Abstract
The hepatitis C virus (HCV) RNA replicates in hepatic cells by forming a replication complex on the lipid raft (detergent-resistant membrane [DRM]). Replication complex formation requires various viral nonstructural (NS) proteins as well as host cellular proteins. In our previous study (C. K. Lai, K. S. Jeng, K. Machida, and M. M. Lai, J. Virol. 82:8838–8848, 2008), we found that a cellular protein, annexin A2 (Anxa2), interacts with NS3/NS4A. Since NS3/NS4A is a membranous protein and Anxa2 is known as a lipid raft-associated scaffold protein, we postulate that Anxa2 helps in the formation of the HCV replication complex on the lipid raft. Further studies showed that Anxa2 was localized at the HCV-induced membranous web and interacted with NS4B, NS5A, and NS5B and colocalized with them in the perinuclear region. The silencing of Anxa2 decreased the formation of membranous web-like structures and viral RNA replication. Subcellular fractionation and bimolecular fluorescence complementation analysis revealed that Anxa2 was partially associated with HCV at the lipid raft enriched with phosphatidylinositol-4-phosphate (PI4P) and caveolin-2. Further, the overexpression of Anxa2 in HCV-nonsusceptible HEK293 cells caused the enrichment of HCV NS proteins in the DRM fraction and increased the colony-forming ability of the HCV replicon. Since Anxa2 is known to induce the formation of the lipid raft microdomain, we propose that Anxa2 recruits HCV NS proteins and enriches them on the lipid raft to form the HCV replication complex.
INTRODUCTION
Lipid rafts are defined as detergent-insoluble cholesterol- and sphingolipid-rich microdomains of cellular membranes that play important roles in signal transduction (54), protein sorting (30), and the budding and assembly of enveloped viruses (7, 37, 42, 51). Our previous studies have demonstrated the involvement of lipid rafts in the formation of the hepatitis C virus (HCV) RNA replication complex (RC) (1, 2, 15, 52).
HCV is the causative agent of hepatitis C, which afflicts more than 170 million people worldwide. Hepatitis C is characterized by an acute, often asymptomatic, onset of infection that proceeds to persistent HCV infection in 70% of cases. HCV is the cause of 27% of liver cirrhosis cases and 25% of cases of hepatocellular carcinoma (3). The virus belongs to the genus Hepacivirus in the family Flaviviridae. The enveloped virus contains a positive-sense, single-stranded, 9.6-kb RNA genome, which is composed of a 5′-untranslated region (UTR), including an internal ribosome entry site (IRES), an open reading frame (ORF) that encodes 10 different structural and nonstructural (NS) proteins (NSPs), and the terminal 3′-UTR. The ORF is cleaved in a mechanistic manner by cellular and viral proteases into structural proteins, including core and enveloped glycoproteins E1 and E2, which together form the viral particle, and NSPs, including the ion channel protein p7, the protease NS2-3, the serine protease and RNA helicase NS3, its cofactor, NS4A, the multifunctional NS4B and NS5A, and the RNA-dependent RNA polymerase (RdRp) NS5B protein. The NS3 to NS5B proteins are hypothesized to be involved in HCV RNA replication. The NS4B protein induces the formation of a specific membrane structure known as the membranous web, which serves as a scaffold for the HCV RC (12).
The mechanism of membranous web biogenesis is hypothesized to be similar to that of the cellular membrane formation. In this process, the HCV proteins are supposedly recruited to these membranous web structures from the endoplasmic reticulum (ER). HCV NS3 is the only soluble protein among all of the NSPs, and its serine protease and RNA helicase activities are crucial to HCV RNA replication. The HCV NS4A protein serves as a cofactor for the NS3 serine protease, and its N-terminal end assists in the anchoring of the NS3-4A complex to the membrane (59). This raises the important question of how the NS3-4A protein is recruited to the replication complex. As viruses rely on the host cell infrastructure for all processes of their life cycle, cellular factors interacting with HCV NSPs may play important roles.
In our previous study, we utilized an immunoprecipitation-proteomics approach to identify target molecules that are associated with NS3/NS4A (32). From several candidate proteins, we initially selected annexin A2 (Anxa2) for further study. Anxa2 is also known as annexin II, calpactin 1, chromobindin 8, lipocortin II, and placental anticoagulant protein IV. Anxa2 belongs to the structurally related calcium- and phospholipid-binding family of proteins known as the annexins, and they are highly expressed in eukaryotic cells. They mediate essential cellular processes, including membrane-trafficking events, mitotic signaling, and cytoskeleton rearrangements (16, 17, 40, 41, 46). Annexins also organize specific membrane microdomains wherein they recruit their interaction partners and thus regulate the formation of compartment-specific signaling platforms (14, 16, 17, 20, 21, 25, 49). These dynamic functions of Anxa2 are executed by influencing the composition and aggregation of membrane proteins. Anxa2 also influences the lateral association and stability of cholesterol-rich microdomains (4, 5, 10) and induces the formation of phosphatidylinositol (4,5)-bisphosphate (PIP2) clusters within the lipid raft (18). This association had been implicated in the HIV-1 viral assembly at the lipid rafts (23). PIP2 also colocalizes with HCV RC (6). Further, Anxa2 has been isolated from the HCV replication complex and identified as an associated host factor (6), and it has been shown to be recruited by NS5A to the HCV replication site. Paradoxically, it was found that Anxa2 was not involved in HCV RNA replication but instead was probably involved in virus assembly (6).
In this study, we show that as a component of the HCV RNA RC, Anxa2 is a regulator of HCV RNA replication. Further, Anxa2 may function as a scaffold protein by interacting with the HCV NSPs NS3 to NS5B and may induce the production of phosphatidylinositol-4-phosphate (PI4P), which is critical for the assembly of HCV RC (26, 47). In addition, we found that Anxa2 is localized to and promotes the structural organization of the membranous web. Finally, the knockdown of Anxa2 suppressed HCV RNA replication, whereas its overexpression allowed HCV to replicate in nonsusceptible HEK293 cells. Thus, Anxa2 is a key cellular protein that facilitates the formation of the viral RNA replication complex.
MATERIALS AND METHODS
Cell culture.
Huh7.5 and HEK293 cells were cultured in complete Dulbecco's modified Eagle's medium (C-DMEM) supplemented with 10% fetal bovine serum, nonessential amino acids, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a 5% CO2 incubator. The HCV subgenomic replicon, HCV-EV71I-Luc, Rep1.1, and HCVrep-HA replicon (11) were derived from HCV N strain 1bneo/delS (genotype 1b) (22) and were maintained in the same medium supplemented with G418 (0.5 mg/ml). Rep1.1 cells were treated with 100 U/ml of gamma interferon for 2 weeks to generate cured Rep 1.1 cells. The cured replicon cells did not form colonies when selected under G418 pressure.
Generation of inducible HEK293 cells.
HEK293 cells were first infected with lentiviruses containing Tet-aOn and Tet-aOFF (a transactivator of the tetracycline inducible system) by following the instructions of the manufacturer (National RNAi Core Facility, Academia Sinica, Taiwan). Infected cells, termed HEK293-TetON and HEK293-TetOff, were selected with blasticidin at a concentration of 5 μg/ml for 1 week. A lentivirus encoding Anxa2 was prepared using pAS4w-Anxa2-Puro vector, as described below and by following the instructions of the manufacturer (National RNAi Core Facility, Academia Sinica, Taiwan). HEK293-TetON and -TetOFF cells were infected with a lentivirus carrying the Anxa2 gene, resulting in tetracycline-regulated TRAnxa2-ON and -OFF cell lines, which were then selected under blasticidin (5 μg/ml) and puromycin (3 μg/ml) pressure for 1 week. The selected cells were first expanded in the absence of selection pressure and subsequently cultured in C-DMEM in the presence of blasticidin and puromycin. They were then tested for Anxa2 expression in response to doxycycline (BD Biosciences) by Western blotting. The expression of Anxa2 was activated (in TRAnxa2-ON cells) or suppressed (in TRAnxa2-OFF cells) by treatment with doxycycline (0.1 μg/ml).
Plasmids and viruses.
The plasmids containing HCV 1b subgenomic replicon, Rep (wild type [wt]), pUC-Rep/S1179I, and Rep/GDD, which had a mutation in the catalytic motif of NS5B (GDD→AAA), have been described previously (35). The plasmids expressing the HCV proteins, pCI-HA-NS3/4A, pCI-NS5A-HA, and pCI-HA-GST, have also been described previously (11, 31, 32). The plasmids expressing other hemagglutinin (HA) epitope-tagged HCV proteins were constructed by inserting HCV core, E1E2, NS3, NS4B, and NS5BΔTM (without the C-terminal 21 amino acids) cDNA of genotype 1b into pUI (pUI-HA-Core, pUI-HA-E1E2, and pUI-HA-NS4B), pCI (pCI-HA-NS3), and pCAG (pCAG-HA-NS5BΔTM) vector backbones. The pUI vector was derived from pCI (Promega) by the replacement of the cytomegalovirus (CMV) promoter with a ubiquitin promoter; the pCAG vector described by Niwa et al. (43) was modified by removing the neomycin resistance gene under the control of the thymidine kinase promoter and inserting a multiple cloning site (MCS) to facilitate cloning. To generate the pUI-Anxa2 vector expressing the Anxa2 protein, total cellular RNA was first amplified using an oligo(dT) primer, and the cDNA was further amplified using the following Anxa2-specific primers: forward primer 5′-ctg aac GCT AGC atg ggc cgc cag cta gcg-3′ and reverse primer 5′-agc ttt GTT TAA Act cag tca tct cca cca cac ag-3′, where the capital letters represent NheI and PmeI restriction enzyme (RE) sites in the forward and reverse primers, respectively. The PCR-amplified DNA then was digested with restriction enzymes NheI and PmeI (New England BioLabs) and cloned into pAS4w.1.pPuro vectors to generate a pAS4w-Anxa2-Puro vector, which was used to express the Anxa2 gene under the Tet-responsive element tight promoter.
The bimolecular fluorescence complementation (BiFC) constructs were based on the pSAT plant vectors described by Lee et al. (36). The BiFC cassette in pSAT vectors is comprised of an autofluorescent protein fragment (cyan fluorescent protein [CFP], Venus, or Cerulean) followed or preceded by an MCS (to generate an N- or C-terminal fusion protein) and a terminal sequence. The BiFC cassette from pSAT vectors was amplified by PCR and cloned between NheI and NotI RE sites in the pCAG plasmid vector. In this manner six vectors were generated, namely, pCAG-cCFP-C or pCAG-cCFP-N, pCAG-nVenus-C or pCAG-nVenus-N, and pCAG-nCerulean-C or pCAG-nCerulean-N, where N and C indicate that the protein of interest is fused to the N or C terminus, respectively, of the autofluorescent protein fragment. The NS5A gene was amplified from the genomic cDNA of HCV 1b, Anxa2 and glutathione S-transferase (GST) were amplified as described above, and Cav2 was a gift from Song-Kun Shyue, Institute of Biomedical Sciences, Academia Sinica. These genes were amplified by PCR and cloned in frame at the EcoRI-KpnI RE site of pCAG BiFC vectors; this strategy resulted in the generation of BiFC vectors with the autofluorescent protein fused to the gene of interest at its N or C terminus. The vectors were named accordingly, i.e., cCFP-NS5A (having NS5A at the C terminus of the autofluorescent protein) and NS5A-cCFP (having NS5A at the N terminus of the autofluorescent protein). Similarly, we named other vectors, i.e., nCerulean-Anxa2, Anxa2-nCerulean, nVenus-Caveolin2, Caveolin2-nVenus, cCFP-GST, and GST-cCFP. All of the clones were verified by DNA sequencing. The HCV JC1 viruses were produced and titrated in Huh7.5 cells based on a previously described method (32).
Antibodies.
The mouse monoclonal antibody (MAb) for JC1 NS5A detection was obtained from Austral Biologicals, whereas that for the genotype 1b replicon NS5A was purchased from Biodesign (Saco, ME). The mouse MAb against the HCV core protein was purchased from Affinity Bioreagents, Inc. (Golden, CO). The rabbit polyclonal antibody against HCV NS4B (RR12) was a kind gift from Michinori Kohara (The Tokyo Metropolitan Institute of Medical Science, Japan). The mouse MAb against Anxa2 used for immunoblotting and immunoprecipitation analysis was obtained from BD Transduction Laboratories; the mouse MAb used for immunofluorescence analysis was a kind gift from Tony Hunter (Molecular and Cellular Biology Laboratory, The Salk Institute for Biological Studies). The rabbit polyclonal antibody against Anxa2, which was used for immunofluorescence analysis, was purchased from Abcam. The mouse MAb against PI4P was purchased from Echelon. Anti-actin mouse MAb was obtained from Santa Cruz Biotechnology, Inc. The mouse MAb for flotillin-1 (Flot1)was purchased from BD Transduction Laboratories. The mouse MAb against vimentin was purchased from Chemicon (Temecula, CA). Rat MAb against the HA epitope was purchased from Roche Diagnostics. Alexa Fluor 488-conjugated anti-mouse, Alexa Fluor 568-conjugated anti-mouse, and anti-rabbit secondary antibodies all were purchased from Invitrogen Molecular Probes.
siRNAs.
Nonspecific control, Anxa2-targeting short interfering RNA (siRNA) and HCV 5′-UTR-targeting siRNAs were purchased from Dharmacon (Thermo Fisher Scientific, Lafayette, CO). ON-TARGETplus, the nontargeting siRNA pool (D-001818-10-20), is referred to as siCtrl RNA in this study. The ON-TARGETplus SMARTpool (L-010741-00-0005) human ANXA2 siRNA, referred to as siAnxa2 RNA in this study, contains a pool of 4 siRNAs (J-010741-07 to J-010741-10). Their sequences are 5′-CGA CGA GGA CUC UCU CAU U-3′, 5′-AUC CAA GUG UCG CUA UUU A-3′, 5′-AAA ACC AGC UUG CGA AUA A-3′, and 5′-GGA AGA AAG CUC UGG GAC U-3′. The HCV siRNA 5′-AGG UCU CGU AGA CCG UGC ATT-3′ targets the 5′-UTR of the HCV genome and inhibits the replication of HCV genotypes 1b and 2a efficiently (6). The siRNA was transfected into cells using DMRIE-C reagent (Invitrogen) per the manufacturer's instructions at a 100 nM final concentration.
Immunofluorescence staining.
The immunofluorescence staining and analysis was performed as described previously (32). For BiFC detection, transfected cells were fixed, followed by permeabilization and blocking. Cells then were incubated with 4′,6-diamidino-2-phenylindole (DAPI) in 5% bovine serum albumin (BSA) for 1 h at 37°C. For combining BiFC with immunostaining, transfected cells were processed for antibody staining as described previously (32). The BiFC signals were observed at 488 nm for the Venus track and at 458 nm for the Cerulean track.
Coimmunoprecipitation.
To test whether endogenous Anxa2 could coimmunoprecipitate HCV proteins, Huh7.5 cells were transfected with plasmid vectors expressing viral proteins or GST with Lipofectamine 2000 transfection reagent (Invitrogen) according to the instructions provided by the manufacturer. Transfected cells were harvested, and the Anxa2 protein was immunoprecipitated from the cell lysate using anti-Anxa2 mouse MAb by following the procedure described previously (15). The immunoprecipitated proteins were separated on a 10% SDS-PAGE gel, and immunoblot analysis was performed for Anxa2, HA-tagged HCV proteins, and GST with respective antibodies as described above.
DRM fractionation assay.
The naïve or transfected Huh7.5, TRAnxa2-OFF or TRAnxa2-ON, and HCVRep-HA replicon cells were cultured in 10-cm dishes and harvested at 48 h posttransfection or 24 h postseeding, respectively. The harvested cells were washed twice with phosphate-buffered saline (PBS) and then subjected to detergent-resistant membrane (DRM) fraction isolation exactly as described by Weaver et al. (58). All steps were conducted on ice or at 4°C. Briefly, following washing, the cells were resuspended in 500 μl PBS with protease inhibitor and were homogenized using a Dounce homogenizer. The total protein content was estimated by Bradford method (Bio-Rad), and equal amounts of protein were brought to a total volume of 1 ml in PBS and centrifuged at 5,000 rpm for 1 h. The pellets were resuspended in TNE buffer (25 mM Tris-HCl [pH 7.6], 150 mM NaCl, 5 mM EDTA) with or without 1% Triton X-100 and subjected to low-speed mixing in an Elmi Intelli-Mixer RM-2 for 20 min. The reaction mixture then was centrifuged at 5,000 rpm for 30 min. The pellet was resuspended in 0.5 ml 40% OptiPrep (Sigma-Aldrich) solution (60% OptiPrep diluted in TNE buffer) and transferred to an ultracentrifuge tube (Beckman). This was overlaid by 3.5 ml 30% OptiPrep solution, and then 0.5 ml 5% OptiPrep solution was layered on top of that. The weight-balanced tubes were spun in a Beckman ultracentrifuge using an SW60Ti swinging bucket rotor at 36,000 rpm for 16 h at 4°C. Following centrifugation, 0.5-ml samples were collected from top to bottom and numbered fractions 1 to 9. These fractions then were measured by the Bradford method (Bio-Rad) for their total protein content, and equal amounts of protein from each fraction were analyzed by 10% SDS-PAGE followed by Western blotting.
In vitro replicase activity assay.
To analyze the effect of siRNA on HCV in vitro replicase activity, the HCVrep-HA replicon cells were seeded in 10-cm dishes and siRNA was transfected. On the sixth day posttransfection, cells were harvested and crude replication complex was isolated. Replicase activity was analyzed on an agarose gel as described by Yang et al. (60). Briefly, cells were washed with PBS and scraped in cold hypotonic buffer (10 mM Tris-HCl [pH 7.5] and 10 mM NaCl). Following incubation for 15 to 20 min on ice, cells were homogenized using a Wheaton Dounce homogenizer, and the lysate was spun down at low speed. The supernatant then was centrifuged at high speed for 20 min at 4°C, and the pellet was resuspended in storage buffer (hypotonic buffer supplemented with 15% glycerol). The resuspended pellet was termed crude replication complex (CRC), which was utilized for in vitro replication in a 60-μl reaction mixture (50 mM HEPES [pH 7.3]; 10 mM KCl; 10 mM MgCl2; 1 U/μl RNase inhibitor; 10 μg/ml actinomycin D; 0.5 mM [each] ATP, GTP, and CTP; 0.2 μM [α-32P]UTP; 0.3 mM MnCl2; 6 μl CRCs; and water to a volume of 60 μl). The reaction mixture was incubated at 30°C for 1 to 2 h. Subsequently, RNA was precipitated by the chloroform-isopropanol-ethanol method and analyzed by nondenaturing gel electrophoresis. The pUC-Rep/S1179I replicon RNA, used as a size marker, was synthesized in vitro with [α-32P]UTP (Perkin-Elmer, NEN Life Science Products, Boston, MA) using the MEGAscript (Ambion) kit by following the manufacturer's instructions.
Protease protection assay.
For protease protection experiments, HCV-EV71I-Luc replicon cells were seeded in 10-cm dishes. Following incubation for 24 h, cells were washed twice with cold PBS, resuspended in ice-cold hypotonic buffer (10 mM Tris-HCl [pH 7.5] and 10 mM NaCl), and incubated for 10 min at 4°C. Cells then were disrupted using a Dounce homogenizer, and the homogenized cell lysate was centrifuged two times briefly for 5 min each at 1,000 × g to remove the nuclei, large debris, and any remaining intact cells. The resulting postnuclear supernatant (PNS) was incubated at 4°C in the absence or presence of 0.5% Triton X-100 for 20 min. The detergent-treated or untreated PNS was incubated for 10 min at 20°C with 20 μg/ml of proteinase K. Thereafter, proteinase K was inactivated by the addition of 2 mM phenylmethylsulfonyl fluoride (PMSF) and fractionated into a 10,000 × g pellet (P) and supernatant (S), and samples were analyzed by Western blotting.
Quantitative detection of HCV and Anxa2 RNA by qRT-PCR.
Quantitative reverse transcription-PCR (qRT-PCR) was performed as described previously (11). The primers for Anxa2 were 5′-CCT GCT CAG TAT GAC GCT TCT-3′ (sense) and 5′-TCT GGA GCA GAT GAT CTC AAT G-3′ (antisense) with universal probe 23 (Roche). The primers for HCV were 5′-CAT GGC GTT AGT ATG AGT GTC G-3′ (sense) and 5′-GGT TCC GCA GAC CAC TAT G-3′ (antisense) with universal probe 75. As an internal control, we amplified human GAPDH (glyceraldehyde 3-phosphate dehydrogenase) with primers 5′-AGC CAC ATC GCT CAG ACA C-3′ (sense) and 5′-GCC CAA TAC GAC CAA ATC C-3′ (antisense) with universal probe 60.
Electron microscopy.
Huh7.5 cells were seeded in 6-well plates and transfected with siRNA the next day. Two days posttransfection, cells were subcultured and infected with HCV JC1 virus at a multiplicity of infection (MOI) of 2 for 3 h. The cells were washed with PBS and incubated in DMEM, as described above, for another 2 days. Cells then were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 30 min and washed in cacodylate buffer, postfixed with 1% osmium tetraoxide for 1 h, washed with water, and then stained with 1% uranyl acetate for 1 h. After staining, cells were washed with water, followed by dehydration in a graded ethanol series and embedding in resin, which was allowed to polymerize for 14 h at 70°C. Ultrathin sections were cut on an ultramicrotome and collected on copper grids. The grids then were observed with a Tecnai Spirit transmission electron microscope (FEI Co.).
The immunoelectron microscopy (IEM) was performed exactly as described previously (32). The ultrathin sections (100 nm) were first incubated with an anti-NS5A mouse MAb and/or rabbit polyclonal anti-Anxa2, followed by incubation with colloidal gold particles of 12 and 6 nm conjugated to anti-mouse and anti-rabbit immunoglobulin G, respectively. Samples were analyzed under a Tecnai Spirit transmission electron microscope at 120 kV.
RNA synthesis, transfection, and HCV colony formation assay.
The in vitro synthesis of HCV RNA from linearized HCV replicon-containing plasmid and electroporation were performed by the methods described previously by Wakita et al. (57), with minor modifications. Cells were mixed with in vitro-transcribed RNA, and the total amount of RNA was adjusted to 20 μg with total RNA isolated from naïve Huh7.5 cells using TRIzol (Invitrogen) reagent by following the manufacturer's instructions. The cells and RNA mixtures were pulsed at 220 V and 975 μF using a BTX ECM630 electroporator. Cells were cultured in DMEM supplemented with 0.5 mg G418/ml for 12 to 15 days; medium was replaced every third day. Finally, the colonies were stained with crystal violet and counted manually.
Statistical analysis.
The statistical analyses for qRT-PCR data were performed using Microsoft Excel software. The graphs represent the means ± standard deviations. P values were determined using Student's t distribution test with one tail. P <0.05 was considered statistically significant.
RESULTS
Anxa2 is involved in HCV RNA replication.
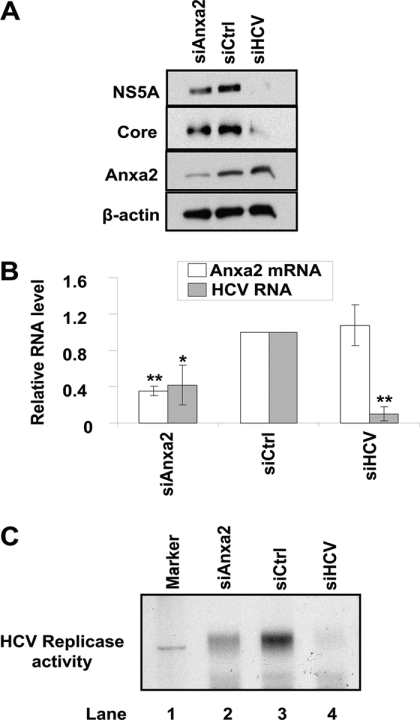
Previously, we paired tandem mass spectrometry (MS/MS) analysis with the MASCOT program to identify proteins that copurified with HA-tagged NS3/NS4A (32). In this study, we focused on the function of the scaffold protein Anxa2, which assists in the organization of membrane microdomains (14, 16, 17, 20, 21, 25, 49). We postulated that Anxa2 functions as a scaffold protein to help build the HCV RC. To test this hypothesis, we analyzed the effect of the siRNA-mediated knockdown of Anxa2 on HCV RNA replication in HCVrep-HA SGR cells or Huh7.5 cells transfected with siRNA followed by infection with JC1 virus. An siRNA directed against the HCV genome, siHCV, served as a positive control, and a nontargeting siRNA, siCtrl, served as a negative control. The results showed that siHCV reduced the production of HCV proteins (Fig. 1A) and RNA (Fig. 1B) to almost undetectable amounts without affecting Anxa2 expression (Fig. 1A and B). Under the same conditions, the siRNA targeting Anxa2 reduced the amount of Anxa2 protein (Fig. 1A) and Anxa2 mRNA (Fig. 1B) by 65% (P = 0.001085). Correspondingly, the amounts of NS5A and core proteins (Fig. 1A) and HCV RNA (Fig. 1B) were reduced by 59% (P = 0.022003). The same experiment was performed using genotype 1b SGR cells, and similar results were obtained (data not shown). To ascertain that HCV RNA replication was directly affected by Anxa2 siRNA, we performed an in vitro HCV replicase activity assay using the endogenous RNA template described by Yang et al. (60). The results showed that the siRNA against Anxa2 significantly reduced the amount of in vitro viral RNA synthesis (Fig. 1C). Thus, we speculate that Anxa2 is involved directly in HCV RNA replication. Furthermore, since we used both genotype 1b and 2a HCV constructs for our assays, this conclusion is not limited to a certain genotype.
Fig 1.
Effect of siRNA-mediated knockdown of Anxa2 on HCV replication. Naïve Huh7.5 (A and B) or HCVrep-HA (C) cells were transfected with the indicated siRNA. siRNA-transfected Huh7.5 cells then were infected with HCV JC1 virus. At 3 days after infection, cells were harvested for immunoblotting and the qRT-PCR determination of RNA. (A) Cell lysates were subjected to 10% SDS-PAGE, followed by Western blotting using anti-NS5A, anti-core, anti-Anxa2, or anti-β-actin antibody. (B) Quantitative RT-PCR of Anxa2 and HCV RNA. The RNA quantity was normalized against GAPDH RNA. Anxa2 and HCV RNA level in cells treated with different siRNA relative to those in siCtrl-treated cells are shown. Representative data (means ± standard deviations) from three independent experiments are shown. *, P < 0.05; **, P < 0.01. (C) In vitro RNA replication of siRNA-transfected HCVrep-HA cells. At 6 days after siRNA transfection, crude replication complex was harvested and analyzed for in vitro replicase activity using [α-32P]UTP as the label as described in Materials and Methods. In vitro-transcribed labeled RNA from the parental replicon pUC-Rep/S1179I served as the RNA size marker (lane 1).
Anxa2 is incorporated into the lipid raft-associated HCV RC.
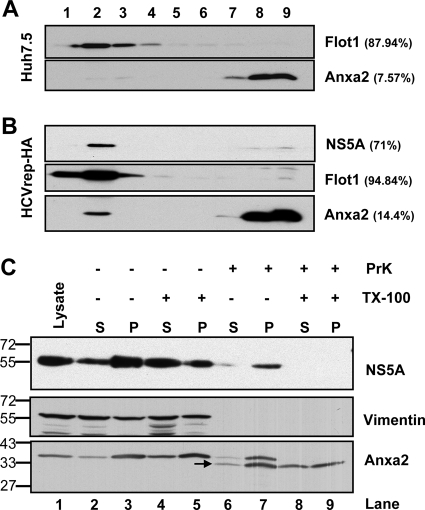
HCV RNA replicates on the membranous web-like structure consisting of detergent-resistant, cholesterol-rich lipid raft membrane domains (1, 2, 15, 52). To determine whether Anxa2 and HCV RC protein components both are present in these membrane domains, we performed the membrane flotation ultracentrifugation of the cellular membrane from HCV SGR cells after treatment with Triton X-100 (1%) at 4°C, which should have dissolved most of the membrane except the DRM. As expected, all of flotillin-1 (Flot1) was fractionated in fractions 1 to 4, which is indicative of DRM (Fig. 2A). In naïve Huh7.5 cells, 7.5% of the total amounts of Anxa2 fractionated in the DRM fraction (Fig. 2A, upper). In comparison, in SGR-harboring Huh7.5 cells, 14.4% of the Anxa2 amount was detected in the DRM fraction (Fig. 2B). Similar to previous studies (2, 15, 52), most of the NS5A was also fractionated in the DRM. These results indicate that the presence of HCV replicon increased the localization of Anxa2 in the DRM, suggesting that Anxa2 is associated with the HCV RC on the DRM.
Fig 2.
Association of Anxa2 protein with detergent-resistant membrane fractions. (A) Huh7.5 naïve cells and (B) HCVrep-HA replicon cells were harvested at 24 h after seeding. Cell lysates were subjected to 1% Triton X-100 treatment and fractionated by discontinuous OptiPrep gradient centrifugation as described in Materials and Methods. Fractions were analyzed on a 10% SDS-PAGE gel, followed by immunoblotting with monoclonal antibodies against Anxa2, Flot1, and NS5A. Fractions are numbered from 1 to 9 in order from top to bottom (light to heavy). Results of Western blotting were quantified by PhosphorImager counting. The percentage of each protein in the DRM fraction (lanes 1 to 4) relative to the total signal intensity of the protein in all 9 fractions is indicated in parentheses. (C) Protection of HCV NSPs and Anxa2 by membranes against protease digestion. The postnuclear supernatant (PNS) from HCV-EV71I-Luc replicon cells were loaded as such (lane 1) or treated with 0.5% Triton X-100 (lanes 4 and 5 and lanes 8 and 9) or left untreated (lanes 2 and 3 and lanes 6 and 7), and this was followed by no digestion (lane 2 to 5) or treatment with proteinase K (lane 6 to 9). Thus, treated PNSs were fractionated into a 10,000 × g pellet (P) and supernatant (S) and analyzed for the presence of HCV NS5A, vimentin, and Anxa2. The positions of protein size markers are indicated on the left, and the arrow indicates the degraded form of Anxa2.
To confirm the association of Anxa2 with the HCV RC, we examined whether Anxa2 is protected by membranes in HCV SGR cells. The cell lysates from the HCV SGR cells were separated into pellet (membrane) and supernatant fractions and tested for their susceptibility to proteinase K in the presence or absence of Triton X-100 (0.5% at 4°C) (Fig. 2C). HCV NS5A in the pellet was partially resistant to proteinase K digestion (Fig. 2C, compare lane 7 to lane 3), in agreement with the notion that NS5A is contained within the membrane structure. NS5A was completely digested by proteinase K after treatment with Triton X-100 (lane 9). Similarly, Anxa2 was partially resistant to proteinase K digestion (lane 7). The lower band of 34 kDa (Fig. 2C, lane 6 to 9) is a digested product of Anxa2. Similar to NS5A, Anxa2 was completely digested by proteinase K after the membrane was treated with Triton X-100. In contrast, vimentin, a soluble protein, was completely digested even without the Triton X-100 treatment. These observations suggest that HCV NS5A and Anxa2 were enclosed and protected by membranes.
Taken together, these results suggest that Anxa2 is associated with the HCV RC that is assembled on the lipid raft.
Anxa2 localizes to the PI4P lipid microenvironment in HCV-infected cells.
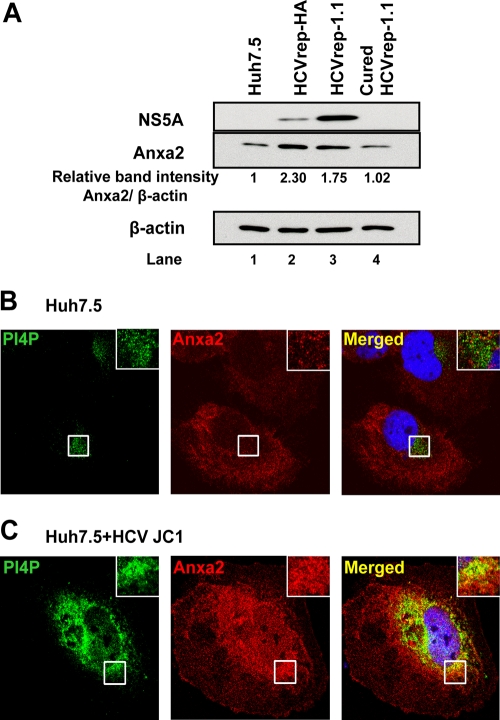
The phosphatidylinositol-4-phosphate (PI4P)-rich lipid microenvironment is required for HCV RNA replication (26, 47). Following HCV infection PI4P is increased, and its subcellular localization is close to the sites of HCV RNA replication (26, 47). We observed that the amount of Anxa2 was also increased by almost 2-fold in two independently derived SGR-harboring Huh7.5 cells (HCVrep-HA and HCVrep-1.1) compared to those in the naïve Huh7.5 cells and in the replicon cells cured with interferon treatment (Fig. 3A, compare lanes 2 and 3 to lanes 1 and 4). Further, the immunofluorescence studies showed that in the naïve Huh7.5 cells, PI4P and Anxa2 did not colocalize (Fig. 3B, inset), whereas in the HCV-JC1-infected cells they were significantly colocalized, particularly in the perinuclear region (Fig. 3C, inset). Both PI4P and Anxa2 also appeared to be more abundant in HCV-infected cells as judged by the intensity of immunofluorescence (Fig. 3B and C). These observations are consistent with our hypothesis that Anxa2, along with PI4P, is partially sequestered to the HCV RC.
Fig 3.
Anxa2 colocalized with the PI4P-rich lipid environment in HCV-infected cells. (A) Huh7.5 naïve, HCVrep-HA, HCVrep-1.1, and cured HCVrep-1.1 cells were harvested at 24 h after seeding, and equal amounts of total protein content were subjected to 10% SDS-PAGE followed by Western blotting with anti-NS5A, anti-Anxa2, and anti-β-actin antibodies. Results of Western blotting were quantified by PhosphorImager counting. Relative band intensities of Anxa2 compared to that of Huh7.5 naïve cells as a ratio of Anxa2 and β-actin are shown. Huh7.5 naïve cells (B) and HCV-JC1-infected cells (C) were costained with an anti-Anxa2 rabbit polyclonal antibody (red) and a PI4P mouse monoclonal antibody (green). Nuclear DNA was stained with DAPI (blue). Images shown were collected sequentially with a confocal laser-scanning microscope and merged to demonstrate colocalization (yellow merge fluorescence). Enlargements of the sections, indicated by the white squares, are shown in the inset panels.
Anxa2 interacts with HCV NS3-NS5B proteins.
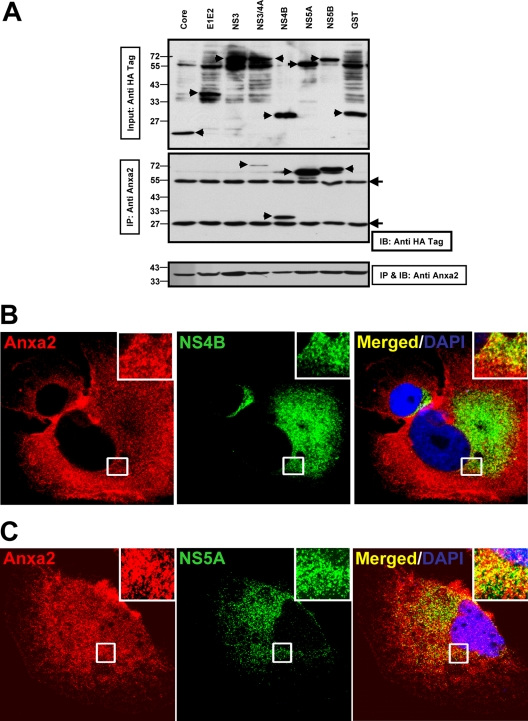
We next investigated whether any viral proteins interact with Anxa2 and thereby affect its distribution. Huh7.5 cells were transfected with plasmid vectors encoding HA-tagged HCV core, E1E2, NS3, NS3/NS4A, NS4B, NS5A, NS5BΔTM (lacking 21 amino acids at the C terminus that form the transmembrane domain), and GST proteins. The proteins were precipitated by monoclonal anti-Anxa2 antibody and analyzed by immunoblotting using an antibody against the HA epitope and a monoclonal anti-Anxa2 antibody. The results showed that Anxa2 specifically coprecipitated NS3/NS4A, NS4B, NS5A, and NS5BΔTM but not core, E1E2, NS3, or GST protein, although the amount of proteins coprecipitated varied (Fig. 4A). These results indicate that Anxa2 interacts with several HCV NSPs but not with the soluble form of NS3 protein. This finding is consistent with the fact that Anxa2 was detected as a binding partner to NS3/4A (32). Furthermore, the immunofluorescence data showed that NS4B (Fig. 4B, inset) and NS5A (Fig. 4C, inset) were colocalized with Anxa2 in the perinuclear region. Taken together, these results suggest that Anxa2 interacts with components of the HCV RC.
Fig 4.
Anxa2 interacts with HCV nonstructural proteins. (A) Huh7.5 cells were transfected with plasmids encoding HA-tagged HCV NS proteins or GST. At 48 h posttransfection, cell lysates were immunoprecipitated (IP) using anti-Anxa2 monoclonal antibodies. The immunoprecipitates and 5% of the total cell lysates were subjected to 10% SDS-PAGE followed by Western blotting with anti-HA and anti-Anxa2 antibodies. (Upper panel) Anti-HA immunoblot showing the input proteins (arrows) (5% total). (Middle panel) Anti-HA immunoblot showing HCV NSP coimmunoprecipitated by anti-Anxa2 antibody. (Lower panel) Anxa2 protein immunoprecipitated by anti-Anxa2 antibody. Protein molecular mass markers are indicated on the left side. Filled arrowheads indicate the IgG heavy (55-kDa) and light (25-kDa) chains. (B and C) Colocalization of NS4B (B) and NS5A (C) with Anxa2 in HCV-JC1-infected Huh7.5 cells. Images shown were collected sequentially with a confocal laser-scanning microscope and merged to demonstrate colocalization. Enlargements of the sections, from different areas of the cells, indicated by the white squares are shown in the inset panels.
Anxa2 is associated with NS5A at HCV-induced membranous web structures.
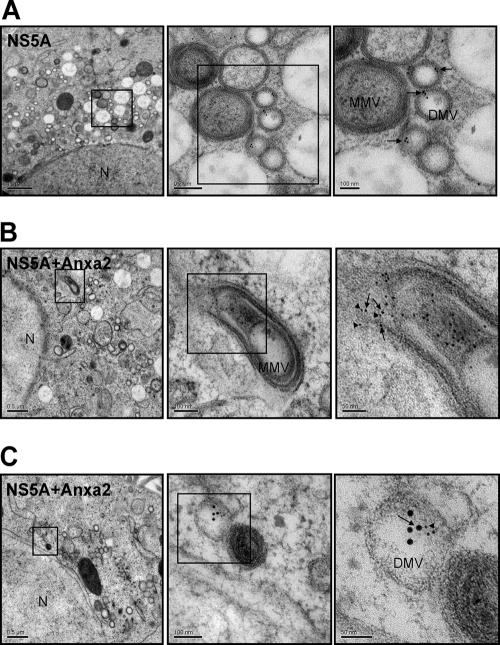
The HCV RC is assembled on the membranous web-like structure. Therefore, we investigated whether Anxa2 and NS5A were colocalized on the membranous web. We first used IEM to localize NS5A in HCV JC1-infected Huh7.5 cells. The NS5A gold particles were detected mainly in the interior surface of double-membrane vesicles (DMVs) (Fig. 5A). Since NS5A is a component of the HCV RNA replication complex, the localization of NS5A suggests that the lumen of the DMV is the site of HCV RNA replication. We next performed double IEM to localize both NS5A and Anxa2. As shown in Fig. 5B and C, many Anxa2 and NS5A gold particles were detected inside the same vesicle, which is suggestive of the role of Anxa2 in HCV RNA replication.
Fig 5.
Immunoelectron microscopy of NS5A and Anxa2 on the membranous web. HCV JC1-infected Huh7.5 cells were labeled with antibody against NS5A alone (A) or with both NS5A and Anxa2 (B and C). NS5A was tethered to 12-nm and Anxa2 to 6-nm gold particles. Shown are consecutively enlarged views of the membranous web. Arrows show NS5A, and arrowheads show Anxa2 in electron-dense granules on the membrane of DMV. N, nucleus. DMV, double-membrane vesicles. MMV, multiple-membrane vesicles.
BiFC demonstrated that NS5A colocalized with Anxa2 and the lipid raft marker caveolin-2 (Cav2).
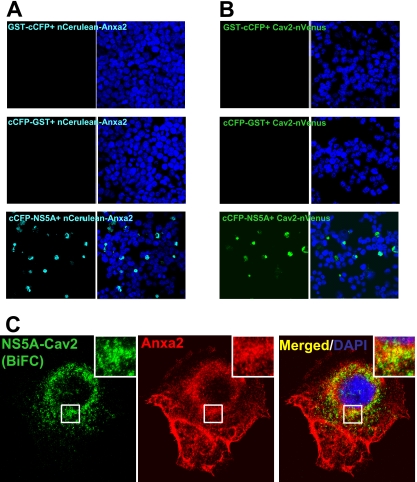
To further demonstrate that HCV proteins and Anxa2 interact with each other at lipid rafts, we employed BiFC in living cells. BiFC is a technique used to study protein-protein interactions in which the tagged proteins are visualized by reconstituting fluorescence from split fluorophores (27, 29, 44, 48). We first performed BiFC assays on the interaction between NS5A and Anxa2. We used the C-terminal half of CFP (amino acids 155 to 238; cCFP) and the N-terminal fragment (amino acids 1 to 173) of Venus (nVenus) or Cerulean (nCerulean), which had been shown to produce strong fluorescence (53). We fused the cCFP fragment to HCV NS5A and GST protein and the nVenus or nCerulean fragment to Cav2 and Anxa2, respectively. All of the BiFC constructs were first confirmed to express the fused proteins by immunoblotting (data not shown). The cCFP-fused proteins were arrayed against the nCerulean-fused proteins to determine their possible interaction. It was found that NS5A-cCFP, but neither GST-cCFP nor cCFP-GST, emitted a fluorescent signal when cotransfected with Anxa2-nCerulean (Fig. 6A), indicating a direct interaction of NS5A with Anxa2. Further, the cotransfection of NS5A-cCFP and nVenus-Cav2 also resulted in fluorescence (Fig. 6B). We next determined the relationship of Anxa2, NS5A, and the lipid raft. Huh7.5 cells were cotransfected with NS5A-cCFP- and nVenus-Cav2-expressing constructs and immunolabeled with Anxa2. NS5A and Cav2 BiFC fluorescence was found to colocalize with Anxa2, mainly in the perinuclear region (Fig. 6C, inset), indicating that the Anxa2-NS5A interaction occurred on the Cav2-containing lipid raft. These observations provide further support that NS5A and Anxa2 interact with each other on the lipid raft, the site of HCV RC formation.
Fig 6.
BiFC showed that Anxa2 interacts with NS5A at the Cav2-containing lipid raft. HEK293 cells were cotransfected with cCFP-fused GST or NS5A with nCerulean-Anxa2 (A) or Cav2-nVenus (B). BiFC constructs were fixed and stained with DAPI and visualized by confocal microscopy at 48 h posttransfection. (C) Huh7.5 cells cotransfected with cCFP-NS5A and Cav2-nVenus BiFC constructs were fixed and immunostained with anti-Anxa2 (red) monoclonal antibody. Cellular DNA was labeled with DAPI (blue). Enlarged views of sections of images are shown (inset).
Anxa2 promotes the enrichment of HCV nonstructural proteins on the lipid raft.
Anxa2 has been reported to influence the lateral association and stability of cholesterol-rich microdomains (4, 5, 10). We next examined whether Anxa2 caused the enrichment of HCV NSP in the lipid raft region.
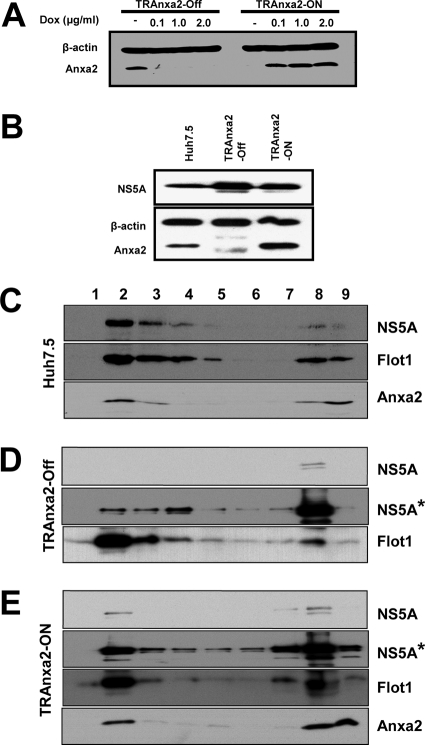
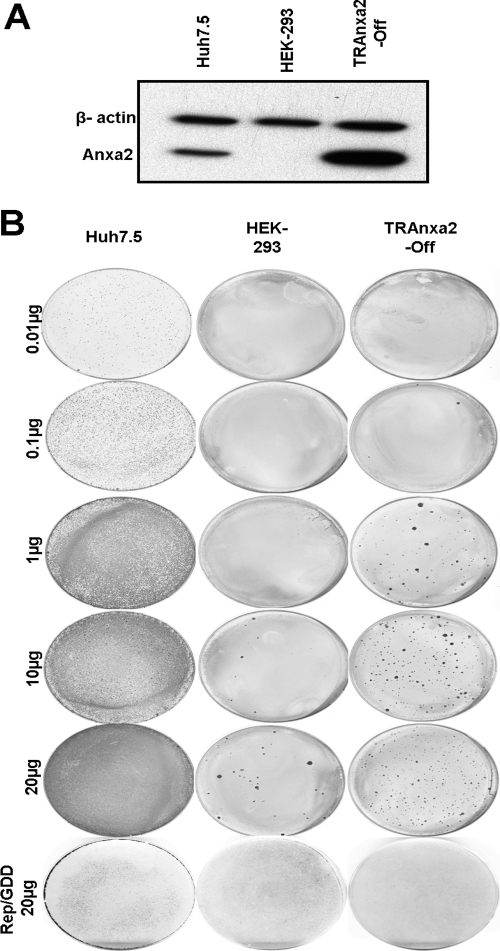
To examine this possibility, we utilized HEK293 cells, which have a very low level of endogenous Anxa2 (23, 61). We established two tetracycline-regulated HEK293 cell lines, TRAnxa2, expressing Anxa2 either in the presence (TRAnxa2-ON) or in the absence (TRAnxa2-OFF) of doxycycline. We found that Anxa2 protein expression was induced or suppressed upon the addition of 0.1 μg/ml of doxycycline (Fig. 7A); very little endogenous Anxa2 was detected in these cells. Further, NS5A was expressed by plasmid transfection in these cells (Fig. 7B). The membrane flotation ultracentrifugation study of these cells revealed that NS5A was fractionated mainly in the DRM fraction (fractions 2 and 3) in Huh7.5 cells (Fig. 7C). Anxa2 was distributed in both the DRM and soluble fractions. In TRAnxa2-OFF cells, the majority of NS5A protein was localized in the soluble fraction when Anxa2 was absent (Fig. 7D); in contrast, in TRAnxa2-ON cells, the expression of Anxa2 increased the proportion of NS5A in the DRM fraction (Fig. 7E). Taken together, these observations indicate that the expression of Anxa2 helps to recruit viral NSPs (e.g., NS5A) to the lipid raft microdomain.
Fig 7.
Anxa2 promotes the enrichment of HCV NS5A in DRM fraction. (A) TRAnxa2-OFF and TRAnxa2-ON cells were cultured in the presence of the indicated amounts of doxycycline (Dox). After 48 h, cell lysates were resolved by SDS-PAGE and then immunoblotted with a monoclonal antibody against Anxa2. Immunoblotting against β-actin serves as a loading control. (B) Huh7.5 cells, TRAnxa2-OFF, and TRAnxa2-ON cells were transfected with a plasmid encoding HA-tagged NS5A protein (NS5A-HA). At 48 h posttransfection, cells were analyzed for the expression of NS5A and Anxa2. Huh7.5 (C), TRAnxa2-OFF (D), and TRAnxa2-ON (E) cells were transfected with NS5A-HA and subjected to 1% Triton X-100 treatment and fractionated by discontinuous OptiPrep gradient centrifugation as described in Materials and Methods. Fractions were separated on a 10% SDS-PAGE gel, followed by immunoblotting with monoclonal antibodies against HA tag (depicting NS5A), Flot1, and Anxa2. Fractions are numbered from 1 to 9 in order from top to bottom (light to heavy). NS5A* represents a longer exposure of the same film shown for NS5A.
Anxa2 helps in the formation of HCV replication complex on the membranous web.
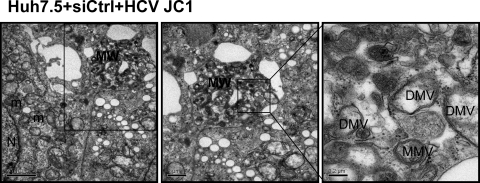
We further examined the importance of Anxa2 in the formation of membranous web induced by HCV. Huh7.5 cells were transfected with siAnxa2 or siCtrl and then infected with HCV JC1. In cells transfected with the control siRNA, abundant membranous web-like structures consisting of an accumulation of a heterogeneous population of double-membrane vesicles (DMVs) and multiple-membrane vesicles (MMVs) were observed (Fig. 8). These vesicles had been reported to represent HCV RC (13, 45). These membranous web-like structures were observed in 29% (9 of 31 cells) of the cells transfected with control siRNA. In comparison, in siAnxa2-transfected cells, of 34 cells only 1 cell was found to contain membranous web-like structures. The uninfected naïve Huh7.5 cells did not contain similar structures (data not shown). These observations suggest that Anxa2 plays a significant role in the formation of the HCV replication complex.
Fig 8.
Silencing of Anxa2 inhibits membranous web formation. Huh7.5 cells were treated with control siRNAs for 2 days and then infected with HCV JC1. Cells were fixed and processed for electron microscopy at the third day postinfection. The consecutive enlargement of the boxed areas are shown from left to right. Note the very heterogeneous membranous web (MW) in control cells. Scale bars are given in the lower left of each panel. N, nucleus; DMV, double-membrane vesicles; MMV, multiple membrane vesicles; ER, endoplasmic reticulum; m, mitochondria.
Anxa2 supports HCV RNA replication in nonhepatic epithelial cells.
HCV has a very narrow host range, infecting only human and chimpanzee. Further, hepatic cells are its preferred site of replication. We asked whether Anxa2 is the key factor missing from nonhepatic cells. For this, we performed a colony formation assay of the HCV replicon on HEK293 cells. HCV1b (wt) SGR RNA or polymerase-defective HCV1b (mutant [mt]) Rep/GDD RNA was electroporated into Huh7.5, HEK293, or TRAnxa2-OFF cells, which express Anxa2 in the absence of doxycycline (Fig. 9A), and selected with G418. The HCV1b (wt) RNA-electroporated Huh7.5 cells formed numerous colonies, with an average of 1.86 × 105 CFU/μg of HCV RNA. In naïve HEK293 cells, very few colonies (1.4 CFU/μg of HCV RNA) were observed. Significantly, TRAnxa2-OFF cells formed more than 50-fold more colonies (82 CFU/μg of HCV RNA) in the presence of Anxa2 than in its absence (Fig. 9B). These observations suggest that Anxa2 is one of the crucial factors for HCV RC formation.
Fig 9.
Colony formation of HCV 1b subgenomic RNA replicon in Huh7.5, HEK293, and TRAnxa2-OFF cell lines. (A) The cell lines were analyzed by immunoblotting for the expression of Anxa2. (B) In vitro-transcribed wild-type or GDD mutant (Rep/GDD) subgenomic replicon RNAs in the indicated amounts were electroporated into each cell line, and cells were cultured with G418 for 12 to 15 days before staining with crystal violet.
DISCUSSION
In the present study, we identified a host cellular protein, Anxa2, as an important factor in the formation of the HCV RC in the lipid microenvironment. We show that HCV NSPs interacted with Anxa2, presumably facilitating the recruitment of various phosphatidylinositides, e.g., PI4P, to the cellular membrane to generate a lipid-enriched membrane microenvironment on which viral RCs could assemble. Further, the ability to form these lipid-enriched membranes correlated, to some extent, with the permissibility of HCV infection.
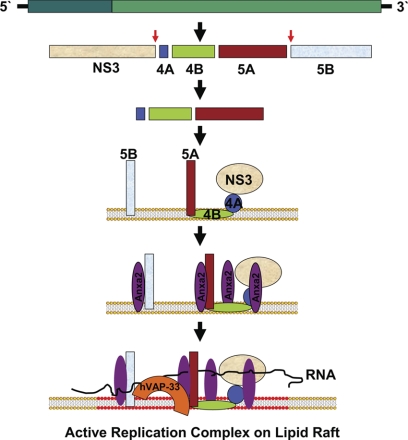
Anxa2 has been identified to be associated with HCV RC in previous studies using different methodologies. Huang et al. (28) used FLAG epitope-tagged NS5A in a functional replicon and identified Anxa2 as its interaction partner. Additionally, Backes and colleagues (6) identified Anxa2 from crude replication complex as an interacting partner of NS5A. These studies, together with our observations, suggest that Anxa2 participates in HCV RNA replication and have led us to propose a model for HCV RC formation on the lipid raft (Fig. 10). We postulate that the NS4AB-NS5A polyprotein is the complex that initiates the formation of RC on the lipid raft membrane (15). Further, since NS3 is a cofactor for NS4A, NS3 is brought into this complex. NS5B, being a membranous protein, is likely also present on the same membrane. Following the organization of this protein complex, Anxa2 may then play a role in rearranging the membrane. Our experimental observations suggest that the interaction of Anxa2 with viral NSPs induces raft clustering on the membrane. The membrane-bound NS5A protein induces calcium ion signaling (19), and Anxa2, in response to this flux, may interact with signal phospholipid, phosphatidylinositol (4,5)-bisphosphate (PIP2), and, as described below, initiate raft clustering (18). Thus, Anxa2 could create lipid rafts around these membrane-anchored HCV NSPs. This lipid raft clustering subsequently will be helpful in recruiting various other factors, which may facilitate viral RNA replication. Further, we have previously shown that hVAP33, another vesicle-associated protein, can interact with NS5A and NS5B (15); therefore, we propose that these interactions bring NS5B into this complex. In this manner, HCV RC may assemble at a lipid raft region. How HCV RNA enters into this complex remains unanswered.
Fig 10.
Hypothetical model for the mechanism of formation of a HCV replication complex on a lipid raft. HCV polyprotein is stepwise processed into intermediate and individual viral proteins. After NS3 and NS5B are proteolytically cleaved from the polyprotein, the uncleaved NS4AB-5A is recruited to the membrane by the intrinsic membrane-anchoring character of NS4B (15). NS3 becomes associated with this membrane by binding to its cofactor, NS4A. NS5B, being a membranous protein, also localizes to the membrane. Anxa2 can interact with NS3/NS4A, NS4B, NS5A, and NS5B. Membrane binding of HCV protein induces calcium ion flux (19), and Anxa2, in response to this flux, interacts with signal phospholipid, which initiates raft clustering. Thus, Anxa2 creates a lipid raft around these membrane-anchored proteins. NS5B, through its interaction with hVAP-33 (15), is brought into this complex, and, in this manner, the HCV RC is established. How HCV RNA enters into this complex is not yet known.
Anxa2 is localized in the membrane through its ability to bind to phospholipids in general (reviewed in reference 49). The membrane-bound Anxa2 specifically interacts with the signal phospholipid PIP2 and thereby is recruited to the lipid raft. This recruitment induces and stabilizes raft clustering, which is probably the prerequisite for HCV RNA RC formation. Previously, phosphatidylinositol-4-kinase III alpha (PI4K-IIIa) was shown to play an important role in HCV RNA replication (8, 9, 55, 56). ER-localized lipid kinase PI4K-IIIa generates a PIP2 precursor lipid, phosphatidylinositol-4-phosphate (PI4P). PI4P was localized to HCV replication sites (6, 26, 47), and we found Anxa2 to be localized on the same PI4P-containing site in HCV-infected cells (Fig. 3C). Further, we found that HCV-infected cells, compared to naïve Huh7.5 cells, had an increased level of Anxa2 (Fig. 3A) as well as PI4P (Fig. 3C), which is suggestive of an interplay in favor of HCV replication. This enhanced level of Anxa2 may explain the relatively larger amount of Anxa2 protein observed in the DRM analysis (compare Fig. 2A, upper panel, with Fig. 2B, middle panel). Also, by using Anxa2-overexpressing stable cell lines, we showed that Anxa2 expression promotes the recruitment of HCV NSPs to the DRM (Fig. 7) and may thereby enhance HCV replication. Even in a nonpermissive, nonhepatic cell line that expresses very little endogenous Anxa2, the overexpression of exogenous Anxa2 promotes colony formation induced by the HCV replicon (Fig. 9B). Thus, Anxa2 is at least a factor that controls HCV replication by assisting in the formation of the HCV RC.
Furthermore, Anxa2 has been shown to interact with several phosphatidylinositides, specifically phosphatidylinositol (PI), PI4P, phosphatidylinositol 5-phosphate (PI5P), phosphatidylinositol 3,4-bisphosphate (PI3,4P2), phosphatidylinositol 3,5-bisphosphate (PI3,5P2), phosphatidic acid (PA), and phosphatidylinositol 4,5-bisphosphate (PI4,5P2), in a calcium ion-dependent manner (24). Therefore, we postulate that the membrane-bound NS5A induces calcium ion flux (19), resulting in the binding of Anxa2 to various phosphatidylinositides and thereby generating a lipid-enriched microenvironment around HCV NSPs.
Anxa2 has been implicated in the regulation of several key processes in HIV (23), influenza virus (34), and more recently in HCV (6). Backes et al. (6) identified Anxa2 from the HCV RC as an associated host factor and further observed the presence of Anxa2 in the active HCV replication site, which is in agreement with our study. These authors report that Anxa2 was colocalized with NS3, NS4B, NS5A, and NS5B, in agreement with our observations (Fig. 4A to C). However, on the knockdown of Anxa2 expression, they found no effect on HCV RNA replication; instead, they observed significant reductions of extra- and intracellular infectious viral titers; they therefore concluded that Anxa2 plays a role in the assembly of infectious HCV particles but not in RNA replication. In contrast to their study, we observed a very clear reduction in viral intracellular proteins (Fig. 1A), viral RNA (Fig. 1B), and in vitro replicase activity (Fig. 1C) following the siRNA-mediated knockdown of Anxa2. These discrepancies may partly be due to the difference in siRNA sequences used by us and Backes et al. Further, Anxa2 is a multifunctional protein, and its role in multivesicular endosome biogenesis (38) and the polymerization of actin (39, 50) have been reported. Recently, we reported that HCV RC is transported through microtubule and actin filaments (32) and that HCV particle release involves endosomal trafficking (33). Thus, the conclusion made by Backes et al., together with our observations, points to the multifaceted role Anxa2 may play in the HCV life cycle, ranging from RC formation to virus particle assembly.
In summary, we have shown that the scaffolding protein Anxa2 assists in the formation of HCV RC by organizing the enrichment of HCV NSPs in the lipid raft region. Further, together with our observations and that of Backes et al., it will be prudent to explore the role of Anxa2 in the possible connection between HCV RC and virion assembly.
ACKNOWLEDGMENTS
We thank Sue-Ping Lee and Sue-Ping Tsai for technical assistance in confocal and electron microscopy and AndreAna Peña for English language editing. We express our sincere thanks to Tony Hunter from the Salk Institute for Biological Studies for mouse monoclonal anti-Anxa2 antibody, Michinori Kohara from The Tokyo Metropolitan Institute of Medical Science, Japan, for rabbit polyclonal anti-NS4B antibody (RR12), Stanton B. Gelvin from Purdue University for the BiFC cassette containing pSAT vector series, Song-Kun Shyue from the Institute of Biomedical Sciences, Academia Sinica, Taiwan, for the caveolin-2-expressing plasmid vector, and Charles M. Rice from the Center for the Study of Hepatitis C, Rockefeller University, for Huh7.5 cells.
Footnotes
Published ahead of print 1 February 2012
REFERENCES
- 1. Aizaki H, Choi KS, Liu M, Li YJ, Lai MM. 2006. Polypyrimidine-tract-binding protein is a component of the HCV RNA replication complex and necessary for RNA synthesis. J. Biomed. Sci. 13:469–480 [DOI] [PubMed] [Google Scholar]
- 2. Aizaki H, Lee KJ, Sung VM, Ishiko H, Lai MM. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450–461 [DOI] [PubMed] [Google Scholar]
- 3. Alter MJ. 2007. Epidemiology of hepatitis C virus infection. World J. Gastroenterol. 13:2436–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ayala-Sanmartin J, Henry JP, Pradel LA. 2001. Cholesterol regulates membrane binding and aggregation by annexin 2 at submicromolar Ca(2+) concentration. Biochim. Biophys. Acta 1510:18–28 [DOI] [PubMed] [Google Scholar]
- 5. Babiychuk EB, Draeger A. 2000. Annexins in cell membrane dynamics. Ca(2+)-regulated association of lipid microdomains. J. Cell Biol. 150:1113–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Backes P, et al. 2010. Role of annexin A2 in the production of infectious hepatitis C virus particles. J. Virol. 84:5775–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bavari S, et al. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195:593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berger KL, et al. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:7577–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Borawski J, et al. 2009. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J. Virol. 83:10058–10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chasserot-Golaz S, et al. 2005. Annexin 2 promotes the formation of lipid microdomains required for calcium-regulated exocytosis of dense-core vesicles. Mol. Biol. Cell 16:1108–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen YC, et al. 2010. Polo-like kinase 1 is involved in hepatitis C virus replication by hyperphosphorylating NS5A. J. Virol. 84:7983–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Egger D, et al. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferraris P, Blanchard E, Roingeard P. 2010. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J. Gen. Virol. 91:2230–2237 [DOI] [PubMed] [Google Scholar]
- 14. Futter CE, White IJ. 2007. Annexins and endocytosis. Traffic 8:951–958 [DOI] [PubMed] [Google Scholar]
- 15. Gao L, Aizaki H, He JW, Lai MM. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gerke V, Creutz CE, Moss SE. 2005. Annexins: linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 6:449–461 [DOI] [PubMed] [Google Scholar]
- 17. Gerke V, Moss SE. 2002. Annexins: from structure to function. Physiol. Rev. 82:331–371 [DOI] [PubMed] [Google Scholar]
- 18. Gokhale NA, Abraham A, Digman MA, Gratton E, Cho W. 2005. Phosphoinositide specificity of and mechanism of lipid domain formation by annexin A2–p11 heterotetramer. J. Biol. Chem. 280:42831–42840 [DOI] [PubMed] [Google Scholar]
- 19. Gong G, Waris G, Tanveer R, Siddiqui A. 2001. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa B. Proc. Natl. Acad. Sci. U. S. A. 98:9599–9604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grewal T, Enrich C. 2009. Annexins–modulators of EGF receptor signalling and trafficking. Cell Signal. 21:847–858 [DOI] [PubMed] [Google Scholar]
- 21. Grewal T, Enrich C. 2006. Molecular mechanisms involved in Ras inactivation: the annexin A6-p120GAP complex. Bioessays 28:1211–1220 [DOI] [PubMed] [Google Scholar]
- 22. Guo JT, Bichko VV, Seeger C. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516–8523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harrist AV, Ryzhova EV, Harvey T, Gonzalez-Scarano F. 2009. Anx2 interacts with HIV-1 Gag at phosphatidylinositol (4,5) bisphosphate-containing lipid rafts and increases viral production in 293T cells. PLoS One 4:e5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hayes MJ, et al. 2004. Annexin 2 binding to phosphatidylinositol 4,5-bisphosphate on endocytic vesicles is regulated by the stress response pathway. J. Biol. Chem. 279:14157–14164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hayes MJ, Rescher U, Gerke V, Moss SE. 2004. Annexin-actin interactions. Traffic 5:571–576 [DOI] [PubMed] [Google Scholar]
- 26. Hsu NY, et al. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu CD, Chinenov Y, Kerppola TK. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9:789–798 [DOI] [PubMed] [Google Scholar]
- 28. Huang H, et al. 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 104:5848–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kerppola TK. 2006. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 7:449–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lafont F, et al. 1999. Raft association of SNAP receptors acting in apical trafficking in Madin-Darby canine kidney cells. Proc. Natl. Acad. Sci. U. S. A. 96:3734–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lai CK, Jeng KS, Machida K, Cheng YS, Lai MM. 2008. Hepatitis C virus NS3/4A protein interacts with ATM, impairs DNA repair and enhances sensitivity to ionizing radiation. Virology 370:295–309 [DOI] [PubMed] [Google Scholar]
- 32. Lai CK, Jeng KS, Machida K, Lai MM. 2008. Association of hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5A. J. Virol. 82:8838–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lai CK, Jeng KS, Machida K, Lai MM. 2010. Hepatitis C virus egress and release depend on endosomal trafficking of core protein. J. Virol. 84:11590–11598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. LeBouder F, et al. 2008. Annexin II incorporated into influenza virus particles supports virus replication by converting plasminogen into plasmin. J. Virol. 82:6820–6828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee KJ, Choi J, Ou JH, Lai MM. 2004. The C-terminal transmembrane domain of hepatitis C virus (HCV) RNA polymerase is essential for HCV replication in vivo. J. Virol. 78:3797–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee LY, Fang MJ, Kuang LY, Gelvin SB. 2008. Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Manie SN, de Breyne S, Vincent S, Gerlier D. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74:305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mayran N, Parton RG, Gruenberg J. 2003. Annexin II regulates multivesicular endosome biogenesis in the degradation pathway of animal cells. EMBO J. 22:3242–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morel E, Parton RG, Gruenberg J. 2009. Annexin A2-dependent polymerization of actin mediates endosome biogenesis. Dev. Cell 16:445–457 [DOI] [PubMed] [Google Scholar]
- 40. Morgan RO, Pilar Fernandez M. 1997. Distinct annexin subfamilies in plants and protists diverged prior to animal annexins and from a common ancestor. J. Mol. Evol. 44:178–188 [DOI] [PubMed] [Google Scholar]
- 41. Moss SE, Morgan RO. 2004. The annexins. Genome Biol. 5:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nguyen DH, Hildreth JE. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199 [DOI] [PubMed] [Google Scholar]
- 44. Nyfeler B, Michnick SW, Hauri HP. 2005. Capturing protein interactions in the secretory pathway of living cells. Proc. Natl. Acad. Sci. U. S. A. 102:6350–6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paul D, et al. 2011. NS4B self-interaction through conserved C-terminal elements is required for the establishment of functional hepatitis C virus replication complexes. J. Virol. 85:6963–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Raynal P, Pollard HB. 1994. Annexins: the problem of assessing the biological role for a gene family of multifunctional calcium- and phospholipid-binding proteins. Biochim. Biophys. Acta 1197:63–93 [DOI] [PubMed] [Google Scholar]
- 47. Reiss S, et al. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Remy I, Michnick SW. 2004. A cDNA library functional screening strategy based on fluorescent protein complementation assays to identify novel components of signaling pathways. Methods 32:381–388 [DOI] [PubMed] [Google Scholar]
- 49. Rescher U, Gerke V. 2004. Annexins–unique membrane binding proteins with diverse functions. J. Cell Sci. 117:2631–2639 [DOI] [PubMed] [Google Scholar]
- 50. Rescher U, Ludwig C, Konietzko V, Kharitonenkov A, Gerke V. 2008. Tyrosine phosphorylation of annexin A2 regulates Rho-mediated actin rearrangement and cell adhesion. J. Cell Sci. 121:2177–2185 [DOI] [PubMed] [Google Scholar]
- 51. Scheiffele P, Rietveld A, Wilk T, Simons K. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038–2044 [DOI] [PubMed] [Google Scholar]
- 52. Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shyu YJ, Liu H, Deng X, Hu CD. 2006. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques 40:61–66 [DOI] [PubMed] [Google Scholar]
- 54. Smart EJ, et al. 1999. Caveolins, liquid-ordered domains, and signal transduction. Mol. Cell. Biol. 19:7289–7304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tai AW, et al. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vaillancourt FH, et al. 2009. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology 387:5–10 [DOI] [PubMed] [Google Scholar]
- 57. Wakita T, et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Weaver AK, Olsen ML, McFerrin MB, Sontheimer H. 2007. BK channels are linked to inositol 1,4,5-triphosphate receptors via lipid rafts: a novel mechanism for coupling [Ca(2+)](i) to ion channel activation. J. Biol. Chem. 282:31558–31568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wolk B, et al. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yang W, Huang M. 2009. Studying HCV RNA synthesis in vitro with replication complexes. Methods Mol. Biol. 510:177–184 [DOI] [PubMed] [Google Scholar]
- 61. Zhou H, et al. 2009. Annexin A2 mediates anti-beta 2 GPI/beta 2 GPI-induced tissue factor expression on monocytes. Int. J. Mol. Med. 24:557–562 [DOI] [PubMed] [Google Scholar]