Abstract
Prasinoviruses infecting unicellular green algae in the order Mamiellales (class Mamiellophyceae) are commonly found in coastal marine waters where their host species frequently abound. We tested 40 Ostreococcus tauri viruses on 13 independently isolated wild-type O. tauri strains, 4 wild-type O. lucimarinus strains, 1 Ostreococcus sp. (“Ostreococcus mediterraneus”) clade D strain, and 1 representative species of each of two other related species of Mamiellales, Bathycoccus prasinos and Micromonas pusilla. Thirty-four out of 40 viruses infected only O. tauri, 5 could infect one other species of the Ostreococcus genus, and 1 infected two other Ostreococcus spp., but none of them infected the other genera. We observed that the overall susceptibility pattern of Ostreococcus strains to viruses was related to the size of two host chromosomes known to show intraspecific size variations, that genetically related viruses tended to infect the same host strains, and that viruses carrying inteins were strictly strain specific. Comparison of two complete O. tauri virus proteomes revealed at least three predicted proteins to be candidate viral specificity determinants.
INTRODUCTION
Planktonic viruses are the most numerous and diversified biological entities on Earth (61), and they are considered to be important players in microbial mortality (52, 59, 60, 73). They may shape the structure of communities by “killing the winner” (64), i.e., by top-down elimination of the most abundant microbes, allowing rare and less competitive species to coexist, and drive the evolution of their hosts via horizontal gene transfers and the evolutionary arms race referred to as the Red Queen theory (68).
More and more research on viruses infecting eukaryotic marine phytoplankton is being done (e.g., see references 5, 12, 34, and 56), and much of this concerns double-stranded DNA viruses, in particular, those of the Phycodnaviridae family. Prasinoviruses of Ostreococcus tauri, the smallest free-living eukaryote known so far (13), are among the best-studied Phycodnaviridae. To date, more than 300 O. tauri viruses (OtVs) have been sampled (5, 6) and 3 complete genomes have been sequenced (15, 71, 72). OtVs were found to be 2 orders of magnitude more abundant in a Mediterranean lagoon than in a coastal marine station (6), perhaps reflecting the distribution of their host species.
Phylogenetic, genomic, and ecological analyses divided the Ostreococcus genus (class Mamiellophyceae [31]) into four clades (called clades A to D [14, 21, 25, 43, 54]) and three ecotypes (coastal, oceanic, and deepwater strains). Ostreococcus tauri strains belong to clade C and are mainly found in Mediterranean lagoons, where the first strain (OTTH0595; now maintained as the clonal cell line RCC745) was isolated (13, 40). So far, 17 O. tauri strains have been described from five coastal locations in the Gulf of Lion (four of these are in lagoons), in the northwest (NW) Mediterranean Sea. Grimsley et al. (20) characterized their nuclear genetic diversity: although these 17 strains have identical 18S and internal transcribed spacer (ITS) rRNA genes, 8 intergenic regions showing a low level of polymorphism (∼0.01) were identified, and their analysis gave evidence for sexual exchanges between these strains. Moreover, despite their low levels of polymorphism, L. Subirana et al. (submitted for publication) observed that two chromosomes (numbers 2 and 19) showed variations in size between strains. These two chromosomes are atypical, carrying numerous transposable elements (14), and their genes are evolving faster than those found on other chromosomes, as measured, for example, by the ratio of nonsynonymous to synonymous base pair substitutions (dN/dS) (25).
In spite of these studies on Ostreococcus tauri and its viruses, few data on the host specificity of OtV are available. Derelle et al. (15) tested the specificity of OtV5 (initially isolated from the Bages-Sigean Lagoon using the strain RCC745 as a host) on different phytoplanktonic host species, including 10 out of the 14 O. tauri strains described above, but found that it was able to infect only RCC745. This observation suggested that such viruses are highly specific. Based on this result, we isolated more viruses from the NW Mediterranean Sea using 8 independent wild-type O. tauri strains in order to (i) gain insight into the host specificity of such viruses, (ii) to determine whether the genetic similarity of viruses reflects similarities in their infection patterns, and (iii) to propose mechanisms controlling the host specificity of OtVs and the susceptibility of O. tauri strains to viruses.
MATERIALS AND METHODS
In addition to 5 previously described OtVs (5, 6, 15), we isolated 35 new OtVs using 8 O. tauri strains, and we tested their host specificity at both an intraspecific level on 13 O. tauri strains and an interspecific level on 7 Mamiellophyceae strains (from 2 other Ostreococcus clades and 2 Mamiellophyceae genera). Here, we consider the different clades of Ostreococcus not only as being different ecotypes but also as being different species (Subirana et al., submitted). To investigate intraspecific host-virus interaction, we looked for links between infectivity/susceptibility patterns and the polymorphic data sets available for OtVs and O. tauri: we used nucleotide sequences for OtVs and atypical chromosome sizes (chromosomes 2 and 19; here named outlier chromosomes) for O. tauri. As OtVs were sampled during the same time period and at different sites, we studied the correlation between their host specificities and their geographical origins. Finally, we compared the two OtV proteomes (OtV1 and OtV5) to look for proteins involved in host recognition.
Host strains.
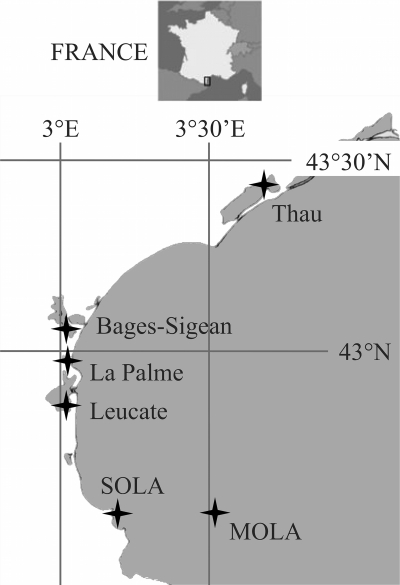
The intraspecific host specificity of OtV was assessed using 13 O. tauri strains (of clade C of the genus Ostreococcus) originating from 3 NW Mediterranean lagoons (Gulf of Lion), except for 2 strains (RCC1112 and RCC1113) isolated from a coastal station in the Gulf of Lion, SOLA (Fig. 1) (20). We also investigated the specificity of the same viruses at an interspecific level and tested if they could infect other Mamiellophyceae strains. For this purpose, we included two Ostreococcus strains sampled in the Mediterranean Sea (the clade D strain RCC789 and the clade A strain RCC371), other Ostreococcus clade A strains (RCC344, RCC356, and CCE9901, isolated from the Atlantic Ocean, English Channel, and Pacific Ocean, respectively), Bathycoccus prasinos (RCC1105, from the marine station MOLA), and a Micromonas sp. (RCC1109, from Leucate Lagoon). All of these strains are available in the Roscoff Culture Collection (http://www.sb-roscoff.fr/Phyto/RCC/).
Fig 1.
Locations of sampling sites. Four locations are lagoons (Leucate, 42°48′24″N, 03°01′27″E; La Palme, 42°57′18.04″, 3°0′3.56″E; Bages-Sigean, 43°03′14.76″N, 2°59′51.63″E; and Thau, 43°26′01.40″N, 03°39′54.49″E), and the two others are open-sea stations, one coastal (SOLA, 42°29′18″N, 3°8′42″E) and one offshore (MOLA, 42°27′11″N, 3°32′36″E). SOLA is a marine station included in the French marine monitoring network SOMLIT.
Isolation of viruses.
Viruses were isolated between January 2006 and March 2009 from the NW Mediterranean Sea (Fig. 1). Thirty-eight out of 40 were from Mediterranean lagoons (Thau, Leucate, Bages-Sigean, and La Palme), one came from a coastal station (SOLA), and another one came from an offshore station (MOLA). Viruses were isolated from 8 O. tauri strains using the plaque assay method: 10 ml of 0.45-μm-pore-size-filtered environmental samples was mixed with 10 ml of each O. tauri strain. After 1 h of incubation at 20°C, 2 ml of agarose (1.5%; type D-5 DNA grade; Euromedex) was added and the whole volume was mixed rapidly, poured into a petri dish, and thereafter incubated under continuous white light (100 μmol photons · m−2 · s−1 at 20°C). One week later, the numbers of PFU were determined and samples were stored at 4°C in 400 μl of a solution of MgSO4 (SM buffer [7]).
Specificity test.
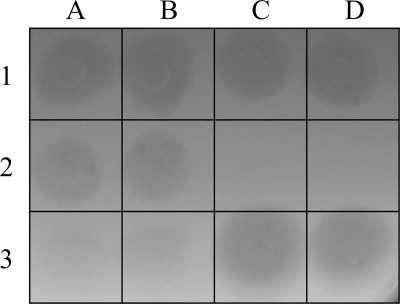
A fresh lysate of each virus was prepared (about 108 particles/ml). Two-microliter drops of each lysate were deposited in duplicate on the host or test strain plate solidified with agarose (final concentration, 0.15%). For each deposit, the result was recorded 10 days later according to the size and turbidity of the lysis plaque: no lysis (Fig. 2, C2 and D2), low lysis (Fig. 2, A3 and B3; plaques that were sometimes much smaller but always more turbid, 10 to 35% of the clarity of fully lysed plaques), intermediate lysis (Fig. 2, A2 and B2; smaller plaques, 50 to 75% of the size and/or clarity of fully lysed plaques), and high lysis (Fig. 2, A1, B1, C1, D1, C3, and D3; clear maximum-sized plaques).
Fig 2.
Specificity test: lysis spots in a suspension of host cells immobilized in soft agarose. No lysis for C2 and D2; low lysis for A3 and B3; intermediate lysis for A2 and B2; high lysis for A1, B1, C1, D1, C3, and D3.
Genetic distances between viruses.
A 600-bp-long fragment of the DNA polymerase gene (polB) of each virus was sequenced after PCR amplification with the primers AVS1 (5′-GARGGIGCIACIGTIYTIGAYGC-3′) and AVS2 (5′-GCIGCRTAICKYTTYTTISWRTA-3′), designed by Chen and Suttle (9). PCRs were set up as follows: 5 μl of virus lysis liquid (with SM buffer) was added to a 95-μl reaction mixture which contained PCR assay buffer (Promega), 0.2 mM each deoxyribonucleoside triphosphate, 1.5 mM MgCl2, 30 pmol of each primer, and 0.5 U of Taq DNA polymerase (Promega). The PCR was conducted in a Mastercycler system (Eppendorf) with an initial step of 95°C (3 min), followed by 35 rounds at 95°C (30 s), 50°C (60 s), and 72°C (90 s) and a final extension at 72°C (4 min). PCR products were electrophoresed in 0.5% TAE (Tris-acetate-EDTA) buffer in a 0.8% agarose gel and examined by UV visualization after ethidium bromide staining. PCR bands were purified directly using a NucleoSpin kit (Macherey-Nagel Company), and amplicons were sequenced. Sequence alignment was carried out with the ClustalW program (66) in BioEdit (version 7.00) software (22). Identical sequences were regrouped to produce a list of haplotypes using the DnaSP software (29). The genetic distance matrices were computed with PAUP 4b10 (63), using the K80+I+G evolution model determined by the ModelTest (version 3.8) program (50) and the Akaike information criterion (AIC).
Host-virus interaction analysis.
We first computed distance matrices based on the infectivity pattern of OtV and the susceptibility pattern of O. tauri. The infectivity and susceptibility patterns were coded 0 and 1, respectively, for no lysis and lysis. Binary codes were preferred, because our experiment did not precisely quantify the nature of lysis intensities. The Jaccard community index was used to calculate the infectivity distances between viruses and the susceptibility distances between O. tauri strains. Distance matrices were also computed for outlier chromosome size distances between O. tauri strains (based on size differences between chromosomes 2 and 19) and geographical distances between sampling sites using the Euclidean metric. All the distance matrices were obtained using normalized data and R (version 2.8.1; R: a language and environment for statistical computing, 2008; R Development Core Team, R Foundation for Statistical Computing, Vienna, Austria [http://www.R-project.org]). Mantel tests (30) were used to test the correlation between virus infectivity distances-virus genetic distances, virus genetic distances-geographical distances, and O. tauri susceptibility distances-outlier chromosome size distances. Mantel tests were computed using the function mantel from the vegan library (J. Oksanen, R. Kindt, P. Legendre, and R. B. O'Hara, vegan: Community Ecology Package, 2007 [http://cran.r-project.org/]) of the R statistical language, version 2.8.1. Significance was assessed using 999 permutations.
Putative specificity markers.
Putative specificity markers were found with a reciprocal best BLAST hit (RBH; E ≤ 10−5 [4]) between OtV5 (a recently corrected version of the nucleotide sequence with GenBank accession number NC_010191.1 [15; unpublished data]) and OtV1 (71) proteomes. The more divergent coding sequences between OtV5 and OtV1 were compared with the sequences in the NCBI database by the BLAST program, and their putative functions were assessed using putative domain and/or first hits. Finally, the protein structure was obtained with the dotmatcher function implemented in EMBOSS software suite (53).
Nucleotide sequence accession numbers.
The sequence data for the following viruses have been submitted to GenBank under the indicated accession numbers: OtV06_1, JQ250705; OtV06_4, JQ250706; OtV06_9, JQ250707; OtV06_11, JQ250708; OtV06_12, JQ250709; OtV09_551, JQ250710; OtV09_552, JQ250711; OtV09_553, JQ250712; OtV09_555, JQ250713; OtV09_556, JQ250714; OtV09_557, JQ250715; OtV09_558, JQ250716; OtV09_559, JQ250717; OtV09_560, JQ250718; OtV09_561, JQ250719; OtV09_562, JQ250720; OtV09_564, JQ250721; OtV09_565, JQ250722; OtV09_566, JQ250723; OtV09_570, JQ250724; OtV09_573, JQ250725; OtV09_574, JQ250726; OtV09_575, JQ250727; OtV09_576, JQ250728; OtV09_577, JQ250729; OtV09_578, JQ250730; OtV09_579, JQ250731; OtV09_580, JQ250732; OtV09_581, JQ250733; OtV09_582, JQ250734; OtV09_584, JQ250735; OtV09_585, JQ250736; OtV09_586, JQ250737; OtV09_587, JQ250738; OtV09_590, JQ250739.
RESULTS
Host specificity.
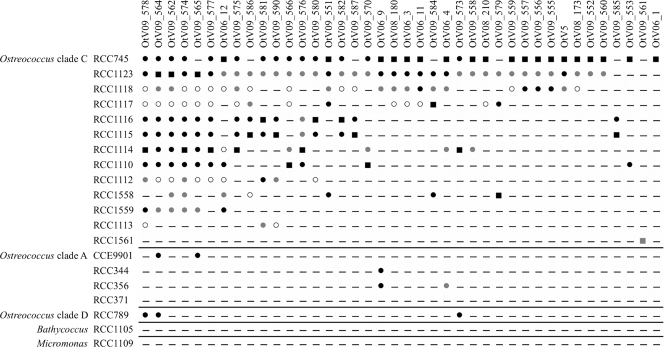
Forty viruses were isolated from 8 O. tauri strains (Tables 1 and 2). Most OtVs were isolated from lagoons: only OtV180 and OtV210 were found at marine locations (SOLA and MOLA, respectively). All of these viruses are considered to be different strains because either they have different polB sequences (see haplotype numbers in Table 1) or they have a distinct infectivity pattern (Fig. 3). Their host range was tested on 18 Ostreococcus strains (13 clade C strains, 4 clade A strains, and 1 clade D strain), 1 Micromonas strain, and 1 Bathycoccus strain (Table 2). Most of them (85%) appear to be specific to the O. tauri species; the remaining 15% are able to infect strains from other clades of the Ostreococcus genus (Fig. 3). No OtV strain could infect Bathycoccus or Micromonas. Among the viruses specific to O. tauri strains, various patterns were observed: from generalists infecting up to 11 host strains (e.g., OtV09_562 and OtV09_574) to highly specific viruses infecting only 1 to 3 strains (e.g., OtV09_561 and OtV5). Similarly, the O. tauri strains showed different levels of susceptibility to viruses. For instance, RCC1561, RCC1113, and RCC1558 are by far less susceptible to viral lysis than RCC745, RCC1123, and RCC1118. In Fig. 3, viruses and hosts are ordered by their infection and susceptibility patterns, according to the number of hosts infected by one virus and to the amount of viruses infecting one host (only for the O. tauri strains). The O. tauri-OtV interaction displayed a nested-like structure, where specialist and generalist viruses infect hosts that are the most susceptible or the most resistant to infection, respectively.
Table 1.
Ostreococcus tauri viruses
| Virusa | Host strain | Clade | Originb | Date of sampling (mo/day/yr) | Haplotype no. | Reference or source |
|---|---|---|---|---|---|---|
| OtV06_1 | RCC745 | C | South Leucate | 01/19/2006 | 1 | This study |
| OtV06_3 | RCC745 | C | La Palme | 01/24/2006 | 2 | 5 |
| OtV06_4 | RCC745 | C | La Palme | 01/24/2006 | 3 | This study |
| OtV5 | RCC745 | C | Bages-Sigean | 01/24/2006 | 4 | 14 |
| OtV06_9 | RCC745 | C | Thau | 02/13/2006 | 4 | This study |
| OtV06_11 | RCC745 | C | Thau | 02/13/2006 | 4 | This study |
| OtV06_12 | RCC745 | C | Thau | 02/13/2006 | 5 | This study |
| OtV08_173 | RCC745 | C | South Leucate | 02/19/2008 | 6 | 6 |
| OtV08_180 | RCC745 | C | SOLA | 02/25/2008 | 3 | 6 |
| OtV08_210 | RCC745 | C | MOLA | 06/10/2008 | 7 | 6 |
| OtV09_551 | RCC745 | C | South Leucate | 03/19/2009 | 4 | This study |
| OtV09_552 | RCC745 | C | South Leucate | 03/19/2009 | 8 | This study |
| OtV09_553 | RCC745 | C | South Leucate | 03/19/2009 | 9 | This study |
| OtV09_555 | RCC745 | C | Thau | 03/23/2009 | 10 | This study |
| OtV09_556 | RCC745 | C | Thau | 03/23/2009 | 4 | This study |
| OtV09_557 | RCC745 | C | La Palme | 03/27/2009 | 3 | This study |
| OtV09_558 | RCC745 | C | La Palme | 03/27/2009 | 8 | This study |
| OtV09_559 | RCC745 | C | La Palme | 03/27/2009 | 11 | This study |
| OtV09_560 | RCC745 | C | La Palme | 03/27/2009 | 12 | This study |
| OtV09_561 | RCC1561 | C | South Leucate | 03/19/2009 | 13 | This study |
| OtV09_562 | RCC1123 | C | La Palme | 03/27/2009 | 14 | This study |
| OtV09_564 | RCC1123 | C | La Palme | 03/27/2009 | 15 | This study |
| OtV09_565 | RCC1123 | C | La Palme | 03/27/2009 | 15 | This study |
| OtV09_566 | RCC1110 | C | La Palme | 03/27/2009 | 3 | This study |
| OtV09_570 | RCC1110 | C | South Leucate | 03/19/2009 | 16 | This study |
| OtV09_573 | RCC1114 | C | La Palme | 03/27/2009 | 10 | This study |
| OtV09_574 | RCC1114 | C | La Palme | 03/27/2009 | 5 | This study |
| OtV09_575 | RCC1114 | C | La Palme | 03/27/2009 | 17 | This study |
| OtV09_576 | RCC1114 | C | La Palme | 03/27/2009 | 18 | This study |
| OtV09_577 | RCC1114 | C | South Leucate | 03/19/2009 | 19 | This study |
| OtV09_578 | RCC1114 | C | South Leucate | 03/19/2009 | 15 | This study |
| OtV09_579 | RCC1558 | C | La Palme | 03/27/2009 | 20 | This study |
| OtV09_580 | RCC1116 | C | La Palme | 03/27/2009 | 21 | This study |
| OtV09_581 | RCC1116 | C | La Palme | 03/27/2009 | 21 | This study |
| OtV09_582 | RCC1116 | C | La Palme | 03/27/2009 | 22 | This study |
| OtV09_584 | RCC1117 | C | South Leucate | 03/19/2009 | 20 | This study |
| OtV09_585 | RCC1115 | C | Thau | 03/23/2009 | 21 | This study |
| OtV09_586 | RCC1115 | C | La Palme | 03/27/2009 | 23 | This study |
| OtV09_587 | RCC1115 | C | La Palme | 03/27/2009 | 22 | This study |
| OtV09_590 | RCC1115 | C | South Leucate | 03/19/2009 | 21 | This study |
In order to avoid duplication with viral isolate numbers that appear in other publications, we prefixed the original numbers by the year in which the strain was collected.
Global Positioning System coordinates are as follow: SOLA, 42°29′18″N, 3°8′42″E; MOLA, 42°27′11″N, 3°32′36″E; Thau, 43°26′01.40″N, 03°39′54.49″E; La Palme, 42°57′18.04″, 3°0′3.56″E; Bages-Sigean, 43°03′14.76″N, 2°59′51.63″E; South Leucate, 42°48′24″N, 03°01′27″E.
Table 2.
Mamiellophyceae strains
| Strain | Genus | Clade | Origina | Date of sampling (mo/day/yr) | Reference |
|---|---|---|---|---|---|
| RCC745 | Ostreococcus | C | Thau | 05/03/1995 | 12 |
| RCC1110 | Ostreococcus | C | South Leucate | 07/28/2006 | 19 |
| RCC1112 | Ostreococcus | C | SOLA | 10/02/2006 | 19 |
| RCC1113 | Ostreococcus | C | SOLA | 10/02/2006 | 19 |
| RCC1114 | Ostreococcus | C | South Leucate | 10/26/2006 | 19 |
| RCC1115 | Ostreococcus | C | North Leucate | 10/26/2006 | 19 |
| RCC1116 | Ostreococcus | C | North Leucate | 10/26/2006 | 19 |
| RCC1117 | Ostreococcus | C | West Leucate | 10/26/2006 | 19 |
| RCC1118 | Ostreococcus | C | West Leucate | 10/26/2006 | 19 |
| RCC1123 | Ostreococcus | C | South Leucate | 11/14/2006 | 19 |
| RCC1558 | Ostreococcus | C | South Leucate | 10/26/2006 | 19 |
| RCC1559 | Ostreococcus | C | South Leucate | 10/26/2006 | 19 |
| RCC1561 | Ostreococcus | C | Thau | 08/21/2006 | 19 |
| CCE9901 | Ostreococcus | A | La Jolla, CA | 01/01/1999 | 40 |
| RCC356 | Ostreococcus | A | Roscoff-Astan | 04/12/2000 | 51 |
| RCC344 | Ostreococcus | A | Maroc-UPW | 09/12/1999 | 51 |
| RCC371 | Ostreococcus | A | Sicile-5 | 09/18/1999 | 51 |
| RCC789 | Ostreococcus | D | Barcelona Harbor | 02/28/2001 | 51 |
| RCC1109 | Micromonas | South Leucate | 07/28/2006 | Unpublished | |
| RCC1105 | Bathycoccus | MOLA | 03/01/2006 | Unpublished |
Global Positioning System coordinates are as follow: Thau, 43°24′N, 3°36′E; South Leucate, 42°48′24″N, 03°01′27″E; North Leucate, 42°54′07″N, 3°02′56″E; West Leucate, 42°52′39″N, 03°02′41″E; SOLA, 42°29′18″N, 03°08′42″E; MOLA, 42°27′11″N, 03°32′36″E; La Jolla, CA, 32°90′N, 117°25′W; Roscoff-Astan, 48°45′N, 03°57′W; Maroc-UPW, 30°8′N, 10°3′W; Sicile-5, 36°29′N, 13°19′E); Barcelona Harbor, unknown.
Fig 3.
Specificity patterns. Lines refer to hosts, and columns refer to viruses. −, no lysis; ■, lysis on the host used for the isolation of the virus; ●, high lysis;  , intermediate lysis; ○, low lysis.
, intermediate lysis; ○, low lysis.
Influence of genetic identity of viruses and size of outlier chromosomes of O. tauri strains.
Surprisingly, both viruses showing strict specificity, i.e., infecting only one host strain, are viruses with an intein inserted in the polB gene (only OtV06_1 and OtV09_561 have an intein-containing polB and infect only one strain). Inteins are selfish genetic elements found within conserved regions of conserved proteins (62) that are translated with the host protein and removed in the maturation of the final protein product (48). In addition, we tried to find a correlation between genetic distances and infection/susceptibility patterns for the whole host-virus interaction. Because of the low level of polymorphism between O. tauri strains, it was not possible to use genetic distances between the available markers (20) and was possible only with the sizes of the outlier chromosomes. The infectivity of viruses was related to their genetic similarity (P = 0.001; Table 3), regardless of their geographical origins (P = 0.881). Moreover, the susceptibility of the O. tauri strains is related to the size of their outlier chromosomes (P = 0.008). In other words, genetically close strains tend to infect similar hosts, and O. tauri strains with outlier chromosomes with related sizes tend to be susceptible to the same viruses.
Table 3.
Correlations between distance matrices (Mantel tests)
| Test | Variable 1 | Variable 2 | P value |
|---|---|---|---|
| A (virus) | Infection patterns | Genetic distances | 0.001 |
| B (virus) | Genetic distances | Geographical origins | 0.881 |
| C (hosts) | Sensitivity patterns | Outlier chromosome sizes | 0.008 |
Putative specificity markers.
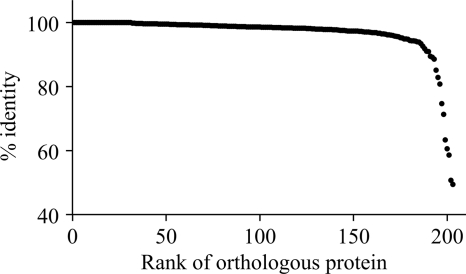
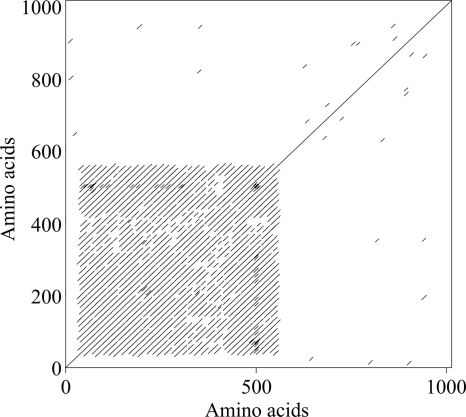
In order to find proteins involved in host recognition, we compared the two available OtV proteomes (OtV1 and OtV5 have, respectively, 232 and 243 proteins) and we looked for the most variable proteins. These viruses are quite similar, possessing 203 orthologous proteins, among which 95% show similarities superior to 88.61% (Fig. 4). Among the 10 more variable proteins, 4 are unknown, 1 has a putative conserved domain of glycosyltransferase (OtV5_183c; Table 4), 2 have putative exonuclease activity (OtV5_008c and OtV5_132; Table 4), and 2 are adhesin-like proteins (OtV5_074 and OtV5_120; Table 4). OtV5_120 is similar to an echinonectin (involved in the attachment of sea urchin larvae to the substrate), and OtV5_074 is similar to a putative Hep-Hag family protein, known to exhibit binding activity. Hep-Hag are residue repeats, and such a structure is visible with a dot plot analysis (Fig. 5). Hence, we analyzed the structure of the 10 more variable proteins and found similar highly repetitive motifs only for OtV5_073 (data not shown).
Fig 4.
Percent identities between OtV5 and OtV1 orthologous proteins. The most divergent proteins could be involved in the adaptive behavior of O. tauri viruses, particularly in their specificity.
Table 4.
Candidate specificity genes of O. tauri viruses
| OtV5 protein | OtV1 protein | % identity | Descriptiona | Putative conserved domainsb |
|---|---|---|---|---|
| OtV5_010c | OtV1_009 | 85.07 | ORFan | No putative conserved domains |
| OtV5_073 | OtV1_072 | 82.87 | Hypothetical protein; similar to predicted protein of Nematostella vectensis (E = 2e−20) | Bactofilin (pfam04519; E = 6.53e−04) |
| OtV5_008c | OtV1_005 | 80.80 | Similar to exonuclease RNase T and DNA polymerase III of Odoribacter splanchnicus (E = 2e−21) | DEDDh 3′-5′ exonuclease domain family (cd06127; E = 3.28e−34) |
| OtV5_048c | OtV1_047 | 74.67 | ORFan | No putative conserved domains |
| OtV5_183c | OtV1_175 | 71.31 | ORFan | Glycosyltransferase family 25 involved in lipooligosaccharide biosynthesis (cd06532; E = 1.69e−08) |
| OtV5_132 | OtV1_126 | 63.33 | Putative exonuclease; similar to ATCV1_Z622R (E = 1e−40) | Putative phage-type endonuclease (TIGR03033; E = 9.84e−30) |
| OtV5_122c | OtV1_115 | 60.60 | ORFan | No putative conserved domains |
| OtV5_074 | OtV1_073 | 58.62 | Putative Hep-Hag family protein of Psychroflexus torquis (E = 3e−25) | No putative conserved domains |
| OtV5_120 | OtV1_113 | 50.72 | Putative echinonectin of Nematostella vectensis (E = 7e−32) | FA58C (cd00057), coagulation factor 5/8 C-terminal domain, cell surface-attached carbohydrate-binding domain (E = 4.87e−16) |
| OtV5_071 | OtV1_070 | 49.42 | ORFan | No putative conserved domains |
ORFan, open reading frame with no clear similarity to known genes.
DEDDh, exoribonucleases which have a characteristic core comprised of four invariant acidic amino acid residues, DEDD, in this case subfamily h.
Fig 5.
OtV5_074 protein structure. The dot plot shows repetitive motifs for the first 600 amino acids of the OtV5_074 protein sequence. Such motifs have also been found for OtV5_073.
DISCUSSION
Our data suggest that all viruses isolated from O. tauri strains are specific to the genus Ostreococcus and most of them infect only the species O. tauri. Moreover, while some viruses tend to be generalists, others are more specific, infecting hosts that are the most resistant or the most susceptible, respectively. Surprisingly, intein-containing viruses showed strict host specificity. Statistically, the more that viruses are genetically close, the more they infect the same type of hosts, and the more that O. tauri strains are close in outlier chromosome sizes, the more they are susceptible to the same viruses. Finally, the more variable viral genes might be adaptive markers for specificity, and in particular, repeated motifs suggest proteins with binding activities.
Host specificity.
Out of 40 isolated viruses, only 2 come from the SOLA and MOLA marine stations. Bellec et al. (6) have shown that viruses infecting O. tauri strain RCC745 are on average 2 orders of magnitude more abundant in the Leucate Lagoon in the NW Mediterranean Sea than at SOLA and MOLA. This high concentration could be explained not only by the eutrophic nature of lagoons, allowing a higher primary production and a high host abundance (40, 75), but also because lagoons are semiclosed environments (connected with the sea) that certainly confine viral particles.
The interspecific host specificities analysis showed that 85% of OtVs were strictly specific to the O. tauri (clade C) species. While some viruses appear to be generalists, infecting up to 11 O. tauri strains out of 13, others can infect only 1 to 3 strains. As seen in the majority of host-phage networks, the structure of the O. tauri-OtV interaction is nested-like (17), where specialists and generalists infect, respectively, hosts that are the most susceptible and the most resistant to infection. This pattern suggests that OtVs evolve to broaden host ranges and O. tauri evolves to increase the number of viruses to which it is resistant (51). Derelle et al. (15) tested the specificity of the virus OtV5 on the same O. tauri hosts and found that it could infect only strain RCC745. In the present study, we have shown that OtV5 can infect two additional strains. The method that we used here to measure specificity differs a little from the one previously used: we inoculated viruses on solidified host cultures, while Derelle et al. (15) observed the complete lysis of hosts in liquid cultures. This highlights the sensitivity of our method and suggests that a viral infection is not always followed by a crash of the host population. Hence, while high lysis intensities on a solidified host corresponded to a complete host lysis liquid culture, low lysis intensities may affect fewer cells of the host population (Fig. 3). Two independent processes might explain these different lysis intensities: (i) different viral virulence and (ii) different host resistance levels (65). Nagasaki and Yamaguchi (35) also observed that viral lysis on the toxic dinoflagellate Heterosigma akashiwo was not always complete, allowing the proliferation of surviving cells.
Although the viruses tested in the present study are mainly specific to O. tauri strains (clade C), six viruses are able to infect one to two strains from other Ostreococcus clades. The different Ostreococcus clades were initially interpreted to be ecotypes adapted to different light intensities (clades C and D, polyvalent strains; clade A, high-light strains [54]), but they probably also represent different species (Subirana et al., submitted). As our result suggests that host switches between Ostreococcus clades are possible, care is required when predicting that a viral sequence obtained from environmental DNA, such as one from metagenomic data (7), may be linked to the presence of a certain host strain. A host clade may be assigned to a viral sequence, but the error rate is about 15%, according to our results.
Finally, since we observed that viruses with the same partial polB sequence exhibited different host ranges, using only polB as a genetic marker for the Phycodnaviridae (6, 10, 57) underestimates the functional diversity of such viruses.
Influence of viral genotypes.
We found that viruses with more closely related sequences of polB showed more similar patterns of infection. Since some of these viruses have been isolated from different lagoons during the same time period (this is not the case for host strains that were mostly sampled in Leucate Lagoon), this link could be the result of geographical isolation. For instance, each lagoon may contain viruses that are genetically similar and have similar infectivity. However, the genetic distance and the geographical origin of viruses appeared to be unlinked (P = 0.881) (7); hence, the four lagoons are connected and two genetically close viruses can have similar infectivity even if they are from different geographical sites. Since polB is used as a neutral marker of evolution (mutational changes occur almost exclusively in synonymous codons [6]), it is surprising to find such a correlation. Indeed, polB genes are well conserved among Phycodnaviridae (11, 16, 24), and adaptive markers (specificity markers) are more likely variable genes (39, 70). Mizumoto et al. (33) observed that the intraspecific host specificity of a Dinophyceae virus, Heterocapsa circularisquama single-stranded RNA virus (HcRNAV), is determined by the upstream events of viral infection (i.e., at the entrance step) and thus more likely by a surface protein located on the capsid. Moreover, because the genome of HcRNAV consists of 4.4 kb containing only four variable regions, the authors assumed that these regions determine the intraspecific host specificity, and a tertiary structure prediction showed that most of the substituted amino acid residues are located on the surface of the virion (36). In another study, Nagasaki et al. (37) noted that 10 Heterosigma akashiwo virus (HaV) strains were very similar for the polB gene but exhibited different host specificities. They also proposed that the determinants of host specificity might be found in variable regions of the genome. Nonetheless, these authors did not look for a correlation between viral genotypes and the infection patterns.
In contrast to our results, the 10 HaVs also had an intein containing polB but were capable of infecting many host strains. Inteins (internal proteins) are selfish genetic elements found in critical conserved proteins (18). Their removal occurs after translation and is required for the host protein to function correctly. Splicing domains are located at the N and C termini of the intein and ensure its excision and ligation of its flanking sequences (extein or external protein, i.e., the host protein) (19, 46, 47, 58). Moreover, inteins can also harbor homing endonuclease domains, particularly the dodecapeptide (DOD) motif EN1, ensuring its horizontal dispersal to intein-less alleles by recognition of a highly conserved homing site (YGDTDS on the polB sequence) (41, 47, 62). Although there is an excision step, experimental studies failed to find any effect of inteins on the host fitness (38, 48). For Ostreococcus tauri viruses, OtV06_1 and OtV09_561 both have an intein with complete homing endonuclease motifs (data not shown) and a strict specificity (we recently found that OtV06_8 has the same features). These results suggest that inteins might have negative effects on OtV fitness, if we assume that the less that viruses infect different host strains, the less they are successful for multiplication. However, our experiment does not shed light on the nature of the fitness cost. We propose three possible explanations for this high specificity: (i) DOD binds and cleaves an unspecific site on the viral genome (55); (ii) intein splicing could slow the viral replication; indeed, the codon usage of polB and the associated intein is slightly different for OtV06_1 and OtV09_561, suggesting recent acquisition (C. Clerissi et al., unpublished data); or (iii) intein splicing could be inhibited by O. tauri. To our knowledge, no organisms are known to synthesize agents inhibiting protein splicing (45). However, as intein splicing is a newly recognized therapeutic target in diseases caused by microbes (1, 44, 45, 74), if O. tauri inhibits intein splicing of OtV, this model could be useful for finding new therapeutic agents.
Is host range influenced by host adaptive response?
Although the 13 O. tauri strains are identical for the 18S and ITS rRNA genes, they contain outlier chromosomes of different sizes and exhibit different susceptibilities to viruses. Hence, similarly to the viral polB gene, the use of these ribosomal genes probably underestimates the real diversity of this picoeukaryotic species (49). Moreover, our study highlights a probable link between the susceptibility of the 13 O. tauri strains and the outlier chromosome sizes (Table 3). Subirana et al. (submitted) observed for 5 Ostreococcus strains (RCC1112, RCC1113, RCC1115, RCC1116, RCC1561) that the size diminution in chromosome 2 was often associated with a size increase in chromosome 19, suggesting a translocation between these two atypical chromosomes. The result of a translocation process could alter gene expression (2), producing adaptive changes (28), such as different susceptibilities to O. tauri viruses. However, for the other O. tauri strains, although the outlier chromosomes exhibit the larger variations in size, other chromosomes show smaller variations. Small variations might arise not only due to differences in the amount of DNA in individual gel tracks (Subirana et al., submitted) but also from other chromosome rearrangements such as inversion and fusion (27). Hence, these two mechanisms could also have implications in O. tauri susceptibilities. Future studies should target the gene expression differences among the 13 O. tauri strains (particularly on genes distributed on chromosomes 2 and 19) to better understand the link found between outlier chromosome sizes and their susceptibilities to viruses.
Putative specificity markers.
The genomes of three O. tauri viruses are available so far: OtV5 (15), OtV1 (71), and OtV2 (72). Because OtV5 and OtV1 are very similar (OtV2 is more divergent, probably because it infects a low-light-adapted Ostreococcus clade B strain), we used them to find the few variable proteins. The strain specificity of OtV1 was not tested, and as a result, we do not know if its infection pattern is different from that of OtV5. However, we found the comparison between their genomes to be important because (i) OtV1 and OtV5 have different geographical origins (the western English Channel and the Bages Lagoon in the Mediterranean Sea, respectively), and (ii) the structure of this host-virus interaction suggests that the specificity of OtV is constantly evolving. We found 10 variable proteins that might be involved in the adaptive behavior (39, 70) of O. tauri viruses, particularly in their specificity. Three could contribute to the specific attachment of viral particles to host receptors, and two putative exonucleases plus one gene harboring a putative glycosyltransferase domain could regulate infectivity at intracellular steps. Although the precise functions of putative exonuclease activities are not known, the present results suggest that they could be implicated in DNA replication, recombination, and repair (26, 69). Furthermore, glycosyltransferases encoded by viruses are considered important in the regulation of host-virus interactions, because they can fix glycans on proteins, lipids, or DNA, and consequently, they can modify the structure and function of proteins or make the viral DNA resistant to host restriction endonucleases (32). Among the proteins that might play a role in the viral adsorption, two are putative adhesin-like proteins: OtV5_074 is similar to a putative Hep-Hag family protein (23, 67) and OtV5_120 is a putative echinonectin, an embryonic substrate adhesion protein found in sea urchin (3). Although OtV5_120 has no repeated motifs, Hep-Hag repeating residues found in bacterial hemagglutinins and invasins are known to exhibit binding activity (67). Onimatsu et al. (42) also found that repetitive motifs in the structural protein Vp130 of chlorovirus (a Chlorella-infecting member of the Phycodnaviridae) are required for host cell wall binding. As there are no proteomic data for OtV, we cannot confirm that the proteins OtV5_074 and OtV5_120 are parts of the virion structure. However, we speculate that their similarity with adhesin-like proteins suggests that they might be. Finally, Thomas et al. (65) showed that the adsorption of OtV5 occurs on O. tauri-resistant cells, even if no viral DNA was observed inside the host. Hence, it is likely that the entry of OtV involves coreceptors. For example, HIV binds to the host CD4 protein, but the coreceptors CXCR4 and CCR5 are necessary for HIV entry into cells (8). As a consequence, in addition to the adhesin-like protein identified by BLAST, the repetitive structures seen in OtV5_074 and OtV5_073, and the presence of intracellular candidates such as exonucleases and glycosyltransferases, the identification and role of the proteins involved in OtV adhesion and entry are worthy of further investigations aimed at elucidating the specificity of O. tauri viruses.
To conclude, we have shown that although most of the tested viruses are specific to the O. tauri species, the O. tauri-virus interaction is more complex than the simple one strain-one virus view, at least at an intraspecific level. In addition, the nested-like structure suggests that specialist viruses are infecting the most susceptible strains. Hence, specialization should not be the result of an adaptive process, but it is more likely a transient state. Only the intein-containing viruses were strictly strain specific, perhaps a result of negative effects of intein on their replication success. Moreover, the rate of infections of other Ostreococcus clades is low (15%), but host switches are possible and are part of the coevolution between hosts and viruses. Statistical analysis suggests that genetically similar OtVs have more similar infectivity patterns. The susceptibility of O. tauri strains also seems to be related to their outlier chromosome sizes, suggesting that chromosome rearrangements could be an adaptive process. Finally, putative specificity markers should be investigated in future studies, particularly with the identification of adhesin-like proteins and/or the presence of repeat motifs among proteins with rapid evolution.
ACKNOWLEDGMENTS
We thank the Genophy team in Banyuls-sur-Mer, France, Martha Clokie, and Ramon Massana for stimulating discussions.
This work was supported by an Agence Nationale de Recherche PICOVIR grant (coordinator, N.G.; grant no. BLAN07-1_200218). Camille Clerissi benefited from a doctoral fellowship from the AXA Research Fund.
Footnotes
Published ahead of print 8 February 2012
REFERENCES
- 1. Adam E, Perler FB. 2002. Development of a positive genetic selection system for inhibition of protein splicing using mycobacterial inteins in Escherichia coli DNA gyrase subunit A. J. Mol. Microbiol. Biotechnol. 4:479–488 [PubMed] [Google Scholar]
- 2. Agopian J, et al. 2009. Agricultural pesticide exposure and the molecular connection to lymphomagenesis. J. Exp. Med. 206:1473–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alliegro MC, Ettensohn CA, Burdsal CA, Erickson HP, McClay DR. 1988. Echinonectin: a new embryonic substrate adhesion protein. J. Cell Biol. 107:2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 5. Bellec L, Grimsley N, Moreau H, Desdevises Y. 2009. Phylogenetic analysis of new prasinoviruses (Phycodnaviridae) that infect the green unicellular algae Ostreococcus, Bathycoccus and Micromonas. Environ. Microbiol. Rep. 1:114–123 [DOI] [PubMed] [Google Scholar]
- 6. Bellec L, Grimsley N, Derelle E, Moreau H, Desdevises Y. 2010. Abundance, spatial distribution and genetic diversity of Ostreococcus tauri viruses in two different environments. Environ. Microbiol. Rep. 2:313–321 [DOI] [PubMed] [Google Scholar]
- 7. Bellec L, Grimsley N, Desdevises Y. 2010. Isolation of prasinoviruses of the green unicellular algae Ostreococcus spp. on a worldwide geographical scale. Appl. Environ. Microbiol. 76:96–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 94:1925–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen F, Suttle CA. 1995. Amplification of DNA polymerase gene fragments from viruses infecting microalgae. Appl. Environ. Microbiol. 61:1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen F, Suttle CA, Short SM. 1996. Genetic diversity in marine algal virus communities as revealed by sequence analysis of DNA polymerase genes. Appl. Environ. Microbiol. 62:2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen F, Suttle CA. 1996. Evolutionary relationships among large double-stranded DNA viruses that infect microalgae and other organisms as inferred from DNA polymerase genes. Virology 219:170–178 [DOI] [PubMed] [Google Scholar]
- 12. Coolen MJL. 2011. 7000 years of Emiliania huxleyi viruses in the Black Sea. Science 333:451–452 [DOI] [PubMed] [Google Scholar]
- 13. Courties C, et al. 1994. Smallest eukaryotic organism. Nature 370:255 [Google Scholar]
- 14. Derelle E, et al. 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. U. S. A. 103:11647–11652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Derelle E, et al. 2008. Life-cycle and genome of OtV5, a large DNA virus of the pelagic marine unicellular green alga Ostreococcus tauri. PLoS One 3:e2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dunigan DD, Fitzgerald LA, Van Etten JL. 2006. Phycodnaviruses: a peek at genetic diversity. Virus Res. 117:119–132 [DOI] [PubMed] [Google Scholar]
- 17. Flores CO, Meyer JR, Valverde S, Farr L, Weitz JS. 2011. Statistical structure of host-phage interactions. Proc. Natl. Acad. Sci. U. S. A. 108:E288–E297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gogarten JP, Senejani AG, Zhaxybayeva O, Olendzenski L, Hilario E. 2002. Inteins: structure, function, and evolution. Annu. Rev. Microbiol. 56:263–287 [DOI] [PubMed] [Google Scholar]
- 19. Gogarten JP, Hilario E. 2006. Inteins, introns, and homing endonucleases: recent revelations about the life cycle of parasitic genetic elements. BMC Evol. Biol. 6:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grimsley N, Pequin B, Bachy C, Moreau H, Piganeau G. 2010. Cryptic sex in the smallest eukaryotic marine green alga. Mol. Biol. Evol. 27:47–54 [DOI] [PubMed] [Google Scholar]
- 21. Guillou L, et al. 2004. Diversity of picoplanktonic prasinophytes assessed by direct nuclear SSU rDNA sequencing of environmental samples and novel isolates retrieved from oceanic and coastal marine ecosystems. Protist 155:193–214 [DOI] [PubMed] [Google Scholar]
- 22. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 23. Holm MM, Vanlerberg SL, Sledjeski DD, Lafontaine ER. 2003. The Hag protein of Moraxella catarrhalis strain O35E is associated with adherence to human lung and middle ear cells. Infect. Immun. 71:4977–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iyer LM, Aravind L, Koonin EV. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720–11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jancek S, Gourbiere S, Moreau H, Piganeau G. 2008. Clues about the genetic basis of adaptation emerge from comparing the proteomes of two Ostreococcus ecotypes (Chlorophyta, Prasinophyceae). Mol. Biol. Evol. 25:2293–2300 [DOI] [PubMed] [Google Scholar]
- 26. Khare V, Eckert KA. 2002. The proofreading 3′ → 5′ exonuclease activity of DNA polymerases: a kinetic barrier to translesion DNA synthesis. Mutat. Res. 510:45–54 [DOI] [PubMed] [Google Scholar]
- 27. King M. 1993. Species evolution: the role of chromosome change. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 28. Larsen PF, et al. 2007. Adaptive differences in gene expression in European flounder (Platichthys flesus). Mol. Ecol. 16:4674–4683 [DOI] [PubMed] [Google Scholar]
- 29. Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451. [DOI] [PubMed] [Google Scholar]
- 30. Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209. [PubMed] [Google Scholar]
- 31. Marin B, Melkonian M. 2010. Molecular phylogeny and classification of the Mamiellophyceae class. nov. (Chlorophyta) based on sequence comparisons of the nuclear- and plastid-encoded rRNA operons. Protist 161:304–336 [DOI] [PubMed] [Google Scholar]
- 32. Markine-Goriaynoff N, et al. 2004. Glycosyltransferases encoded by viruses. J. Gen. Virol. 85:2741–2754 [DOI] [PubMed] [Google Scholar]
- 33. Mizumoto H, Tomaru Y, Takao Y, Shirai Y, Nagasaki K. 2007. Intraspecies host specificity of a single-stranded RNA virus infecting a marine photosynthetic protist is determined at the early steps of infection. J. Virol. 81:1372–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moreau H, et al. 2010. Marine prasinovirus genomes show low evolutionary divergence and acquisition of protein metabolism genes by horizontal gene transfer. J. Virol. 84:12555–12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nagasaki K, Yamaguchi M. 1998. Intra-species host specificity of HaV (Heterosigma akashiwo virus) clones. Aquat. Microb. Ecol. 14:109–112 [Google Scholar]
- 36. Nagasaki K, et al. 2005a. Comparison of genome sequences of single-stranded RNA viruses infecting the bivalve-killing dinoflagellate Heterocapsa circularisquama. Appl. Environ. Microbiol. 71:8888–8894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nagasaki K, Shirai Y, Tomaru Y, Nishida K, Pietrokovski S. 2005b. Algal viruses with distinct intraspecies host specificities include identical intein elements. Appl. Environ. Microbiol. 71:3599–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Naor A, Lazary R, Barzel A, Papke RT, Gophna U. 2011. In vivo characterization of the homing endonuclease within the polB gene in the halophilic archaeon Haloferax volcanii. PLoS One 6:e15833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nielsen E, et al. 2009. Genomic signatures of local directional selection in a high gene flow marine organism; the Atlantic cod (Gadus morhua). BMC Evol. Biol. 9:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Not F, et al. 2004. A single species, Micromonas pusilla (Prasinophyceae), dominates the eukaryotic picoplankton in the western English Channel. Appl. Environ. Microbiol. 70:4064–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ogata H, Raoult D, Claverie JM. 2005. A new example of viral intein in mimivirus. Virol. J. 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Onimatsu H, Suganuma K, Uenoyama S, Yamada T. 2006. C-terminal repetitive motifs in Vp130 present at the unique vertex of the Chlorovirus capsid are essential for binding to the host Chlorella cell wall. Virology 353:433–442 [DOI] [PubMed] [Google Scholar]
- 43. Palenik B, et al. 2007. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. U. S. A. 104:7705–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paulus H. 2003. Inteins as targets for potential antimycobacterial drugs. Front. Biosci. 8:S1157–S1165 [DOI] [PubMed] [Google Scholar]
- 45. Paulus H. 2007. Protein splicing inhibitors as a new class of antimycobacterial agents. Drugs Future 32:973–984 [Google Scholar]
- 46. Pietrokovski S. 1998. Identification of a virus intein and a possible variation in the protein-splicing reaction. Curr. Biol. 8:R634–R638 [DOI] [PubMed] [Google Scholar]
- 47. Pietrokovski S. 1998. Modular organization of inteins and C-terminal autocatalytic domains. Protein Sci. 7:64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pietrokovski S. 2001. Intein spread and extinction in evolution. Trends Genet. 17:465–472 [DOI] [PubMed] [Google Scholar]
- 49. Piganeau G, Eyre-Walker A, Grimsley N, Moreau H. 2011. How and why DNA barcodes underestimate the diversity of microbial eukaryotes. PLoS One 6:e16342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Posada D. 2006. ModelTest server: a web-based tool for the statistical selection of models of nucleotide substitution online. Nucleic Acids Res. 34:W700–W703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Poullain V, Gandon S, Brockhurst MA, Buckling A, Hochberg ME. 2008. The evolution of specificity in evolving and coevolving antagonistic interactions between a bacteria and its phage. Evolution 62:1–11 [DOI] [PubMed] [Google Scholar]
- 52. Proctor LM, Fuhrman JA. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature 343:60–62 [Google Scholar]
- 53. Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 54. Rodriguez F, et al. 2005. Ecotype diversity in the marine picoeukaryote Ostreococcus (Chlorophyta, Prasinophyceae). Environ. Microbiol. 7:853–859 [DOI] [PubMed] [Google Scholar]
- 55. Saves I, et al. 2002. Investigating the endonuclease activity of four Pyrococcus abyssi inteins. Nucleic Acids Res. 30:4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schroeder DC, et al. 2009. Genomic analysis of the smallest giant virus—Feldmannia sp. virus 158. Virology 384:223–232 [DOI] [PubMed] [Google Scholar]
- 57. Short SM, Suttle CA. 2002. Sequence analysis of marine virus communities reveals that groups of related algal viruses are widely distributed in nature. Appl. Environ. Microbiol. 68:1290–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Starokadomskyy PL. 2007. Protein splicing. Mol. Biol. 41:278–293 [Google Scholar]
- 59. Suttle CA, Chan AM, Cottrell MT. 1990. Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347:467–469 [Google Scholar]
- 60. Suttle CA. 1994. The significance of viruses to mortality in aquatic microbial communities. Microb. Ecol. 28:237–243 [DOI] [PubMed] [Google Scholar]
- 61. Suttle CA. 2005. Viruses in the sea. Nature 437:356–361 [DOI] [PubMed] [Google Scholar]
- 62. Swithers K, Senejani A, Fournier G, Gogarten JP. 2009. Conservation of intron and intein insertion sites: implications for life histories of parasitic genetic elements. BMC Evol. Biol. 9:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Swofford D. 2003. PAUP*: phylogenetic analysis using parsimony, version 4.0b10. Sinauer Associates, Sunderland, MA [Google Scholar]
- 64. Thingstad TF, Lignell R. 1997. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat. Microb. Ecol. 13:19–27 [Google Scholar]
- 65. Thomas R, et al. 2011. Acquisition and maintenance of resistance to viruses in eukaryotic phytoplankton populations. Environ. Microbiol. 13:1412–1420 [DOI] [PubMed] [Google Scholar]
- 66. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Valle J, et al. 2008. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J. Bacteriol. 190:4147–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Van Valen L. 1973. A new evolutionary law. Evol. Theory 1:1–30 [Google Scholar]
- 69. Vellani TS, Myers RS. 2003. Bacteriophage SPP1 Chu is an alkaline exonuclease in the SynExo family of viral two-component recombinases. J. Bacteriol. 185:2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Voolstra CR, et al. 2011. Rapid evolution of coral proteins responsible for interaction with the environment. PLoS One 6:e20392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Weynberg KD, Allen MJ, Ashelford K, Scanlan DJ, Wilson WH. 2009. From small hosts come big viruses: the complete genome of a second Ostreococcus tauri virus, OtV-1. Environ. Microbiol. 11:2821–2839 [DOI] [PubMed] [Google Scholar]
- 72. Weynberg KD, Allen MJ, Gilg IC, Scanlan DJ, Wilson WH. 2011. Genome sequence of Ostreococcus tauri Virus OtV-2 throws light on the role of picoeukaryote niche separation in the ocean. J. Virol. 85:4520–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wilhelm SW, Suttle CA. 1999. Viruses and nutrient cycles in the sea. Bioscience 49:781–788 [Google Scholar]
- 74. Zhang L, Zheng Y, Callahan B, Belfort M, Liu Y. 2011. Cisplatin inhibits protein splicing, suggesting inteins as therapeutic targets in mycobacteria. J. Biol. Chem. 286:1277–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhu F, Massana R, Not F, Marie D, Vaulot D. 2005. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol. Ecol. 52:79–92 [DOI] [PubMed] [Google Scholar]