Abstract
To be clinically relevant, drug-resistant mutants must both evade drug action and retain pathogenicity. Many acyclovir-resistant herpes simplex virus mutants from clinical isolates have one or two base insertions (G8 and G9) or one base deletion (G6) in a homopolymeric run of seven guanines (G string) in the gene encoding thymidine kinase (TK). Nevertheless, G8 and G9 mutants express detectable TK activity and can reactivate from latency in mice, a pathogenicity marker. On the basis of studies using cell-free systems, ribosomal frameshifting can explain this ability to express TK. To investigate frameshifting in infected cells, we constructed viruses that express epitope-tagged versions of wild-type and mutant TKs. We measured TK activity by plaque autoradiography and expression of frameshifted and unframeshifted TK polypeptides using a very sensitive immunoprecipitation-Western blotting method. The G6 mutant expressed ∼0.01% of wild-type levels of TK polypeptide. For the G9 mutant, consistent with previous results, much TK expression could be ascribed to reversion. For the G8 mutant, from these assays and pulse-labeling studies, we determined the ratio of synthesis of frameshifted to unframeshifted polypeptides to be 1:100. The effects of stop codons before or after the G string argue that frameshifting can initiate within the first six guanines. However, frameshifting efficiency was altered by stop codons downstream of the string in the 0 frame. The G8 mutant expressed only 0.1% of the wild-type level of full-length TK, considerably lower than estimated previously. Thus, remarkably low levels of TK are sufficient for reactivation from latency in mice.
INTRODUCTION
Resistance of herpes simplex virus (HSV) to acyclovir (ACV) and the related more orally available drugs valacyclovir and famciclovir is a critical concern as it occurs in 5 to 10% of immunocompromised patients and 15 to 30% of bone marrow transplant patients with HSV disease (9, 23, 28). To confer clinically significant drug resistance, a viral mutant must both evade drug action and retain pathogenicity. The vast majority of ACV-resistant (ACVr) clinical isolates have mutations in the viral gene encoding thymidine kinase (TK) (2), which is the enzyme required to phosphorylate and thus activate the drug in infected cells. Although ACVr mutations can arise in many different sites in the tk gene, about half of the ACVr tk mutants from clinical isolates have insertions or deletions at homopolymeric stretches of Cs and Gs (26). Of these, about half have an insertion of one or two Gs (G8 or G9) and 5% have a deletion of one G (G6) in a run of 7 Gs (the G string) (Fig. 1) (2, 5, 11, 27). These mutants should produce polypeptides with no TK activity since the sequences downstream of the insertion or deletion are out of frame. However, G8 and G9 mutants are not truly TK negative (reviewed in reference 13). Both G8 and G9 exhibit low but higher-than-background TK activities, as seen by plaque autoradiography (14, 16, 17), which can suffice to permit some reactivation of the viruses from latent infection in mice, a marker of pathogenicity (14). In contrast, isogenic truly TK-negative viruses fail to reactivate from latency (4, 7, 10, 29, 30). To our knowledge, no one has characterized the TK activity or polypeptide expression of G6 mutants.
Fig 1.
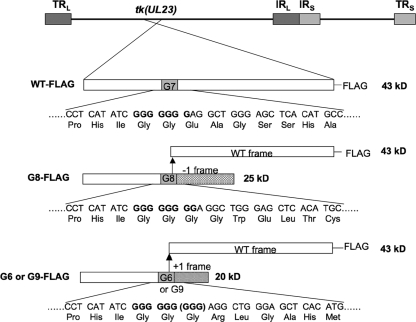
Diagram of TK expression from FLAG-tagged viruses. At the top of the panel is the HSV genome showing the position of the tk (UL23) gene, with long repeat sequences (TRL and IRL) and short repeat sequences (IRS and TRS) shown as dark and light gray boxes, respectively. Below, for each virus, the tk locus is expanded to show the expected protein products. To the left of the panel are the names of the viruses. The bars represent the tk coding sequence, with the open segments representing the WT reading frame; the solid gray segments, the position of the G string; the hatched segment, the reading frame created by the 1-base insertion mutation; and the striped segment, the reading frame created by the 1-base deletion or 2-base insertion mutations. The arrow shows that a frameshifting event could occur at that position to reenter the WT reading frame. FLAG tags at the C terminus are indicated, and the molecular masses to the right of the bars were calculated using the online Protein Calculator tool (Scripps Institute). Below the bars are shown the nucleotide and amino acid sequences surrounding the G string. For G9-FLAG, an additional GGG codon that encodes Gly is shown in parentheses.
In the case of G9 mutants, plaque autoradiography studies show that much of their TK activity and ability to reactivate from latency result from reversion to a TK-positive phenotype, often by addition of a 10th G (12, 16). Reversion can also account for some of the ability of G8 mutants to reactivate from latency (14, 25, 26). Nevertheless, there is considerable evidence that much of the TK activity of G8 mutants is a result of ribosomal frameshifting (12, 16, 20). Plaque autoradiography provides no evidence for reversion of G8 mutants in cell culture (14, 17). Full-length TK expression has been observed in G8 mutant-infected cells and has been recapitulated in an in vitro rabbit reticulocyte lysate translation system, where it was shown to be due to translational rather than other mechanisms (20). In vitro, the frequency of frameshifting has been estimated to be ∼1% (19, 20). The mechanism of ribosomal frameshifting on the G8 sequence in reticulocyte lysates has been studied using a dual-reporter system and found to be unusual, involving the G richness of the frameshifting signal rather than slippage and pausing (19). However, frameshifting has not been quantified and its mechanisms have not been explored in G8 mutant-infected cells.
To investigate TK expression in G-string insertion and deletion mutant-infected cells, we constructed viruses expressing epitope-tagged TK, which allowed us to very sensitively detect and quantify the amount of full-length and truncated TK polypeptides arising from mutant viruses and thus assess the frameshifting efficiency of the G8 virus. We also used this system to address certain mechanistic questions about frameshifting in infected cells. Finally, previous measurements of TK expression from G8 and G9 mutants have been indirect, relying on plaque autoradiography assays in which TK activity was compared to that of mutants with various tk mRNA levels (6, 8, 17, 21). Our assays permitted us to measure the amount of TK protein expressed by G8, G9, and G6 mutants directly and have led us to considerably revise downward estimates of the amount of TK that is sufficient to permit reactivation from latency in a mouse model.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) and TK-negative human osteosarcoma (143B) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% newborn calf serum and 10% fetal bovine serum, respectively, at 37°C in 5% CO2. A bacterial artificial chromosome (BAC) was developed containing wild-type (WT) HSV-1 strain KOS in which Cre-expressing vector sequences flanked by a pair of loxP sites are deleted from the viral genome by Cre recombinase upon transfection into cells, leaving a 36-bp in-frame sequence (34 bp loxP + 2 bp) ∼380 bp downstream of the G string within sequences encoding a loop on the surface of the TK structure. The insertion does not affect TK activity. A more detailed description of the BAC will be presented in a separate paper (I. Jurak et al., unpublished data). Mutations were introduced into the BAC using two-step Red-mediated recombination (31) with a kanamycin-resistant gene as a positive selection marker. Briefly, mutations were introduced together with the selection marker, which was later removed by I-SceI cleavage and intramolecular Red recombination utilizing a previously introduced sequence duplication, leaving no additional sequences besides the mutations. The primers used are listed in supplemental Table 1, which can be found at https://coen.med.harvard.edu. The integrity of BAC DNAs was verified by restriction digestion, and the presence of the engineered mutations and the absence of unwanted mutations in the tk gene were verified by sequencing the tk region both in BACs and in the viruses derived from them. The untagged G8 mutant was described in reference 14.
Plaque autoradiography.
Plaque autoradiography was performed by modifying a previously described protocol (14). The modifications were that the dishes were seeded with 4 × 105 143B cells 24 h prior to addition of an inoculum of 200 PFU of virus/dish and that radiolabeling was carried out for 20 h in the presence of 5 μCi/ml of [3H]thymidine.
Immunoprecipitation (IP).
143B cells were cultured in 100-mm-diameter dishes at 107 cells/dish, infected at a multiplicity of infection (MOI) of 10, and incubated at 37°C for the times indicated. Monolayers were washed with phosphate-buffered saline (PBS), scraped into 0.5 ml of lysis buffer (50 mM HEPES-KOH [pH 7.4], 1% Triton X-100, 150 mM NaCl, 10% glycerol, 0.1 mM dithiothreitol, and 2 mM EDTA plus one Complete EDTA-free protease inhibitor tablet [Roche] per 50 ml), and rocked at 4°C for 1 h before being centrifuged. After setting aside an aliquot for subsequent Western blot analysis, the supernatant was incubated with 20 μl of settled EZview anti-FLAG M2 affinity resin (Sigma) at 4°C with rotation for 4 h. The beads were then washed twice by rotating in 800 μl of lysis buffer for 20 min at 4°C before being incubated in 30 μl of 2× SDS-PAGE sample buffer (62.5 mM Tris-HCl [pH 6.8], 25% glycerol, 2% SDS, 0.01% bromophenol blue, and 5% β-mercaptoethanol) at 95°C for 5 min to elute proteins.
Western blot analyses.
Total proteins from infected cells, prepared by lysing cells in each well of a 12-well plate with 100 μl of SDS-PAGE sample buffer, aliquots of lysates used for IPs, prepared by mixing with equal volumes of 2× SDS-PAGE sample buffer, and samples eluted from affinity resin following IP were resolved on 12% SDS-polyacrylamide gels. Proteins from gels were transferred to polyvinylidene fluoride membranes (Pall Corporation) using a Trans-Blot semidry transfer cell (Bio-Rad). For probing the blots, the following antibodies were used: anti-FLAG antibody conjugated to horseradish peroxidase (Sigma) at a dilution of 1:1,000, anti-TK rabbit polyclonal antibody (generously provided by Bill Summers, Yale University) at 1:1,000, antihemagglutinin (anti-HA) rabbit polyclonal antibody (Sigma) at 1:1,000, anti-ICP8 rabbit polyclonal antibody (a kind gift of David Knipe, Harvard University) at 1:10,000, and to detect bound rabbit antibodies, goat anti-rabbit IgG conjugated to horseradish peroxidase (Southern Biotech) at 1:2,000. Signals were visualized by applying enhanced chemiluminescence reagents (Pierce) and exposing the membranes to films. Western blot images were quantified by densitometry using Quantity One, version 4.5, software (Bio-Rad).
Northern blot hybridization.
143B cells in 6-well plates were infected at an MOI of 10 and incubated for 5 h at 37°C. RNAs were extracted using an RNeasy minikit (Qiagen) and separated on a 1% agarose gel in NorthernMax-Gly running buffer (Ambion). Ethidium bromide staining showed that the same amount of RNA was loaded in each well (data not shown). The gel was transferred to a Nytran nylon membrane (Whatman) using a Turboblotter kit (Whatman) following the manufacturer's instructions, UV cross-linked, hybridized with a tk riboprobe, and washed using buffers provided in the NorthernMax-Gly kit (Ambion) following the manufacturer's instructions. After the image was obtained using a phosphorimager, the membrane was then hybridized with an ICP0 probe without stripping. After visualizing that image, the same membrane was hybridized with an actin probe without stripping and visualized. The tk, ICP0, and actin gene riboprobes were generated by in vitro transcription of linearized pTKprobe, pICP0probe, and pActinprobe plasmids, respectively. pTKprobe was generated by inserting a 251-bp tk gene fragment (positions 47395 to 47646, based on the sequence with accession number NC_001806 [NCBI]) into a pSL301 vector (Invitrogen); pICP0probe was generated by inserting a 333-bp BamHI-XhoI ICP0 gene fragment from pcDNAICP0 into pSL301 (pcDNAICP0 is a plasmid containing the spliced cDNA of ICP0 transcripts inserted into EcoRI-XbaI sites of pcDNA3.1 [Invitrogen]); pActinprobe was generated by inserting a 140-bp fragment of exon 4 of the human β-actin gene (nucleotides 682 to 821, based on the sequence with accession number NM_001101 [NCBI]) into pIDTBblue (Integrated DNA Technologies).
Pulse-chase protocol.
Infections were performed as described above for IP in 100-mm-diameter dishes. Cells were incubated for 5 h at 37°C before the media were replaced with 2 ml/dish fresh media containing 100 μCi/ml [35S]methionine (Perkin Elmer). Thirty minutes later, the radioactive media were removed and the monolayers were washed twice with PBS and supplied with fresh media (which contain unlabeled methionine at 30 mg/liter). At the time points indicated, the monolayers were washed twice with PBS, scraped into 0.6 ml of lysis buffer, and lysed at 4°C for 1 h. A portion of the cleared lysate was set aside, and the rest was used for IP following the above-described procedure, except that EZview anti-HA affinity gel (Sigma) was used and beads were washed 5 times instead of 2 times. IP and lysate samples were resolved on 12% SDS-polyacrylamide gels. Gels were then dried and exposed to a phosphorimager screen, and the band intensities were quantified by densitometry using Quantity One, version 4.5, software (Bio-Rad) and corrected for different methionine contents (full-length TK, 13 methionines; truncated G8 TK, 7 methionines).
RESULTS
Levels of full-length TK polypeptides in G-string mutant-infected cells.
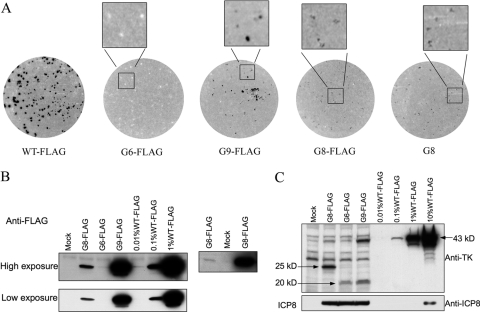
We wished to analyze the expression of full-length TK by G8 and G9 mutant viruses in infected cells and also investigate whether G6 viruses express TK activity and full-length TK. These studies required a sensitive and quantitative assay of TK polypeptide. Although we had previously observed radiolabeled full-length TK from G8-infected cells using IP with an anti-TK antibody (20), the sensitivity was too low for the assay to be quantitative. Our initial attempts using Western blotting with a commercially available anti-TK antibody also failed due to insufficient sensitivity (data not shown). To more quantitatively detect low levels of TK from the mutants, we constructed viruses in which WT HSV-1 and G8, G9, and G6 mutant TKs included a FLAG tag at the C terminus of the TK open reading frame (Fig. 1), using a BAC derived from WT HSV-1 strain KOS (Jurak et al., unpublished). Addition of the FLAG tag did not affect virus replication; TK polypeptide expression, as monitored by comparing WT and WT-FLAG using Western blotting (data not shown); or enzyme activity, as monitored by comparing G8 and G8-FLAG using plaque autoradiography (Fig. 2A). As had been seen previously with untagged G9 mutants (16, 17), plaque autoradiography revealed a heterogeneous population from G9-FLAG, with some plaques exhibiting high levels of TK activity and some exhibiting low levels (Fig. 2A). This variability is consistent with a high frequency of phenotypic reversion for this mutant, with reversion occurring at different times during plaque formation, as has been confirmed with untagged G9 mutants (16, 17). In contrast, G8-FLAG formed plaques with similar TK activities (Fig. 2A). G6-FLAG formed plaques without detectable TK activity (Fig. 2A).
Fig 2.
TK expression in WT and mutant virus-infected cells. (A) Plaque autoradiography showing in situ TK activity in plaques. Black or gray dots represent plaques with TK activity, and white dots represent plaques without detectable TK activity. The enlarged images of selected areas are shown at the top of autoradiographs, and the viruses assayed are indicated below each autoradiograph. (B) (Left) Two exposures (high and low) of IP-Western (FLAG) analysis of full-length TK expression in mock-, mutant-, and WT-infected cells compared to a dilution series of an IP from WT-infected cells (12 hpi, MOI = 10), with the viruses and dilutions used indicated at the top; (right) high exposure of IP-Western (FLAG) analysis of full-length TK expression in G6-FLAG-, mock-, and G8-FLAG-infected cells. (C) Western blot analysis using anti-TK antibody showing full-length as well as N-terminal TK polypeptides in infected cells (top) or using anti-ICP8 antibody (bottom). The samples analyzed were lysates reserved in the experiment whose results are shown in panel B and dilutions of lysates of WT-infected cells, with the viruses and dilutions used indicated at the top. The arrows point to bands corresponding to N-terminal (20-kDa and 25-kDa) or full-length (43-kDa) TK polypeptides. The molecular masses indicated were interpolated from molecular mass markers run alongside the samples.
To detect full-length TK expression in infected cells, we optimized a procedure in which FLAG-tagged TK polypeptide was efficiently immunoprecipitated with anti-FLAG antibody (∼99% of FLAG-TK immunoprecipitated [data not shown]) and was then detected by Western blotting, also with anti-FLAG antibody. The optimized procedure, denoted IP-Western (FLAG), was sensitive enough to detect 0.01% of the WT level of TK polypeptide, based on a dilution series in which the WT-FLAG-infected lysate was diluted in mock-infected cell lysate prior to IP (Fig. 2B, left). The assay revealed that G8-FLAG, G9-FLAG, and G6-FLAG express about 0.1%, 0.5%, and 0.01% of the WT level of full-length TK, respectively. The detection of TK from G6-FLAG was reproducible and was not due to contamination from adjacent samples, because when the G6-FLAG sample was electrophoresed adjacent to a sample from mock-infected cells, full-length TK was still clearly detectable (Fig. 2B, right). Nevertheless, the undetectable TK activity of G6-FLAG in plaque autoradiography correlated with an ∼10,000-fold decrease in full-length TK expression.
For G8-FLAG and G9-FLAG, similar results were obtained in three additional independent experiments in which WT-FLAG samples were diluted after IP. In all experiments, full-length TK levels for G8-FLAG and G9-FLAG were in the range of 0.08 to 0.15% and 0.3 to 1%, respectively. The latter range of values is consistent with amounts of TK enzyme activity measured in lysates of cells infected with a different G9 mutant (12).
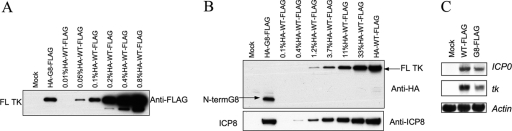
We also measured TK polypeptide expression using a polyclonal TK antibody generously provided by Bill Summers (Yale University) and a direct Western blotting procedure. Based on a dilution series made by diluting cell lysates in SDS-PAGE sample buffer, we detected about 0.3% of the WT level of full-length TK from G9-FLAG, about 0.1% of the WT level from G8-FLAG (and from an untagged G8 virus [data not shown]), and background levels for G6-FLAG (Fig. 2C), consistent with our results using IP-Western (FLAG). This Western blot also revealed bands from N-terminal TK products with the expected mobilities (Fig. 1). These products appear to be less abundant than full-length TK from WT-FLAG because they contained only a portion of the epitopes recognized by the polyclonal antibody and because of reduced synthesis (see below). A similar, relatively low abundance of N-terminal products was also observed with untagged G8 and G9 viruses that were not derived from the BAC (see supplemental Fig. 1 at https://coen.med.harvard.edu; data not shown). The direct Western blotting procedure also allowed us to measure ICP8 expression as an internal control for infection and loading. All viruses expressed similar amounts of ICP8 (Fig. 2C). Thus, using both the IP-Western (FLAG) and direct Western blotting methods, the G8 mutant virus expressed ∼0.1% of WT levels of full-length TK polypeptide. These levels are 5- to 10-fold lower than previous estimates of TK expression obtained using less direct methods (3, 15–18) (see Discussion).
Does reversion account for TK expression from G8 and G9 mutants?
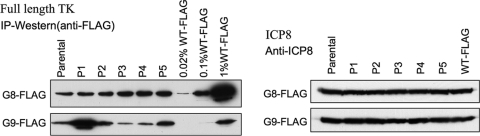
Our plaque autoradiography results suggested that the G9 mutant reverted to high-level TK expression, while TK expression by the G8 mutant was more stable. To examine this further, we purified single plaques from G9-FLAG- or G8-FLAG-infected Vero cells, amplified them, and used the amplified stock to infect 143B cells. Stocks derived from individual plaques of G9-FLAG showed great variation in the levels of expression of full-length TK polypeptide shown by IP-Western (FLAG) (0.3 to 3% in the five plaques selected) (Fig. 3), consistent with the phenotypic reversion occurring during plaque formation resulting in mixed populations of virus. This variation was not due to differences in infection, as the lysates used for IP-Western (FLAG) expressed similar levels of ICP8 (Fig. 3). In contrast, stocks derived from five different G8-FLAG plaques showed uniform full-length TK expression (Fig. 3), supporting the notion that reversion plays at most a limited role in the expression of active TK by G8. This result is consistent with our never having observed high levels of TK activity in G8 plaques in assays of thousands of such plaques (3, 14–17, 20) (data not shown).
Fig 3.
Analysis of G8 and G9 mutant reversion. (Left) IP-Western (FLAG) analysis of full-length TK expression in cells infected with the parental stock and plaque isolates (P1 to P5) from G8-FLAG (upper image) and G9-FLAG (lower image) (12 hpi, MOI = 10); (right) Western blot analysis using anti-ICP8 antibody of the lysates used for the IP assay whose results are shown in the left panel. The viruses and dilutions used are indicated at the top.
Frameshifting efficiency in G8 mutant-infected cells.
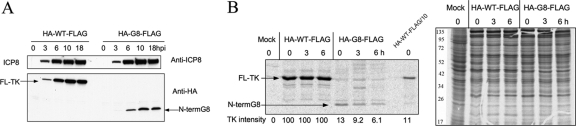
We next wished to determine the frameshifting efficiency for the G8 mutant, which entailed measuring the ratio of the full-length frameshifted product to the N-terminal unframeshifted product. To this end, an HA tag was introduced near the N terminus of TK, which was already FLAG tagged at its C terminus, in viruses containing either the WT or the G8 mutant tk gene. The tag was not added at the very N terminus of TK to avoid perturbation of the important viral gene UL24, which overlaps tk for 138 bp (22). The added HA tag did not affect TK activity, as shown by plaque autoradiography (data not shown). After cells were infected with HA-WT-FLAG and HA-G8-FLAG viruses, the levels of the N-terminal and full-length proteins were determined by reference to a dilution series in Western blotting (HA) and IP-Western (FLAG) assays (Fig. 4A and B). The level of full-length protein in HA-G8-FLAG-infected cells was about 0.1% of that in HA-WT-FLAG-infected cells, the same value that we had obtained using viruses with FLAG tags alone (Fig. 2B), suggesting that the added HA tag did not affect TK expression. Interestingly, the N-terminal product expressed by HA-G8-FLAG was present at only about 10% the level of full-length TK from HA-WT-FLAG, as judged by comparison with a dilution series (Fig. 4B). In contrast, the level of tk mRNA produced by the mutant was reduced less than 2-fold, as assayed by Northern blot hybridization and densitometric scanning (Fig. 4C).
Fig 4.
Frameshifting efficiency. (A) IP-Western (FLAG) analysis of full-length TK expression of HA-WT-FLAG and HA-G8-FLAG mutant viruses with a FLAG tag at the C terminus and an HA tag near the N terminus of TK (12 hpi, MOI = 10). (B) Western blot analysis using anti-HA antibody (top) or anti-ICP8 antibody (bottom) to measure the level of the N-terminal product from HA-G8-FLAG mutant-infected cells (12 hpi, MOI = 10) compared to that from a dilution series derived from HA-WT-FLAG-infected cells. For panels A and B, the names of the viruses and dilutions used are indicated at the top of the panel, and the positions of full-length (FL) TK and truncated (N-terminal [N-term]) G8 are indicated. (C) Northern blot analysis of tk mRNA, and, as controls, actin and ICP0 mRNA (as indicated to the right) in mock-infected (left lane), WT-FLAG-infected (middle lane), and G8-FLAG mutant-infected (right lane) cells (5 hpi, MOI = 10). By densitometric scanning, there was 52% as much tk mRNA and 73% as much ICP0 mRNA in the G8-FLAG-infected sample as in the WT-FLAG-infected sample. There were no meaningful differences among the amounts of actin mRNA in the three different samples.
To test whether the reduced level of N-terminal product was due to reduced synthesis or increased degradation, we first analyzed the time course of expression of the N-terminal product by Western blotting (HA), which showed that levels of both full-length TK and the N-terminal product increased until about 10 h postinfection (hpi) and then plateaued, similar to ICP8 (Fig. 5A). We then performed a pulse-chase experiment by pulse-labeling infected cells with [35S]methionine for 30 min at 5 hpi, followed by a chase in the presence of unlabeled methionine for 6 h. HA-tagged products were then immunoprecipitated using anti-HA antibody for analysis. During the pulse phase, the N-terminal product from HA-G8-FLAG was labeled less efficiently than the full-length protein from HA-WT-FLAG. Using densitometric scanning and correcting for different methionine contents, the level of labeled N-terminal product was 13% of that of full-length protein, similar to the steady-state level observed by Western blotting. During the chase period, HA-WT-FLAG TK remained stable (Fig. 5B). There was some reduction in the amount of labeled N-terminal product from HA-G8-FLAG during the chase (Fig. 5B). However, the reduction was only 2-fold over a 6-h period. This reduction is not sufficient to explain the 10-fold lower level of the N-terminal product and explains why we saw no decrease of steady-state protein levels in the time course experiment (Fig. 5A).
Fig 5.
Stability of full-length and truncated TK products. (A) Western blot analyses of time courses of expression of ICP8 using anti-ICP8 antibody (top) and TK products using anti-HA antibody (bottom) in cells infected with doubly tagged HA-WT-FLAG (left) and HA-G8-FLAG (right). The positions of ICP8, full-length (FL) TK, and N-terminal product (N-term G8) are indicated. The time points (hours) postinfection at which cells were harvested are indicated at the top of each lane. (B) Pulse-chase analysis showing SDS-PAGE of [35S]methionine-labeled N-terminal products in samples immunoprecipitated using anti-HA antibody (left) and lysate samples (right). Cells were either mock infected (left-most lane) or infected with HA-WT-FLAG or HA-G8-FLAG (as indicated at the top) at an MOI of 10. At 5 hpi, cells were pulsed for 30 min and then chased for the times indicated at the top of each lane. The numbers at the bottom of the left panel show the intensities of the TK bands determined by densitometric analysis, after correcting for different methionine contents in the full-length and truncated (N-terminal G8) proteins. The rightmost lane of the left panel is a 10-fold dilution of the zero time point sample for HA-WT-FLAG. The numbers to the left of the right panel show the positions of molecular mass markers.
Thus, with a level of full-length TK of ∼0.1% that of WT and a level of N-terminal product of ∼10% that of WT full-length TK, the frameshifting efficiency in G8 mutant-infected cells is about 1%, which is consistent with previous in vitro results using a dual-reporter assay (19).
Analysis of nonsense mutations.
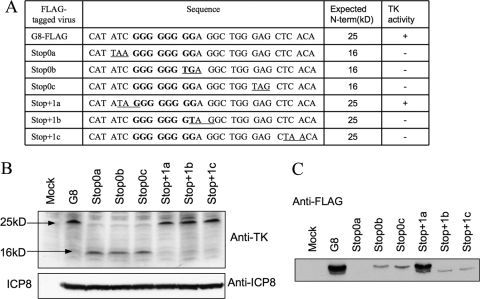
To address where frameshifting occurs, we introduced nonsense mutations into sequences surrounding the G string. Our reasoning was that if frameshifting occurs on the G string, a stop codon immediately before it in the 0 reading frame (N-terminal frame) of the mutant or immediately after it in the +1 frame (the frame used after frameshifting) of the mutant should block expression of full-length TK. We therefore constructed three mutant viruses: Stop 0a, with a stop codon in the 0 reading frame immediately before the G string; Stop+1b, with a stop codon in the +1 reading frame after the 7th G of the G string; and Stop+1c, with a stop codon in the +1 reading frame four codons further downstream (Fig. 6A and 7A, top). Each virus contained a FLAG tag at the C terminus of TK. Plaque autoradiography showed no TK activity from any of these three nonsense mutants (data not shown). Lysates of cells infected with these viruses or with G8-FLAG contained similar amounts of N-terminal TK product, as assessed by Western blotting with anti-TK antiserum (Fig. 6B). The lysates were subjected to IP-Western (FLAG). In this case, we electrophoresed the immunoprecipitated proteins further into the SDS gel. This resolved two immunoreactive species in the G8-FLAG sample, a major species at the position of full-length TK and a minor species with higher mobility (Fig. 6C). (We discuss the possible origin of the higher-mobility species in the Discussion.) The Stop0a mutant expressed no detectable immunoreactive species (Fig. 6C). The Stop+1b and Stop+1c mutants expressed the minor, higher-mobility species but none migrating at the position of full-length TK, consistent with plaque autoradiography results (data not shown). Thus, the behavior of these three mutants was consistent with our hypothesis that frameshifting occurs on the G string (Fig. 7A, top), as mutations predicted to abolish full-length TK synthesis abolished expression of the polypeptide that comigrated with full-length TK and abolished TK activity.
Fig 6.
Effects of nonsense mutations on frameshifting. (A) Table summarizing the names of viruses, sequences surrounding the G string in the N-terminal reading frame (underlined codons are introduced stop codons, and the G strings are in bold), expected sizes of N-terminal products, and TK activity (+, detectable; −, not detectable), as shown by plaque autoradiography (data not shown). (B) Western blot analysis of the expression of the N-terminal products using anti-TK antibody (top) and ICP8 using anti-ICP8 antibody (bottom). The viruses used (12 hpi, MOI = 10) are indicated at the top. The molecular mass numbers to the left are derived from molecular mass markers run alongside the samples. (C) IP-Western (FLAG) analysis of the expression of full-length TK in G8-FLAG- and nonsense mutant-infected cells (12 hpi, MOI = 10). The names of the viruses are indicated at the top.
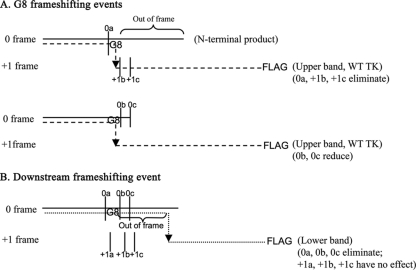
Fig 7.
Proposed origins of TK polypeptides expressed by G8 and nonsense mutants. The horizontal solid lines, dashed lines, and dotted lines represent the N-terminal products, the G8 frameshifting events, which give rise to the upper bands in Fig. 6C, and the hypothetical downstream frameshifting event, which gives rise to the lower bands in Fig. 6C, respectively. The arrows indicate the frameshifting sites. The vertical solid lines indicate the positions of the introduced stop codons. The upper schematic shows that the Stop0a, Stop+1b, and Stop+1c mutations result in premature termination of the G8 frameshifting event. The middle schematic shows that although the G8 frameshifting event is permitted by Stop0b and Stop0c, its efficiency is reduced by the nucleotide changes. The lower schematic shows that the downstream frameshifting event can be prematurely terminated by Stop0a, -b, or -c but not by Stop+1a, -b, or -c. This event would also result in an out-of-frame amino acid sequence in the frameshifted product, as indicated, which could explain its increased mobility during SDS-PAGE relative to that of WT full-length TK.
Using a similar approach, we also investigated the effects of a stop codon in the +1 frame immediately upstream of the G string (Stop+1a), in the 0 frame after the sixth G in the G string (Stop0b), or three codons downstream of the G string (Stop0c). We reasoned that these mutations should not abolish frameshifting if that occurs on the G string. Again, lysates of cells infected with these viruses expressed amounts of N-terminal TK product similar to the amount expressed by G8-FLAG (Fig. 6B). The Stop+1a mutant, as predicted, exhibited TK activity by plaque autoradiography (data not shown) and expressed amounts of full-length TK polypeptide and also the minor immunoreactive species similar to those expressed by G8-FLAG (Fig. 6C). The Stop0b and Stop0c mutants did express full-length TK, in line with our hypothesis, although at reduced levels (Fig. 6C and 7A, bottom). Scanning densitometry relative to a dilution series indicated that full-length TK expression was reduced ∼5-fold. Interestingly, these mutants failed to express the higher-mobility immunoreactive species (Fig. 6C). These data are consistent with frameshifting occurring on the G string, with at least some frameshifting occurring within the first 6 Gs of the G string. However, they also indicate that sequences that are altered by the Stop0b and Stop0c mutations are important for frameshifting efficiency.
DISCUSSION
To both evade drug therapy and cause disease, HSV ACVr mutants often use mechanisms to compensate for the effects of the mutations on TK activity. In this study, we examined the effects of three tk frameshift mutations found in clinical ACVr isolates on TK expression in HSV-infected cells using plaque autoradiography and a highly sensitive IP-Western blotting technique. For one mutation, G9, our results confirm previous findings (12, 16) that reversion can account for much of the TK activity of these viruses (although it is likely that frameshifting also contributes to this [13, 16, 17]) and extend those findings by detecting and quantifying full-length TK polypeptide synthesis from the mutant with this mutation. For a second mutation, G6, we found that the mutation abolished detectable TK enzyme activity in a plaque autoradiography assay and reduced full-length TK expression ∼10,000-fold. For the third mutation, G8, we not only quantified its full-length TK expression but also explored the mechanisms that account for this expression. Using pulse-chase protocols, we determined that the ratio of synthesis of full-length TK to the N-terminal product resulting from the frameshift mutation—i.e., the frameshifting efficiency—was ∼1%. By introducing nonsense mutations before and after the G string, we found that frameshifting most likely occurs on the G string. Of note, the G8 mutation reduced full-length TK expression substantially more than previously estimated (3, 15–18). We discuss our results with reference to mechanisms that compensate for tk frameshift mutations for TK expression and for latency and pathogenicity.
The G6 mutation.
To our knowledge, the TK activity of G6 mutants has not been previously assessed. We found that G6 exhibited no detectable TK activity in plaque autoradiography assays and expressed ∼10-fold less TK than the G8 mutant. In cultured cells, the G6 deletion mutation arises at roughly one-third the frequency of the G8 and G9 insertion mutations together (C. B. C. Hwang, personal communication). However, in clinical isolates, the G6 mutation occurs roughly 1/10 as frequently as the G8 and G9 mutations together (2, 5, 11, 27). We suggest that these differences in frequencies of isolation in cells versus patients are because the G8 and G9 mutants express more TK, which can abet their replication and pathogenesis in vivo. We speculate that clinical G6 isolates compensate for their loss of TK by alleles in genes other than tk, as has been suggested for certain other clinical isolates (14, 18).
Interestingly, the G6 motif is sufficient to mediate net +1 frameshifting in vitro (19), but it clearly mediates much less net −1 frameshifting in infected cells (this study) and very little if any in vitro (D.P. and D.M.C., unpublished results). In contrast, a mutant with a run of 7 Gs and a deletion of one G downstream (referred to as G7dG), which required −1 frameshifting for full-length TK expression, exhibited detectable TK activity (albeit lower than that of the G8 mutant) and could reactivate in latently infected trigeminal ganglia with a low frequency (17). Our results thus suggest that the seventh G strongly enhances net −1 frameshifting on the G string.
Mechanism of full-length TK expression in G8 mutant-infected cells.
The wild-type reading frame can be restored in cells infected with an insertion mutant such as G8 in three ways: DNA replication errors (reversion), transcription errors, and ribosomal frameshifting. The uniformity in the G8 population shown by both plaque autoradiography and IP-Western (FLAG) assays argues against a reversion mechanism for this mutant. We attempted to assess whether transcription errors could account for full-length TK expression by using reverse transcription-PCR, followed by deep sequencing and comparing the number of sequence reads containing G8 to the number containing G7. However, we found that applying this approach to a synthetic RNA template containing G8 resulted in a frequency of G7-containing reads that was substantially greater than 0.1%, the level of full-length TK expressed by G8, making this assay difficult to interpret. Thus, we cannot exclude the possibility that transcription errors contribute to frameshifting in infected cells. However, in rabbit reticulocyte lysates, G7-containing transcripts occurred with a frequency of <0.2%, much lower than the ∼1% frameshifting efficiency observed in the same system, implying that full-length TK expression occurs by ribosomal frameshifting in that system (20). Additionally, we found that the frameshifting efficiency in infected cells, calculated by the ratio of full-length TK to the N-terminal product, was ∼1%, similar to that in rabbit reticulocyte lysates. We note that this ratio was ∼10-fold higher than the ratio of full-length TK expressed by the mutant to that expressed by WT virus. This was due to ∼10-fold lower expression of the N-terminal product by the mutant relative to TK expression by WT virus but was not due to any obvious decrease in tk mRNA expression by the mutant. The weak expression of the N-terminal product may be related to the finding in Saccharomyces cerevisiae that mRNAs containing premature stop codons are translated more poorly (24). Regardless, based on the findings in rabbit reticulocyte lysates and the similarities in frameshifting efficiencies in vitro and in infected cells, we favor the interpretation that ribosomal frameshifting accounts for much of the full-length TK expression by the G8 mutant in infected cells.
Our results from experiments in which we introduced nonsense mutations around the G string in HSV confirm in vitro studies in which nonsense mutations in the 0 reading frame upstream of the G string inhibited downstream reporter gene expression (19) and extend them by showing that the effects of mutations in the +1 frame upstream or downstream of the G string are also consistent with frameshifting on the G string. Moreover, we found that a nonsense mutation in the 0 frame after just 6 Gs still permitted frameshifting, albeit at a reduced frequency. This finding is consistent with the sequences G6CGA, G6TGA, and AG6AG permitting net +1 frameshifting in vitro at reduced frequency (19). The reduced frequency could be due to fewer opportunities to frameshift on 6 versus 8 Gs or to alteration of residues important for frameshifting.
Our studies of frameshifting in infected cells also produced two unexpected findings. First, we found that a nonsense mutation in the 0 frame considerably downstream of the G string resulted in reduced frameshifting, suggesting that the sequences altered by the mutation are important for frameshifting efficiency. This is surprising because, in vitro, the sequence G8AG resulted in frameshifting as efficient as that in any longer sequence (19). This discrepancy may reflect differences in factors important for frameshifting between rabbit reticulocyte lysates in vitro and in infected human cells in culture.
Second, we detected a novel FLAG-reactive species with slightly higher mobility than full-length TK which was expressed at ∼10% the level of full-length TK in G8-infected cells. We hypothesize that this species is the result of a low-frequency, net +1 frameshifting event downstream of the G string. This hypothesis is illustrated in Fig. 7B. Expression of this species was abolished by nonsense mutations in the 0 frame around the G string but not by nonsense mutations in the +1 frame in similar locations, consistent with our hypothesis (Fig. 7B). The slightly higher mobility of the species could be due to differences in amino acid composition of the out-of-frame residues between the G string and the novel frameshifting event. Whether this novel frameshifting event has any biological relevance remains to be seen.
Pathogenicity of ACVr HSV mutants can be supported by even lower levels of full-length TK than thought.
We have shown that the G8 and G9 mutations and certain other ACVr HSV tk mutations permit some reactivation from latency in a mouse model, while an isogenic TK-negative mutant does not reactivate (1, 7, 14, 16, 17). In the case of G8, this ability to reactivate from latency is due to expression of low levels of TK (14). The levels of TK expressed by G8 and G9 were previously estimated to be 1% and 3%, respectively, using a quantitative plaque autoradiography assay that was normalized by comparison with a series of mutants expressing different amounts of TK (16, 17). However, there are several reasons why this comparison series may have yielded inaccurate estimates of TK expression. The viruses in the comparison series were tk promoter mutants linked to a mutant tk gene that encodes a temperature-sensitive TK (KG111). This mutant tk gene contains a stop codon between the first and second AUG codons, and the temperature-sensitive TK is synthesized via initiation of translation at the second AUG codon in tk mRNA (6, 8, 21). With a wild-type promoter, this mutant's TK activity was taken to be 10% that of WT, but some estimates of TK expression from this mutant are 2-fold lower (21). In addition, the TK levels of the different promoter mutants were estimated using their respective tk mRNA levels, assuming a linear relationship between TK mRNA and protein levels and between protein levels and TK activity. These assumptions may not be correct. Moreover, this mutant gene results in TK that is active and stable at 34°C but is unstable at 39°C (6, 21). A time course study of the KG111 mutant-infected cells carried out at 37°C showed a significant decrease in the level of truncated protein between 10 and 18 hpi (data not shown), suggesting that the mutant protein is also less stable at 37°C. In some cases, the plaque autoradiography assays were conducted at 34°C (3, 16, 18), where frameshifting may differ from that at 37°C, and in other cases, the plaque autoradiography assays were conducted at 37°C (14, 15), where the comparison series of mutants likely expressed less active TK than thought.
The present method of IP-Western (FLAG) compares the amounts of full-length TK from mutant-infected cells to those from a dilution series of TK from WT-FLAG-infected cells and can detect less than 0.01% of WT TK. It therefore is a more direct measure of TK expression and more sensitive than the plaque autoradiography assay and does not rely on the assumptions used in the plaque autoradiography assay. Data from Western blotting assays using an anti-TK antibody were consistent with the IP-Western (FLAG) data. The results of these assays reveal that G8 and G9 express only about 0.1 and 0.5% of WT levels of TK (Fig. 2B), respectively, 5- to 10-fold lower than previous estimates. This further indicates that a level of the enzyme much lower than previously thought is sufficient for detectable TK activity by plaque autoradiography. Our data indicate that plaque autoradiography has a sensitivity of about 0.05% of WT TK. Considering the labor involved in deriving the mutants expressing epitope-tagged TK, plaque autoradiography remains an excellent tool to assess TK activities from ACVr mutants, especially when analyzing mixed populations.
More importantly, the 0.1% TK activity of G8 is sufficient to permit some reactivation from latently infected mouse trigeminal ganglia. Remarkably two other mutants, C6 + 1C (with one C insertion in the run of 6 Cs in positions 548 to 553) and C5 − 1C (with one C deletion in the run of 5 Cs in positions 460 to 464), showed undetectable TK activity in plaque autoradiography, but both reactivate from latently infected mouse trigeminal ganglia, albeit with lower frequencies (1, 17), suggesting that even less than 0.1% TK can provide the virus with some pathogenic potential. It is truly amazing how so little TK expression, generated by frameshifting, can be biologically relevant.
ACKNOWLEDGMENTS
We thank Bill Summers and David Knipe for generously providing antibodies, Charles Hwang for kindly communicating results regarding the frequencies of isolation of G-string mutants, Anthony Griffiths for helpful advice on plaque autoradiography and valuable discussions, and Blair Strang for valuable discussions.
This work was supported by National Institutes of Health grant RO1 AI026126.
Footnotes
Published ahead of print 1 February 2012
REFERENCES
- 1. Besecker MI, Furness CL, Coen DM, Griffiths A. 2007. Expression of extremely low levels of thymidine kinase from an acyclovir-resistant herpes simplex virus mutant supports reactivation from latently infected mouse trigeminal ganglia. J. Virol. 81:8356–8360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burrel S, Deback C, Agut H, Boutolleau D. 2010. Genotypic characterization of UL23 thymidine kinase and UL30 DNA polymerase of clinical isolates of herpes simplex virus: natural polymorphism and mutations associated with resistance to antivirals. Antimicrob. Agents Chemother. 54:4833–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen SH, Cook WJ, Grove KL, Coen DM. 1998. Human thymidine kinase can functionally replace herpes simplex virus type 1 thymidine kinase for viral replication in mouse sensory ganglia and reactivation from latency upon explant. J. Virol. 72:6710–6715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen SH, Pearson A, Coen DM. 2004. Failure of thymidine kinase-negative herpes simplex virus to reactivate from latency following efficient establishment. J. Virol. 78:520–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chibo D, Druce J, Sasadeusz J, Birch C. 2004. Molecular analysis of clinical isolates of acyclovir resistant herpes simplex virus. Antiviral Res. 61:83–91 [DOI] [PubMed] [Google Scholar]
- 6. Coen DM, Irmiere AF, Jacobson JG, Kerns KM. 1989. Low levels of herpes simplex virus thymidine-thymidylate kinase are not limiting for sensitivity to certain antiviral drugs or for latency in a mouse model. Virology 168:221–231 [DOI] [PubMed] [Google Scholar]
- 7. Coen DM, et al. 1989. Thymidine kinase-negative herpes simplex virus mutants establish latency in mouse trigeminal ganglia but do not reactivate. Proc. Natl. Acad. Sci. U. S. A. 86:4736–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coen DM, Weinheimer SP, McKnight SL. 1986. A genetic approach to promoter recognition during trans induction of viral gene expression. Science 234:53–59 [DOI] [PubMed] [Google Scholar]
- 9. Danve-Szatanek C, et al. 2004. Surveillance network for herpes simplex virus resistance to antiviral drugs: 3-year follow-up. J. Clin. Microbiol. 42:242–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Efstathiou S, Kemp S, Darby G, Minson AC. 1989. The role of herpes simplex virus type 1 thymidine kinase in pathogenesis. J. Gen. Virol. 70:869–879 [DOI] [PubMed] [Google Scholar]
- 11. Gaudreau A, Hill E, Balfour HH, Jr, Erice A, Boivin G. 1998. Phenotypic and genotypic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J. Infect. Dis. 178:297–303 [DOI] [PubMed] [Google Scholar]
- 12. Grey F, et al. 2003. Characterization of a neurovirulent aciclovir-resistant variant of herpes simplex virus. J. Gen. Virol. 84:1403–1410 [DOI] [PubMed] [Google Scholar]
- 13. Griffiths A. 2011. Slipping and sliding: frameshift mutations in herpes simplex virus thymidine kinase and drug-resistance. Drug Resist. Updat. 14:251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Griffiths A, Chen SH, Horsburgh BC, Coen DM. 2003. Translational compensation of a frameshift mutation affecting herpes simplex virus thymidine kinase is sufficient to permit reactivation from latency. J. Virol. 77:4703–4709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Griffiths A, Coen DM. 2005. An unusual internal ribosome entry site in the herpes simplex virus thymidine kinase gene. Proc. Natl. Acad. Sci. U. S. A. 102:9667–9672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Griffiths A, Coen DM. 2003. High-frequency phenotypic reversion and pathogenicity of an acyclovir-resistant herpes simplex virus mutant. J. Virol. 77:2282–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Griffiths A, Link MA, Furness CL, Coen DM. 2006. Low-level expression and reversion both contribute to reactivation of herpes simplex virus drug-resistant mutants with mutations on homopolymeric sequences in thymidine kinase. J. Virol. 80:6568–6574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Horsburgh BC, et al. 1998. Recurrent acyclovir-resistant herpes simplex in an immunocompromised patient: can strain differences compensate for loss of thymidine kinase in pathogenesis? J. Infect. Dis. 178:618–625 [DOI] [PubMed] [Google Scholar]
- 19. Horsburgh BC, Kollmus H, Hauser H, Coen DM. 1996. Translational recoding induced by G-rich mRNA sequences that form unusual structures. Cell 86:949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hwang CB, et al. 1994. A net +1 frameshift permits synthesis of thymidine kinase from a drug-resistant herpes simplex virus mutant. Proc. Natl. Acad. Sci. U. S. A. 91:5461–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Irmiere AF, Manos MM, Jacobson JG, Gibbs JS, Coen DM. 1989. Effect of an amber mutation in the herpes simplex virus thymidine kinase gene on polypeptide synthesis and stability. Virology 168:210–220 [DOI] [PubMed] [Google Scholar]
- 22. Jacobson JG, Martin SL, Coen DM. 1989. A conserved open reading frame that overlaps the herpes simplex virus thymidine kinase gene is important for viral growth in cell culture. J. Virol. 63:1839–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morfin F, et al. 2004. HSV excretion after bone marrow transplantation: a 4-year survey. J. Clin. Virol. 30:341–345 [DOI] [PubMed] [Google Scholar]
- 24. Muhlrad D, Parker R. 1999. Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping. Mol. Biol. Cell 10:3971–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sasadeusz JJ, Sacks SL. 1996. Spontaneous reactivation of thymidine kinase-deficient, acyclovir-resistant type-2 herpes simplex virus: masked heterogeneity or reversion? J. Infect. Dis. 174:476–482 [DOI] [PubMed] [Google Scholar]
- 26. Sasadeusz JJ, et al. 1997. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J. Virol. 71:3872–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sauerbrei A, Deinhardt S, Zell R, Wutzler P. 2010. Phenotypic and genotypic characterization of acyclovir-resistant clinical isolates of herpes simplex virus. Antiviral Res. 86:246–252 [DOI] [PubMed] [Google Scholar]
- 28. Stranska R, et al. 2005. Survey of acyclovir-resistant herpes simplex virus in the Netherlands: prevalence and characterization. J. Clin. Virol. 32:7–18 [DOI] [PubMed] [Google Scholar]
- 29. Tenser RB, Edris WA. 1987. Trigeminal ganglion infection by thymidine kinase-negative mutants of herpes simplex virus after in vivo complementation. J. Virol. 61:2171–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tenser RB, Miller RL, Rapp F. 1979. Trigeminal ganglion infection by thymidine kinase-negative mutants of herpes simplex virus. Science 205:915–917 [DOI] [PubMed] [Google Scholar]
- 31. Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197 [DOI] [PubMed] [Google Scholar]