig. 8 model).
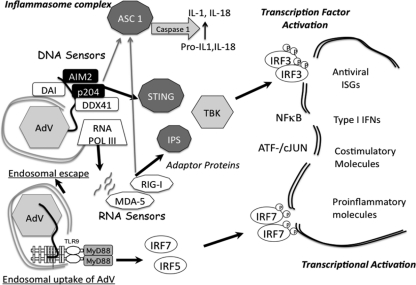
Fig 8.
Summary model of rAdV-sensing cascades in RAW 264.7 cell line. Following endosomal entry of rAdV into the cell, viral DNA can trigger the TLR9 pathway, which, through the MyD88 adaptor protein, leads to activation of IRF7 and IRF5 but not IRF3. Alternatively, adenovirus escapes the endosomal compartment, revealing virus and viral DNA complexes to the cytosolic compartment. In RAW 264.7 cells, an array of DNA sensors are potentially available to bind viral DNA. Depending on the complexes formed, DNA sensor complexes engage STING (MITA) either directly or indirectly, leading to TBK1 activation and phosphorylation of IRF3. IRF3 dimerizes and translocates to the nucleus, where it associates with transcription units (IFN-β or ISG56). In collaboration with additional transcription factors (NF-κB, ATF/cJUN), gene expression is induced. AIM2 and p204 are Hin-200 proteins that have been shown to complex with ASC through Pyrin domains to form an inflammasome complex, leading to IL-1/IL-18 secrestion. DNA template either in the cytosol or in the nucleus can be transcribed by RNA polymerase II (not depicted) and RNA polymerase III. Small viral RNAs have been implicated in stimulating the antiviral response through activation of RNA sensors (MDA-5, RIG-I), which in turn complex with IPS (MAVS) and activate TBK1. RIG-I has also been shown to complex with ASC to form an inflammasome complex.
One major alternative to a direct DNA sensing mechanism is detection of viral RNA transcripts leading to IRF3 activation. Two methods, psoralen/UV inactivation and ML60218 inhibition of RNA polymerase III, were used to inhibit viral transcription; neither impacted antiviral activation responses at the level of early IRF3 phosphorylation. UV/psoralen-inactivated virus stimulates IRF3, IRF3-inducible transcripts, and secondary phosphorylation of STAT1/2 to the same degree as untreated virus. This is in agreement with previous studies in primary BMMO cells (32). Although levels of viral transcript are extremely low in a RAW 264.7 infection, the quality of antiviral IRF3 activation was not affected by the Ps-inactivated vector.
The overall effect of ML60218 was more complicated than simple inhibition of viral RNA polymerase III transcripts. The RNA polymerase III transcriptome plays a critical role in regulation of host cell metabolism (reviewed in reference 12), and stress response pathways may be affected by changes in steady-state RNA polymerase III transcript pools. We found that inhibition of RNA polymerase III by ML60218 was not absolute; steady-state levels of M6 tRNA were diminished, and, in our estimation, the effect on transcription of viral VAI was marginal—in part due to inherently low levels of transcript. This incomplete inhibition of RNA polymerase III is consistent with the 50% inhibitory concentration (IC50) calculated for human Pol III (27 μM) (45). Nevertheless, treatment of cells with ML60218 has consequences. Although ML60218 did not compromise IRF3 phosphorylation or levels of IRF3-dependent transcripts (IFN-β or ISG56) at 6 h postinfection, type I IFN-mediated activation of STAT1/2 was inhibited. Disruption of antiviral amplification cascades at 6 h (resulting from the lack of STAT1/2 activation) contributes to the expanded loss of antiviral transcripts, including IFN-β, TNF-α, and IL-6, at 24 h postinfection. Further study might be considered to determine how ML60218 impacts amplification cascades that contribute to establishing a complete antiviral response in macrophage.
An array of DNA sensors is present in RAW 264.7 cells (Fig. 8). Each, in theory, is available to interact with infecting virus. They may provide redundant, complementary, or antagonistic activities that contribute to the antiviral response (30). We asked how knockdown of 4 DNA sensors influenced IRF3 activation following rAd5CiG infection, and found two basic response profiles. The knockdown of DAI or p204 did not compromise levels of pser388IRF3 following rAdV infection; rather, levels of activated IRF3 were often elevated compared to shscrambled-infected cells. In contrast, knockdown of Aim2 and DDX41 resulted in diminished levels of pser388IRF3. We believe that these results demonstrate the complexity and redundancy of the DNA sensing response present in this cell line.
The availability of the array of DNA sensors, including the TLR9 pathway, imparts options that define the nature of an antiviral response. Cell-specific differences in concentrations, locations, and interactions of these proteins are anticipated to influence the signaling cascades activated. When we use the shRNA knockdown strategy, we are artificially shifting the balance of competing proteins. The response of the cell includes a loss of activity due to the primary target gene (p204, DAI, AIM2, or DDX41) but a possible gain of activity by proteins that have increased access to the viral DNA (p204, DAI, AIM2, or DDX41), adaptor proteins, and other proteins (e.g., TBK1) involved in the signaling cascade. The modest pIRF3 enhancement following knockdown of DAI (or p204) is reminiscent of the signaling flux redistribution found at TLR pathway junctions, where knockdown of MyD88 enhanced signaling responses through TRAM and vice versa (39). Another example of flux distribution is that represented by increased levels of IKKε following knockdown of TBK1 (Fig. 2). Not only are proteins competing for the viral ligand, but there is also competition for activation of signaling cascades. IFI16, the human ortholog of murine p204, was shown to stimulate an IRF3/IFN-β response following DNA transfection or HSV1 infection (43). In a recent study, IFI16 functioned as a nuclear sensor to KSHV infection generating a caspase I/IFI16/ASC inflammasome (20). IFI16 participation in both IFN and inflammasome cascades provides a precedent for our observation that AIM2 acts in the IRF3/IFN pathway in the RAW 264.7 model. RIG-I, an established dsRNA sensor leading to IRF3 activation, can also form with ASC to generate a RIG-I-dependent inflammasome complex (35).
Nucleic acid DNA sensor complexes may serve as branch points leading toward either inflammasome- or IRF3-biased responses (7, 24). In the case of naïve macrophage exposed to rAdV, the inflammasome activation pathway is modest compared to that of LPS-primed cells (2). In unprimed cells, several DNA sensors may be situated in a manner that feeds into the IRF3 type I IFN induction cascade (Fig. 8). Under conditions of sustained IFN induction where inflammasome priming occurs (DAI, AIM2, and p204 are all IFN-inducible genes), DNA sensor branch points may switch to formation of inflammasome complexes. A balance between these two pathways, type I IFN induction/APC maturation and inflammasome/pyroptosis, may be strongly influenced through type I (and type II) IFN signaling. Further characterization of rAdV infection of APCs should provide critical insight into how these putative sensor-switch junctions are established and controlled in a cell-dependent manner.
ACKNOWLEDGMENTS
E.F.-P is a recipient of a Hearst Foundation Award. This work was supported by Public Health Service grants RO1 AI094050 and R56 AI063142 to E.F.-P.
We thank Tyler Janovitz for critical reading of the manuscript.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Ablasser A, et al. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 10:1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barlan AU, Griffin TM, McGuire KA, Wiethoff CM. 2011. Adenovirus membrane penetration activates the NLRP3 inflammasome. J. Virol. 85:146–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bürckstümmer T, et al. 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10:266–272 [DOI] [PubMed] [Google Scholar]
- 4. Chiu YH, Macmillan JB, Chen ZJ. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cotten M, et al. 1994. Psoralen treatment of adenovirus particles eliminates virus replication and transcription while maintaining the endosomolytic activity of the virus capsid. Virology 205:254–261 [DOI] [PubMed] [Google Scholar]
- 6. Di Paolo NC, et al. 2009. Virus binding to a plasma membrane receptor triggers interleukin-1 alpha-mediated proinflammatory macrophage response in vivo. Immunity 31:110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duan X, et al. 2011. Differential roles for the interferon-inducible IFI16 and AIM2 innate immune sensors for cytosolic DNA in cellular senescence of human fibroblasts. Mol. Cancer Res. 9:589–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fejer G, et al. 2008. Key role of splenic myeloid DCs in the IFN-alphabeta response to adenoviruses in vivo. PLoS Pathog. 4:e1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458:509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fitzgerald KA, et al. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491–496 [DOI] [PubMed] [Google Scholar]
- 11. Gastaldelli M, et al. 2008. Infectious adenovirus type 2 transport through early but not late endosomes. Traffic 9:2265–2278 [DOI] [PubMed] [Google Scholar]
- 12. Goodfellow SJ, White RJ. 2007. Regulation of RNA polymerase III transcription during mammalian cell growth. Cell Cycle 6:2323–2326 [DOI] [PubMed] [Google Scholar]
- 13. Greber UF, Webster P, Weber J, Helenius A. 1996. The role of the adenovirus protease on virus entry into cells. EMBO J. 15:1766–1777 [PMC free article] [PubMed] [Google Scholar]
- 14. Hornung V, et al. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458:514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ishii KJ, et al. 2006. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 7:40–48 [DOI] [PubMed] [Google Scholar]
- 16. Ishii KJ, et al. 2008. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 451:725–729 [DOI] [PubMed] [Google Scholar]
- 17. Ishikawa H, Barber GN. 2011. The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cell. Mol. Life Sci. 68:1157–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ishikawa H, Ma Z, Barber GN. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461:788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaiser WJ, Upton JW, Mocarski ES. 2008. Receptor-interacting protein homotypic interaction motif-dependent control of NF-kappa B activation via the DNA-dependent activator of IFN regulatory factors. J. Immunol. 181:6427–6434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kerur N, et al. 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe 9:363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krieg AM. 2009. AIMing 2 detect foreign DNA. Sci. Signal. 2:pe39. [DOI] [PubMed] [Google Scholar]
- 22. Lamkanfi M. 2011. Emerging inflammasome effector mechanisms. Nat. Rev. Immunol. 11:213–220 [DOI] [PubMed] [Google Scholar]
- 23. Lamkanfi M, Dixit VM. 2010. Manipulation of host cell death pathways during microbial infections. Cell Host Microbe 8:44–54 [DOI] [PubMed] [Google Scholar]
- 24. Li S, Wang L, Berman M, Kong YY, Dorf ME. 2011. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity 35:426–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 26. McIntyre GJ, Fanning GC. 2006. Design and cloning strategies for constructing shRNA expression vectors. BMC Biotechnol. 6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McWhirter SM, et al. 2004. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 101:233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moffat J, et al. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124:1283–1298 [DOI] [PubMed] [Google Scholar]
- 29. Muruve DA, et al. 2008. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 452:103–107 [DOI] [PubMed] [Google Scholar]
- 30. Nish S, Medzhitov R. 2011. Host defense pathways: role of redundancy and compensation in infectious disease phenotypes. Immunity 34:629–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nociari M, Ocheretina O, Murphy M, Falck-Pedersen E. 2009. Adenovirus induction of IRF3 occurs through a binary trigger targeting Jun N-terminal kinase and TBK1 kinase cascades and type I interferon autocrine signaling. J. Virol. 83:4081–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nociari M, Ocheretina O, Schoggins JW, Falck-Pedersen E. 2007. Sensing infection by adenovirus: Toll-like receptor-independent viral DNA recognition signals activation of the interferon regulatory factor 3 master regulator. J. Virol. 81:4145–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pétrilli V, Dostert C, Muruve DA, Tschopp J. 2007. The inflammasome: a danger sensing complex triggering innate immunity. Curr. Opin. Immunol. 19:615–622 [DOI] [PubMed] [Google Scholar]
- 34. Philpott NJ, Nociari M, Elkon KB, Falck-Pedersen E. 2004. Adenovirus-induced maturation of dendritic cells through a PI3 kinase-mediated TNF-alpha induction pathway. Proc. Natl. Acad. Sci. U. S. A. 101:6200–6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poeck H, et al. 2010. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat. Immunol. 11:63–69 [DOI] [PubMed] [Google Scholar]
- 36. Roberts TL, et al. 2009. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 323:1057–1060 [DOI] [PubMed] [Google Scholar]
- 37. Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 38. Schoggins JW, Falck-Pedersen E. 2006. Fiber and penton base capsid modifications yield diminished Ad5 transduction and proinflammatory gene expression with retention of antigen-specific humoral immunity. J. Virol. 80:10634–10644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Selvarajoo K, et al. 2008. Signaling flux redistribution at toll-like receptor pathway junctions. PLoS One 3:e3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Servant MJ, et al. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 278:9441–9447 [DOI] [PubMed] [Google Scholar]
- 41. Stetson DB, Medzhitov R. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24:93–103 [DOI] [PubMed] [Google Scholar]
- 42. Takaoka A, et al. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448:501–505 [DOI] [PubMed] [Google Scholar]
- 43. Unterholzner L, et al. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11:997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van Helden SF, van Leeuwen FN, Figdor CG. 2008. Human and murine model cell lines for dendritic cell biology evaluated. Immunol. Lett. 117:191–197 [DOI] [PubMed] [Google Scholar]
- 45. Wu L, et al. 2003. Novel small-molecule inhibitors of RNA polymerase III. Eukaryot. Cell 2:256–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yamaguchi T, et al. 2010. Induction of type I interferon by adenovirus-encoded small RNAs. Proc. Natl. Acad. Sci. U. S. A. 107:17286–17291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yuan B, Latek R, Hossbach M, Tuschl T, Lewitter F. 2004. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 32:W130–W134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Z, et al. 2011. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12:959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhu J, Huang X, Yang Y. 2007. Innate immune response to adenoviral vectors is mediated by both Toll-like receptor-dependent and -independent pathways. J. Virol. 81:3170–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]