Abstract
We report that type I interferons (IFNs) upregulate latent membrane protein 1 (LMP-1) expression by direct activation of the ED-L1 promoter in several Epstein-Barr virus (EBV)-carrying Burkitt's lymphoma lines. In EBV-infected primary B cells, IFN-α transiently upregulates LMP-1 mRNA, but not protein levels, followed by downregulation of both, suggesting a novel antiproliferative mechanism of type I IFNs. Furthermore, our results may explain the expression of LMP-1 in memory B cells of systemic lupus erythematosus patients.
TEXT
Epstein-Barr virus (EBV) is a ubiquitous gammaherpesvirus associated with a wide variety of neoplasms, including Burkitt's lymphoma (BL), nasopharyngeal carcinoma, posttransplant lymphoproliferative disease, and Hodgkin's disease. Only a subset of viral genes is transcribed from latent episomal EBV genomes in lymphoblastoid cell lines (LCLs) and in EBV-associated neoplasms. Besides EBV-encoded RNAs (EBERs) and BamHI-A transcripts, in type I latency only EBV nuclear antigen 1 (EBNA-1) is expressed, while in type III latency all six EBNAs and three EBV-encoded latent membrane proteins (LMPs) are expressed. In type II latency, which is observed in Hodgkin T cell and NK cell lymphomas, in the lymphoid tissues of healthy virus carriers and infectious mononucleosis patients one or all of the LMPs are expressed in addition to the type I latency gene products (39). LMP-1 plays a central role in EBV biology, since it acts in part as a constitutively active CD40 receptor analog and is essential for B cell proliferation and transformation by EBV (24). In type III latency, EBNA-2 is the major transactivator of the LMP promoters, while in type II latency, depending on the cellular context, different cytokines (interleukin-4 [IL-4], IL-10, -13, -15, and -21) are responsible for the activation of LMP-1 transcription (20, 26, 27, 28).
Type I interferons (IFNs) are produced in relatively large amounts in response to pathogen sensing by the innate immune system (46). In addition to their direct antiviral activities, these proteins also have antiproliferative and immunomodulatory properties. Consequently, type I IFNs find diverse clinical application in the treatment of certain forms of cancer, as well as in the therapy of viral infections or immunological disorders (31). On the other hand, type I IFNs have a major pathophysiological role in human diseases, such as systemic lupus erythematosus (SLE), with characteristic high IFN-α levels (46).
Several interactions have been described between EBV and the type I IFN system. EBV virions and/or EBER1 (secreted in complex with lupus erythematosus-associated antigen or added exogenously in an in vitro-synthesized form) induces type I IFN production in several cell types, including B cells and plasmacytoid dendritic cells (21, 25, 38). Conversely, IFN-α treatment of adult B cells prior to or at the time of in vitro infection nearly completely prevents EBV-mediated B cell proliferation and outgrowth into LCLs (30, 47), at least partially through the inhibition of the capping of EBV-CD21 complexes (6). However, EBV-infected B cells become progressively resistant to the effects of type I IFNs within a few days postinfection (30, 47), possibly through the inhibitory effect of LMP-1 on IFN-α-induced Tyk2 and subsequent STAT2 phosphorylation (12). On the other hand, the mechanism of the partial inhibition of B cell transformation by type I IFNs added within the first 48 h after infection (30, 47) is still unidentified. Furthermore, despite the observation of this complex network of interactions, no direct effect of type I IFNs has been shown on the regulation of latent EBV gene expression. Using EBV-positive BL lines and freshly infected peripheral blood B cells, we show now that type I IFNs can directly modulate LMP-1 expression.
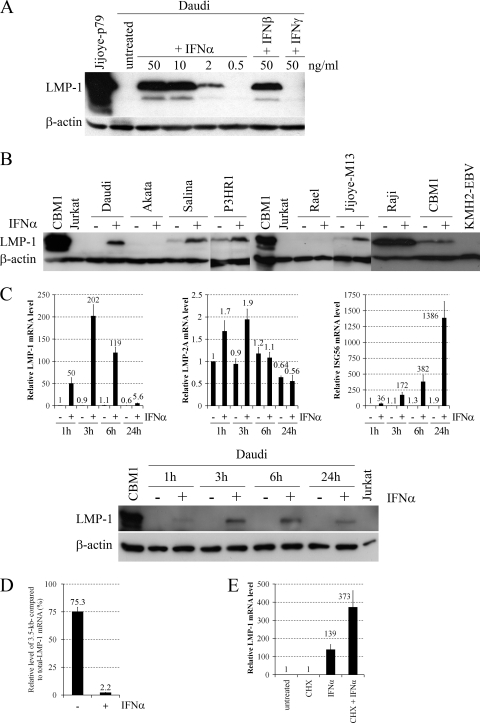
For initial experiments, we chose the highly IFN-α-sensitive, EBV-positive BL line Daudi (29), in which IFN-α treatment inhibits cell proliferation and concomitantly induces plasmacytoid differentiation (8). Daudi cells were treated with different concentrations of IFN-α, IFN-β, and IFN-γ (Peprotech) for 24 h, and the level of LMP-1 protein was analyzed by Western blotting (Fig. 1A). Type I IFNs strongly upregulated LMP-1 protein expression in a dose-dependent manner, while IFN-γ did not. Since the antiproliferative effect of IFN-α already reaches its maximum at 0.3 ng/ml (data not shown), while LMP-1 expression is not induced even at 0.5 ng/ml (Fig. 1A), the growth-inhibitory effect of IFN-α on Daudi cells is not a consequence of LMP-1 upregulation.
Fig 1.
Effects of IFNs on LMP-1 expression in BL lines. (A) Immunoblot analysis of LMP-1 (S12 supernatant) and β-actin (AC-15 mouse anti-human β-actin [Sigma-Aldrich]) protein expression in total cell extracts of Daudi cells left untreated or treated with the indicated amounts of IFN-α, IFN-β, or IFN-γ for 24 h. Total cell extract of Jijoye-p79 (1) was used as a positive control for LMP-1 protein expression. (B) Immunoblot analysis of LMP-1 and β-actin protein expression in total cell extracts of BL lines and CBM1-Ral-STO (7) left untreated (−) or treated with 10 ng/ml IFN-α (+) for 24 h. Total cell extract of CBM1-Ral-STO was used as a positive control, while Jurkat (42) and KMH2-EBV (26) were the negative controls for LMP-1 protein expression. (C) Relative levels of LMP-1, -2A, and ISG56 mRNAs normalized to GAPDH, quantified by real-time RT-PCR (upper panels), and immunoblot analysis of LMP-1 and β-actin protein expression (lower panel) in Daudi cells left untreated (−) or treated with 20 ng/ml IFN-α (+) for the hours indicated. CBM1-Ral-STO was used as a positive control, and Jurkat cells were the negative controls for LMP-1 protein expression. (D) Relative levels of the 3.5-kb LMP-1 mRNA compared to total LMP-1 mRNA, quantified by real-time RT-PCR in Daudi left untreated (−) or treated with 20 ng/ml IFN-α (+) for 3 h. (E) Relative levels of LMP-1 mRNA normalized to GAPDH, quantified by real-time RT-PCR in Daudi cells preincubated for 30 min without or with 50 μg/ml CHX and then left untreated or treated with 50 ng/ml IFN-α for an additional 1 h. Primers are listed in Table 1, and PCR conditions are described in the text. CBM1, CBM1-Ral-STO cells.
Next, we analyzed the LMP-1-inducing effect of IFN-α on a panel of EBV-positive BL lines, including Daudi, Salina, and P3HR1 (lines carrying a virus strain that has a deletion involving EBNA-2, and therefore the cells use the W-promoter for the transcription of the EBNAs and do not express or express only minimal amounts of LMPs [1, 4, 22, 23]), the type I BL lines Akata (45), Rael (32), and Jijoye-M13 (27), the type III BL line Raji (28), and the cord blood-derived LCL CBM1-Ral-STO, transformed with the Rael EBV strain (7). IFN-α treatment for 24 h led to the upregulation of LMP-1 protein expression in Jijoye-M13 cells and in all the BL lines carrying EBNA-2-deleted virus, while LMP-1 mRNA and protein levels did not increase in the other cell lines (Fig. 1B and data not shown). Since IFN-stimulated gene 56 (ISG56; a well-characterized target of the IFN-α-induced IFN-stimulated gene factor 3 [ISGF3] transactivation complex [STAT-1 and -2 and IFN regulatory factor 9] [10]) is highly upregulated upon IFN-α treatment (41), the type I IFN receptor and at least the classical ISGF3 signaling pathway are functional and cannot be responsible for the lack of LMP-1 upregulation in type I BL lines. However, the presence of highly methylated CpG dinucleotides at the LMP-1 regulatory sequences may explain the failure of LMP-1 induction in Rael cells (32, 33, 44). Furthermore, the different inducibility of LMP-1 in type I BL cell lines and BL lines carrying an EBNA-2-deleted virus suggest that the presence of EBNA-3 proteins and/or EBNA-LP may play a role in the upregulation of LMP-1 by IFN-α. The failure of LMP-1 induction in Raji and CBM1-Ral-STO cells is in line with previous observations on the relative insensitivity of type III cells to type I IFNs (12, 30), although upregulation of ISG56 mRNA was only partially reduced in the type III cell lines (5- and 52-fold ISG56 upregulation after 24 h in Raji and CBM1-Ral-STO cells, respectively [data not shown]), compared to its high level of induction in all other lines (41).
To analyze the kinetics and specificity of LMP-1 upregulation, LMP-1 mRNA and protein levels were measured together with LMP-2A and ISG56 mRNA expression in Daudi cells treated with IFN-α for 1, 3, 6, and 24 h (Fig. 1C). Strong (50-fold) upregulation of LMP-1 mRNA was detected already after 1 h, with a maximum increase of more than 200-fold after 3 h, and then progressively decreased at 6 and 24 h. In contrast, LMP-2A mRNA was only slightly (2.1-fold) upregulated during the kinetic analysis, while ISG56 mRNA was rapidly and progressively upregulated to a maximum of 729-fold after 24 h. Upregulation of LMP-1 protein was detected already after 1 h, with a maximum after 6 h and a lower level after 24 h. These results show that IFN-α rapidly and specifically upregulates LMP-1 through transcriptional activation and with a kinetics different from the IFN-α-induced upregulation of the classical target ISG56.
To analyze the role of the ED-L1 (2.8-kb mRNA [18]) and L1-TR (3.5-kb mRNA [40]) promoters in the IFN-α-induced upregulation of LMP-1, the relative levels of total (2.8- and 3.5-kb mRNAs) and 3.5-kb LMP-1 mRNA were measured with real-time reverse transcription-PCR (RT-PCR) in Daudi cells left untreated or treated with IFN-α for 3 h (Fig. 1D). The primers used for PCR analysis are listed in Table 1. Total cellular RNA was isolated using the Quick RNA miniprep kit (Zymo Research), treated with Turbo DNase (Ambion), and then cleaned up with the RNA Clean and Concentrator kit (Zymo Research) according to the manufacturer's instructions. One microgram of RNA was reverse transcribed using the SuperScript VILO cDNA synthesis kit (Invitrogen) according to the manufacturer's instructions. The relative level of each transcript was determined with the LC FastStart DNA Master SYBR green I kit in a LightCycler 1.2 instrument (Roche) using the standard curve method. Each PCR mixture was initially denatured at 95°C for 10 min and then cycled 40 times at 95°C for 8 s, 60°C for 5 s, and 72°C for 8 s. Target genes were measured and normalized simultaneously with the endogenous control glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The analysis showed that, in untreated Daudi cells 75% of LMP-1 mRNAs, while in IFN-α-treated Daudi cells only 2.2% of LMP-1 mRNAs were transcribed from the L1-TR promoter.
Table 1.
Primers used in real-time RT-PCR
| Primer | Sequencea |
|---|---|
| Total LMP-1 | 5′-GCAGGAGGGTGATCATCAGT-3′ |
| 5′-GTCCTCTATTCCTTTGCTCTCATG-3′ | |
| 3.5-kb LMP-1 | 5′-TACGTAGCCGCCCTACATAAG-3′ |
| 5′-CCTCTCAAGGTCCAGGTCCAT-3′ | |
| LMP-2A | 5′-CAGGCAGGCATACTGGATTC-3′ |
| 5′-CGATGAGGAACGTGAATCTAATG-3′ | |
| EBNA-2 | 5′-GGACACAAGAGCCATCACCT-3′ |
| 5′-CAAAGCATTCGCATAGCAGA-3′ | |
| ISG56 | 5′-GTGACATCTCAATTGCTCCAGAC-3′ |
| 5′-GGAGCCTGGCTAAGCAAAAC-3′ | |
| GAPDH | 5′-GGAAGGTGAAGGTCGGAGTCA-3′ |
| 5′-ATGGGTGGAATCATATTGGAACA-3′ |
Sequences of forward and reverse primers are provided. The LMP-1 coding and regulatory regions containing the primer binding sites were sequenced in Daudi cells. Primers (Sigma-Aldrich) for LMP-1 were designed on the basis of these sequences.
To test whether de novo protein synthesis is required for the rapid induction of LMP-1 by type I IFNs, Daudi cells were preincubated with or without cycloheximide (CHX; Sigma-Aldrich), a protein synthesis inhibitor, for 30 min, after which the cells were left untreated or treated with IFN-α for an additional 1 h, and LMP-1 mRNA levels were measured (Fig. 1E). CHX treatment did not block IFN-α-induced upregulation of LMP-1 but strongly enhanced it, proving that LMP-1 transcription is directly induced by type I IFN signaling.
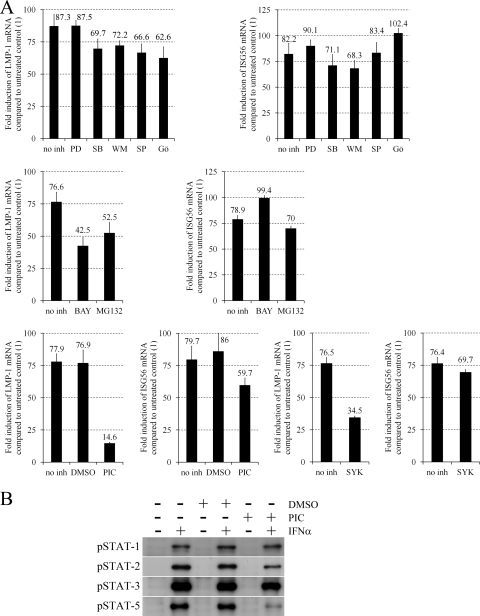
Type I IFNs activate the classical JAK/STAT and several alternate pathways, including the extracellular signal-regulated kinase 1 and 2 (ERK-1/2), p38 mitogen-activated protein kinase (MAPK), insulin receptor substrate (IRS)–phosphatidylinositol (PI) 3′-kinase, Jun N-terminal kinase (JNK), Crk, protein kinase C (PKC), and NF-κB pathways (31, 35, 36, 37, 43). First, we analyzed the role of the alternate pathways by measuring LMP-1 and ISG56 mRNA levels in Daudi cells treated with IFN-α or left untreated for 90 min in the presence or absence of inhibitors of MEK1 (PD98059; Calbiochem), p38 MAPK (SB203580; Calbiochem), PI 3′-kinase (wortmannin; Calbiochem), JNK-1, -2, and -3 (SP600125; Calbiochem), PKCα, -β, -γ, -δ, and -ε (Gö6850; Calbiochem), NF-κB (BAY117082 [Calbiochem] and MG132 [Enzo]) (Fig. 2A). PD98059 did not, while all other inhibitors slightly (SB203580, wortmannin, SP600125, Gö6850, and MG132) or moderately (BAY117082) prevented LMP-1 upregulation. SB203580, wortmannin, and MG132 minimally inhibited, while all other inhibitors did not affect or slightly enhanced ISG56 upregulation. These results suggest a partial role for the NF-κB, PKC, and JNK pathways in the type I IFN-induced transcription of LMP-1.
Fig 2.
Specific inhibition of IFN-α-induced LMP-1 mRNA upregulation by NF-κB inhibitors piceatannol and the Syk inhibitor BAY613606. (A) Fold induction of LMP-1 and ISG56 mRNA expression normalized to GAPDH, quantified by real-time RT-PCR in Daudi cells preincubated for 45 min without inhibitor (no inh) or with 10 μM PD98059 (PD), 5 μM SB203580 (SB), 1 μM wortmannin (WM), 10 μM SP600125 (SP), 5 μM Gö6850 (Gö) (upper panels), 2 μM BAY117082 (BAY), 5 μM MG132 (middle panels), 50 μM piceatannol (PIC), vehicle control (dimethyl sulfoxide [DMSO]), or 500 nM BAY613606 (SYK) (lower panels) and then left untreated or treated with 20 ng/ml IFN-α for an additional 90 min. Primers are listed in Table 1, and PCR conditions are described in the text. (B) Immunoblot analysis of phospho(Tyr701)–STAT-1, phospho(Tyr690)–STAT-2, phospho(Tyr705)–STAT-3, and phospho(Tyr694)–STAT-5 (all antibodies obtained from Cell Signaling) expression in total cell extracts of Daudi cells preincubated for 45 min without (−) or with (+) vehicle control (DMSO) or 50 μM PIC and then left untreated (−) or treated with 10 ng/ml IFN-α (+) for 20 min.
IFN-α activates STAT-1, -2, -3, -5, and -6 in human BL lines, mostly through the classical JAK/STAT pathway (9, 14, 43). The tyrosine kinase inhibitor piceatannol has been reported to selectively prevent the IFN-α-induced tyrosine phosphorylation of STAT-3 and -5, but not that of STAT-1 and -2 through the specific inhibition of Tyk2 kinase activity in the Ramos BL and Jurkat T cell lines (43). Our analysis using Daudi cells showed that piceatannol (Sigma-Aldrich) strongly inhibits phosphorylation of STAT-5, while it only slightly prevents STAT-2 and -3 phosphorylation and does not affect STAT-1 phosphorylation upon IFN-α treatment (Fig. 2B), providing an efficient tool for the selective analysis of the role of STAT-5 in the IFN-α-induced upregulation of LMP-1. Since piceatannol is a specific inhibitor of Syk tyrosine kinase as well, the effect of BAY613606, a highly selective inhibitor of Syk (49), was also tested. Daudi cells were treated with IFN-α or left untreated for 90 min in the presence or absence of piceatannol or BAY613606 (Sigma-Aldrich), and LMP-1 and ISG56 mRNA levels were measured (Fig. 2A). Piceatannol strongly inhibited while BAY613606 moderately inhibited LMP-1 upregulation, although both compounds only slightly blocked ISG56 upregulation. These results suggest that STAT-1, -2, and -3 tyrosine phosphorylation is not needed or is not sufficient for the direct upregulation of LMP-1 mRNA by IFN-α, while Tyk2 and Syk tyrosine kinase activities and STAT-5 tyrosine phosphorylation may play a role in it.
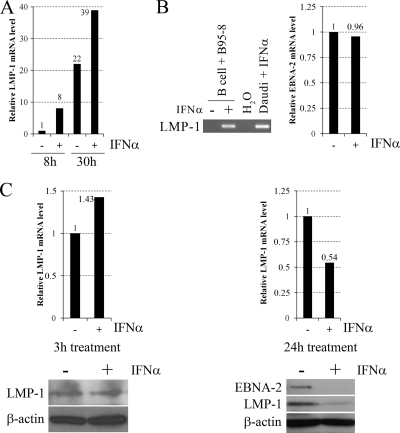
In order to validate the LMP-1-inducing effect of type I IFNs in primary B cells, peripheral blood B cells (purified by positive selection with CD19 Dynabeads [Invitrogen]) of healthy adult donors were infected with B95-8 virus and 8 or 30 h after infection were left untreated or treated with IFN-α for 90 min, when LMP-1 mRNA levels were measured. In B cells of donor 1 (Fig. 3A), IFN-α treatment upregulated LMP-1 mRNA levels by 8- and 1.8-fold at the 8- and 30-h time points, respectively. In B cells of donor 2 at the 8-h time point (Fig. 3B), LMP-1 mRNA could be detected only after IFN-α treatment, while EBNA-2 mRNA levels were nearly equal in both the untreated and treated samples. To analyze LMP-1 upregulation at the protein level, peripheral blood B cells were treated 24 h after EBV infection with IFN-α for 3 h (donor 3) or 24 h (donor 4), when LMP-1 mRNA and protein levels, together with the EBNA-2 protein level (only after the 24-h treatment), were measured (Fig. 3C). Three hours of IFN-α treatment upregulated LMP-1 mRNA expression by 1.43-fold, while the LMP-1 protein level did not change, suggesting inhibition of LMP-1 translation. Interestingly, 24 h of IFN-α treatment moderately reduced (by 46%) LMP-1 mRNA, while it strongly downregulated LMP-1 (by 80%) and EBNA-2 protein levels. Since these proteins play an essential role in EBV-induced B cell transformation (24), their downregulation may explain the previously reported partial inhibition of transformation by IFN-α when added 24 h postinfection (30, 47). Previous publications also reported the downregulation of EBNAs upon type I IFN treatment in EBV-infected peripheral blood lymphocytes (17), T cell-depleted mononuclear cells (5, 11), and B cells (2). However, in those studies, type I IFNs were added prior to or at the time of in vitro infection, when inhibition of EBV-mediated CD21 capping by IFN-α (6) may completely prevent later steps of the infection. Since viral entry and the transition to the circular form have already occurred by 24 h postinfection (19), inhibition of these steps cannot be responsible for the observed downregulation of EBNA-2 and LMP-1 in our experiments, suggesting a novel inhibitory mechanism of type I IFNs on EBV-induced B cell transformation. According to previous publications (2′–5′), oligoadenylates may play a role in this mechanism (17).
Fig 3.
Effect of IFN-α on LMP-1 expression in EBV-infected peripheral blood B cells. (A) Relative levels of LMP-1 mRNA normalized to GAPDH, quantified by real-time RT-PCR in B95-8 virus-infected peripheral blood B cells of donor 1, left untreated (−) or treated with 30 ng/ml IFN-α (+) for 90 min, 8 or 30 h after infection. (B) LMP-1 (left panel) and EBNA-2 (right panel) mRNA expression in B95-8 virus-infected peripheral blood B cells of donor 2, left untreated (−) or treated with 30 ng/ml IFN-α (+) for 90 min, 8 h after infection. LMP-1 mRNA expression was analyzed by RT-PCR and visualized on an ethidium bromide-stained agarose gel (Daudi cells treated with 20 ng/ml IFN-α for 1 h were used as a positive control). Relative levels of EBNA-2 mRNA normalized to GAPDH were quantified by real-time RT-PCR. (C) Relative levels of LMP-1 mRNA normalized to GAPDH, quantified by real-time RT-PCR (upper panels), and immunoblot analysis (lower panels) of EBNA-2 (only in B cells of donor 4), LMP-1 and β-actin protein expression in B95-8 virus-infected peripheral blood B cells of donor 3, left untreated (−) or treated with 30 ng/ml IFN-α (+) for 3 h (left panels), or of donor 4, left untreated (−) or treated with 30 ng/ml IFN-α (+) for 24 h (right panels), 24 h after infection. Quantification of immunoblots was performed by using Image J software (W. Rasband, NIH, Bethesda, MD). Primers are listed in Table 1, and the PCR conditions are described in the text. H2O, negative water control.
SLE is a complex, multifactorial disease involving genetic, epigenetic, and environmental risk factors, but it is now well-established that elevated levels of type I IFNs have a central role in its pathophysiology (46). Several lines of evidence suggest that EBV infection also plays an important role in the pathophysiology of SLE, possibly by antigenic cross-reactivity between viral antigens and self-antigens, and/or the rescue of preexisting autoreactive B cells from apoptosis (15, 34). LMP-1 mRNA is the most frequently detected latent EBV product in the increased population of EBV-infected memory B cells in the blood of SLE patients, although it is never detected in the blood of healthy individuals (13). Since LMP-1 induces B cells to express B cell-activating factor (BAFF) and a proliferation-inducing ligand (APRIL), which are mediators of B cell survival, T cell-independent antibody production, and immunoglobulin class switching (16), LMP-1 appears to be an important link between EBV infection and SLE. Since EBV-positive BLs originate from and therefore represent either late germinal center B cells or memory B cells (3, 48), our observations on the LMP-1-inducing effect of IFN-α in these lines and in primary B cells may explain the expression of LMP-1 mRNA in the memory B cells of SLE patients and suggest a direct connection between immune dysfunction and altered regulation of EBV latency in SLE. However, the lack of IFN-α-induced LMP-1 protein upregulation, despite its transcriptional induction in primary B cells, calls for careful analysis of LMP-1 protein expression in the memory B cells of SLE patients in future studies.
Nucleotide sequence accession numbers.
The LMP-1 coding and regulatory regions containing the primer binding sites were sequenced in Daudi cells and deposited in GenBank/EMBL/DDJB with accession numbers HE653895 and HE653896.
ACKNOWLEDGMENTS
This work was supported by grants from the The Swedish Cancer Society (Cancerfonden) and Karolinska Institutet. D.S., M.A., L.W., L.L.K., H.M., and N.N. are or were recipients of cancer research fellowships from the Cancer Research Institute (New York)/Concern Foundation (Los Angeles).
D.S., L.L.K., and E.K. designed the research; D.S., M.A., D.U., L.W., L.L.K., H.M., and N.N. performed research; D.S. and E.K. analyzed and interpreted data; D.S. created the figures; D.S., G.K., and E.K. wrote the manuscript. All authors read and commented on the manuscript. D.S. has access to all study materials and data and is responsible for the integrity of the study.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Altiok E, et al. 1992. Host-cell phenotype-dependent control of the BCR2/BWR1 promoter complex regulates the expression of Epstein-Barr virus nuclear antigens 2–6. Proc. Natl. Acad. Sci. U. S. A. 89:905–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andersson JP, Andersson UG, Ernberg IT, Britton SF, DeLey M. 1985. Effects of pure interferons on Epstein-Barr virus infection in vitro. J. Virol. 54:615–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bellan C, et al. 2005. Immunoglobulin gene analysis reveals 2 distinct cells of origin for EBV-positive and EBV-negative Burkitt lymphomas. Blood 106:1031–1036 [DOI] [PubMed] [Google Scholar]
- 4. Bornkamm GW, Hudewentz J, Freese UK, Zimber U. 1982. Deletion of the nontransforming Epstein-Barr virus strain P3HR-1 causes fusion of the large internal repeat to the DSL region. J. Virol. 43:952–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang RS. 1984. Interferons on the transformation of non-T blood mononuclear leukocytes by Epstein-Barr virus. J. Interferon Res. 4:173–177 [DOI] [PubMed] [Google Scholar]
- 6. Delcayre AX, Lotz M, Lernhardt W. 1993. Inhibition of Epstein-Barr virus-mediated capping of CD21/CR2 by alpha interferon (IFN-alpha): immediate antiviral activity of IFN-alpha during the early phase of infection. J. Virol. 67:2918–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ernberg I, et al. 1989. The role of methylation in the phenotype dependent modulation of Epstein-Barr nuclear antigen 2 and latent membrane protein genes in cells latently infected with Epstein-Barr virus. J. Gen. Virol. 70:2989–3002 [DOI] [PubMed] [Google Scholar]
- 8. Exley R, Gordon J, Nathan P, Walker L, Clemens MJ. 1987. Anti-proliferative effects of interferons on Daudi Burkitt lymphoma cells: induction of cell differentiation and loss of response to autocrine growth factors. Int. J. Cancer 40:53–57 [DOI] [PubMed] [Google Scholar]
- 9. Fasler-Kan E, Pansky A, Wiederkehr M, Battegay M, Heim HM. 1998. Interferon-alpha activates signal transducers and activators of transcription 5 and 6 in Daudi cells. Eur. J. Biochem. 254:514–519 [DOI] [PubMed] [Google Scholar]
- 10. Fensterl V, Sen GC. 2011. The ISG56/IFIT1 gene family. J. Interferon Cytokine Res. 31:71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garner JG, Hirsch MS, Schooley RT. 1984. Prevention of Epstein-Barr virus-induced B-cell outgrowth by interferon alpha. Infect. Immun. 43:920–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Geiger TR, Martin JM. 2006. The Epstein-Barr virus-encoded LMP-1 oncoprotein negatively affects Tyk2 phosphorylation and interferon signaling in human B cells. J. Virol. 80:11638–11650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gross AJ, Hochberg D, Rand WM, Thorley-Lawson DA. 2005. EBV and systemic lupus erythematosus: a new perspective. J. Immunol. 174:6599–6607 [DOI] [PubMed] [Google Scholar]
- 14. Gupta S, Jiang M, Pernis AB. 1999. IFN-alpha activates Stat6 and leads to the formation of Stat2:Stat6 complexes in B cells. J. Immunol. 163:3834–3841 [PubMed] [Google Scholar]
- 15. Harley JB, Harley IT, Guthridge JM, James JA. 2006. The curiously suspicious: a role for Epstein-Barr virus in lupus. Lupus 15:768–777 [DOI] [PubMed] [Google Scholar]
- 16. He B, Raab-Traub N, Casali P, Cerutti A. 2003. EBV-encoded latent membrane protein 1 cooperates with BAFF/BLyS and APRIL to induce T cell-independent Ig heavy chain class switching. J. Immunol. 171:5215–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henderson EE, Doetsch PW, Charubala R, Pfleiderer W, Suhadolnik RJ. 1982. Inhibition of Epstein-Barr virus-associated nuclear antigen (EBNA) induction by (2′,5′)oligoadenylate and the cordycepin analog: mechanism of action for inhibition of EBV-induced transformation. Virology 122:198–201 [DOI] [PubMed] [Google Scholar]
- 18. Hudson GS, Farrell PJ, Barrell BG. 1985. Two related but differentially expressed potential membrane proteins encoded by the EcoRI Dhet region of Epstein-Barr virus B95-8. J. Virol. 53:528–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hurley EA, Thorley-Lawson DA. 1988. B cell activation and the establishment of Epstein-Barr virus latency. J. Exp. Med. 168:2059–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ishii H, et al. 2012. Monocytes enhance cell proliferation and LMP1 expression of nasal natural killer/T-cell lymphoma cells by cell contact-dependent interaction through membrane-bound IL-15. Int. J. Cancer 130:48–58 [DOI] [PubMed] [Google Scholar]
- 21. Iwakiri D, et al. 2009. Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. J. Exp. Med. 206:2091–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones MD, Foster L, Sheedy T, Griffin BE. 1984. The EB virus genome in Daudi Burkitt's lymphoma cells has a deletion similar to that observed in a non-transforming strain (P3HR-1) of the virus. EMBO J. 3:813–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kelly G, Bell A, Rickinson A. 2002. Epstein-Barr virus-associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat. Med. 8:1098–1104 [DOI] [PubMed] [Google Scholar]
- 24. Kieff E, Rickinson AB. 2007. Epstein-Barr virus and its replication, p 2603–2654 In Fields BN, Knipe DM, Howley PM. (ed), Fields virology, 5th ed Lippincott-Williams & Wilkins Publishers, Philadelphia, PA [Google Scholar]
- 25. Kikuta H, Mizuno F, Yano S, Osato T. 1984. Interferon production by Epstein-Barr virus in human mononuclear leukocytes. J. Gen. Virol. 65:837–841 [DOI] [PubMed] [Google Scholar]
- 26. Kis LL, et al. 2011. STAT6 signaling pathway activated by the cytokines IL-4 and IL-13 induces expression of the Epstein-Barr virus-encoded protein LMP-1 in absence of EBNA-2: implications for the type II EBV latent gene expression in Hodgkin lymphoma. Blood 117:165–174 [DOI] [PubMed] [Google Scholar]
- 27. Kis LL, et al. 2010. IL-21 imposes a type II EBV gene expression on type III and type I B cells by the repression of C- and activation of LMP-1-promoter. Proc. Natl. Acad. Sci. U. S. A. 107:872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kis LL, Takahara M, Nagy N, Klein G, Klein E. 2006. IL-10 can induce the expression of EBV-encoded latent membrane protein-1 (LMP-1) in the absence of EBNA-2 in B lymphocytes and in Burkitt lymphoma- and NK lymphoma-derived cell lines. Blood 107:2928–2935 [DOI] [PubMed] [Google Scholar]
- 29. Klein E, et al. 1968. Surface IgM-kappa specificity on a Burkitt lymphoma cell in vivo and in derived culture lines. Cancer Res. 28:1300–1310 [PubMed] [Google Scholar]
- 30. Lotz M, Tsoukas CD, Fong S, Carson DA, Vaughan JH. 1985. Regulation of Epstein-Barr virus infection by recombinant interferons. Selected sensitivity to interferon-gamma. Eur. J. Immunol. 15:520–525 [DOI] [PubMed] [Google Scholar]
- 31. Maher SG, Romero-Weaver AL, Scarzello AJ, Gamero AM. 2007. Interferon: cellular executioner or white knight? Curr. Med. Chem. 14:1279–1289 [DOI] [PubMed] [Google Scholar]
- 32. Masucci MG, et al. 1989. 5-Azacytidine up regulates the expression of Epstein-Barr virus nuclear antigen 2 (EBNA-2) through EBNA-6 and latent membrane protein in the Burkitt's lymphoma line Rael. J. Virol. 63:3135–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Minarovits J, Hu LF, Minarovits-Kormuta S, Klein G, Ernberg I. 1994. Sequence-specific methylation inhibits the activity of the Epstein-Barr virus LMP 1 and BCR2 enhancer-promoter regions. Virology 200:661–667 [DOI] [PubMed] [Google Scholar]
- 34. Niller HH, Wolf H, Ay E, Minarovits J. 2011. Epigenetic dysregulation of Epstein-Barr virus latency and development of autoimmune disease. Adv. Exp. Med. Biol. 711:82–102 [DOI] [PubMed] [Google Scholar]
- 35. Pfeffer LM. 2011. The role of nuclear factor κB in the interferon response. J. Interferon Cytokin Res. 31:553–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pfeffer LM, et al. 1991. Transmembrane signaling by interferon alpha involves diacylglycerol production and activation of the epsilon isoform of protein kinase C in Daudi cells. Proc. Natl. Acad. Sci. U. S. A. 88:7988–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Platanias LC. 2005. Mechanisms of type-I- and type-II-interferon-mediated signaling. Nat. Rev. Immunol. 5:375–386 [DOI] [PubMed] [Google Scholar]
- 38. Quan TE, Roman RM, Rudenga BJ, Holers VM, Craft JE. 2010. Epstein-Barr virus promotes interferon-alpha production by plasmacytoid dendritic cells. Arthritis Rheum. 62:1693–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rickinson AB, Kieff E. 2007. Epstein-Barr virus, p 2655–2700 In Fields BN, Knipe DM, Howley PM. (ed), Fields virology, 5th ed Lippincott-Williams & Wilkins Publishers, Philadelphia, PA [Google Scholar]
- 40. Sadler RH, Raab-Traub N. 1995. The Epstein-Barr virus 3.5-kilobase latent membrane protein 1 mRNA initiates from a TATA-less promoter within the first terminal repeat. J. Virol. 69:4577–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salamon D, et al. 2012. Type I interferons directly down-regulate BCL-6 in primary and transformed germinal center B cells: differential regulation in B cell lines derived from endemic or sporadic Burkitt's lymphoma. Cytokine 57:360–371 [DOI] [PubMed] [Google Scholar]
- 42. Schneider U, Schwenk HU, Bornkamm G. 1977. Characterization of EBV-genome negative “null” and “T” cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int. J. Cancer 19:621–626 [DOI] [PubMed] [Google Scholar]
- 43. Su L, David M. 2000. Distinct mechanisms of STAT phosphorylation via the interferon-alpha/beta receptor. Selective inhibition of STAT3 and STAT5 by piceatannol. J. Biol. Chem. 275:12661–12666 [DOI] [PubMed] [Google Scholar]
- 44. Takacs M, et al. 2001. Epigenetics of latent Epstein-Barr virus genomes: high resolution methylation analysis of the bidirectional promoter region of latent membrane protein 1 and 2B genes. Biol. Chem. 382:699–705 [DOI] [PubMed] [Google Scholar]
- 45. Takada K, et al. 1991. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus Genes 5:147–156 [DOI] [PubMed] [Google Scholar]
- 46. Theofilopoulos AN, Baccala R, Beutler B, Kono DH. 2005. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 23:307–336 [DOI] [PubMed] [Google Scholar]
- 47. Thorley-Lawson DA. 1981. The transformation of adult but not newborn human lymphocytes by Epstein Barr virus and phytohemagglutinin is inhibited by interferon: the early suppression by T cells of Epstein Barr infection is mediated by interferon. J. Immunol. 126:829–833 [PubMed] [Google Scholar]
- 48. Thorley-Lawson DA, Allday MJ. 2008. The curious case of the tumour virus: 50 years of Burkitt's lymphoma. Nat. Rev. Microbiol. 6:913–924 [DOI] [PubMed] [Google Scholar]
- 49. Yamamoto N, et al. 2003. The orally available spleen tyrosine kinase inhibitor 2-[7-(3,4-dimethoxyphenyl)-imidazo[1,2-c]pyrimidin-5-ylamino]-nicotinamide dihydrochloride (BAY 61-3606) blocks antigen-induced airway inflammation in rodents. J. Pharmacol. Exp. Ther. 306:1174–1181 [DOI] [PubMed] [Google Scholar]