Abstract
To investigate the requirements of herpesvirus entry and fusion, the four homologous glycoproteins necessary for herpes simplex virus (HSV) fusion were cloned from herpes B virus (BV) (or macacine herpesvirus 1, previously known as cercopithecine herpesvirus 1) and cercopithecine herpesvirus 2 (CeHV-2), both related simian simplexviruses belonging to the alphaherpesvirus subfamily. Western blots and cell-based enzyme-linked immunosorbent assay (ELISA) showed that glycoproteins gB, gD, and gH/gL were expressed in whole-cell lysates and on the cell surface. Cell-cell fusion assays indicated that nectin-1, an HSV-1 gD receptor, mediated fusion of cells expressing glycoproteins from both BV and CeHV-2. However, herpesvirus entry mediator (HVEM), another HSV-1 gD receptor, did not facilitate BV- and CeHV-2-induced cell-cell fusion. Paired immunoglobulin-like type 2 receptor alpha (PILRα), an HSV-1 gB fusion receptor, did not mediate fusion of cells expressing glycoproteins from either simian virus. Productive infection with BV was possible only with nectin-1-expressing cells, indicating that nectin-1 mediated entry while HVEM and PILRα did not function as entry receptors. These results indicate that these alphaherpesviruses have differing preferences for entry receptors. The usage of the HSV-1 gD receptor nectin-1 may explain interspecies transfer of the viruses, and altered receptor usage may result in altered virulence, tropism, or pathogenesis in the new host. A heterotypic cell fusion assay resulting in productive fusion may provide insight into interactions that occur to trigger fusion. These findings may be of therapeutic significance for control of deadly BV infections.
INTRODUCTION
The family Herpesviridae is a large, diverse family of double-stranded enveloped DNA viruses. B virus (BV) (or macacine herpesvirus 1, herpesvirus simiae, monkey B virus, or cercopithecine herpesvirus 1) and cercopithecine herpesvirus 2 (CeHV-2) (or simian agent 8) are primate herpesviruses belonging to the alphaherpesvirus subfamily and as such are closely related to herpes simplex virus 1 (HSV-1). The HSV-1 entry process and virus-induced cell fusion require glycoproteins B (gB), D (gD), H (gH), and L (gL). Binding of gD to a cellular entry receptor is required for triggering membrane fusion. To date, four gD receptors, herpesvirus entry mediator (HVEM) (38), nectin-1 (6, 16, 36, 37, 48), nectin-2 (31, 62), and modified heparan sulfate (49, 50), have been identified. More recently, three gB receptors, the paired immunoglobulin-like type 2 receptor alpha (PILRα) (46), myelin-associated glycoprotein (MAG) (2), and nonmuscle myosin heavy chain IIA (NMHC-IIA) (2), have been identified. HVEM is a member of the tumor necrosis factor receptor family (61). Nectin-1 and nectin-2 are cell adhesion molecules that belong to the immunoglobulin superfamily and are widely expressed by a variety of cell types, including epithelial cells and neurons (53). Modified heparan sulfate generated by particular 3-O-sulfotransferases can also serve as a gD-binding entry receptor (50). PILRα was identified as an entry receptor that binds to gB (46). PILRα is expressed on cells of the immune system, including monocytes, dendritic cells, NK cells, B cells, macrophages, neutrophils, eosinophils, mast cells, and megakaryocytes/platelets and neurons (15, 28, 39, 40, 46, 47, 57). MAG is a cell surface molecule that is preferentially expressed in neural tissues, especially on myelin sheath, and plays an important role in the regulation of axonal growth (3, 30, 35, 60). NMHC-IIA is expressed in a wide variety of cultured cell lines and in various tissues and cell types in vivo (17, 58).
HSV-1 causes recurrent mucocutaneous lesions on the mouth, face, or genitalia and occasionally meningitis or encephalitis. BV naturally infects macaques but is one of the most deadly viruses for humans. Previous studies have shown that only BV is known to be pathogenic for humans among the 35 herpesviruses identified in nonhuman primates (20). BV naturally infects macaque monkeys, and infections in foreign hosts often result in encephalitis, encephalomyelitis, and death (11, 21, 63). CeHV-2 was initially isolated from an African green monkey in 1958 but has subsequently been recognized as a pathogen of baboons and classified as a herpesvirus on the basis of its characteristics in cell culture and neurotropism in monkeys and in experimentally inoculated rabbits (32). CeHV-2 is not known to cause diseases in primates outside the natural hosts. The virulence patterns displayed by HSV-1, BV, and CeHV-2 in humans encouraged us to explore the role of known HSV-1 receptors in the infectious cycle of these viruses. The complete genome sequences of BV (42) and CeHV-2 (56) provided further impetus, allowing the investigation of the relevant glycoproteins as well as a tool for comparinge the entry and fusion of these alphaherpesviruses at the molecular level.
Here we report the receptor usage of BV and CeHV-2 herpesviruses using cell-cell fusion and infection. We also examined fusion activity of the various glycoproteins by using substitution, which may provide a basis for identifying functionally interactive domains of HSV-1, BV, and CeHV-2 entry glycoproteins.
MATERIALS AND METHODS
Cells.
Chinese hamster ovary (CHO-K1; ATCC) cells, Vero cells, and CHO cells stably expressing human HVEM (38), nectin-1 (16), or PILRα were used in this study. The human PILRα cell line was isolated after transfection of pQF003 (12), a plasmid expressing human PILRα. The CHO-K1 cell line and its derivatives were grown in Ham's F12 medium supplemented with 10% fetal bovine serum (FBS). The Vero cell line was grown in Dulbecco minimal essential medium containing 10% fetal bovine serum.
BV and biosafety.
BV strain E2490 (NCBI reference sequence no. NC_004812) was used for the in vitro experiments in this study. BV is classified as a risk group 4 agent (5a) and may be propagated only in a maximum-containment laboratory. All experiments using infectious virus were performed in the ABSL-4 (animal biosafety level 4) laboratory at the Texas Biomedical Research Institute (certified by the CDC) by trained personnel wearing protective biosafety suits with the approval of the Texas Biomedical Research Institute Biohazard and Recombinant DNA Committees. Plasmid-based cell-cell fusion experiments at Northwestern University were done using subgenomic expression constructs of the relevant glycoproteins and with the approval of the Northwestern University Institutional Biosafety and Recombinant DNA Committees.
Plasmids.
Plasmids expressing HSV-1(KOS) gB (pPEP98), gD (pPEP99), gH (pPEP100), and gL (pPEP101) were previously described (43), as were plasmids expressing human nectin-1, pBG38 (16), HVEM, pBEC10 (38), African green monkey (AGM) HVEM cloned from Vero cells (14), and PILRα (pQF003) (12). Glycoproteins from open reading frames UL27 (gB), US6 (gD), UL22 (gH), and UL1 (gL) were cloned using viral DNA from BV and CeHV-2 (Table 1). The BV gD, gB, gH, and gL genes were amplified using viral DNA from strain E2490, harvested from Vero-infected cells under biosafety level 4 (BSL-4) conditions. BV genes were first cloned into a TA cloning vector (pCR2.1TOPO; Invitrogen) and were subsequently used to generate the FLAG-tagged and untagged constructs listed in Table 1. FLAG-tagged BV gB, gD, gH, and gL were generated by subcloning the glycoproteins (without the native signal sequence from amino acids 1 to 34 for gB, 1 to 24 for gD, 1 to 18 for gH, and 1 to 19 for gL) into the pFLAG-myc-CMV-21 expression vector (E5776; Sigma) downstream of the FLAG peptide.
Table 1.
Plasmids generated for this study
| Construct | Protein | Commenta |
|---|---|---|
| pQF087 | CeHV-2 gH | PCR product of CeHV-2 gH ORF from CeHV-2 viral DNA was cloned into pCDNA3 |
| pQF088 | CeHV-2 gL | PCR product of CeHV-2 gL ORF from CeHV-2 viral DNA was cloned into pCDNA3 |
| pQF089 | CeHV-2 FLAG-gB | PCR product of CeHV-2 gB ORF from CeHV-2 viral DNA was cloned into pFLAG-myc-CMV-21 (sigma) |
| pQF090 | CeHV-2 FLAG-gD | PCR product of CeHV-2 gD ORF from CeHV-2 viral DNA was cloned into pFLAG-myc-CMV-21 |
| pQF091 | CeHV-2 FLAG-gH | PCR product of CeHV-2 gH ORF from CeHV-2 viral DNA was cloned into pFLAG-myc-CMV-21 |
| pQF092 | CeHV-2 FLAG-gL | PCR product of CeHV-2 gL ORF from CeHV-2 viral DNA was cloned into pFLAG-myc-CMV-21 |
| pQF097 | BV gH | PCR product of BV gH ORF from BV viral DNA was cloned into pCDNA3 |
| pQF098 | BV gL | PCR product of BV gL ORF from BV viral DNA was cloned into pCDNA3 |
| pQF0106 | BV FLAG gB | PCR product of BV gB ORF from BV viral DNA was cloned into pFLAG-myc-CMV-21 |
| pQF100 | BV FLAG-gD | PCR product of BV gD ORF from BV viral DNA was cloned into pFLAG-myc-CMV-21 |
| pQF101 | BV FLAG gH | PCR product of BV gH ORF from BV viral DNA was cloned into pFLAG-myc-CMV-21 |
| pQF102 | BV FLAG gL | PCR product of BV gL ORF from BV viral DNA was cloned into pFLAG-myc-CMV-21 |
ORF, open reading frame.
CeHV-2 (SA8) strain B264 (ATCC no. VR-936; GenBank accession no. AY714813) was cultured in a Vero cell line (ATCC number Cl-81). Viral DNA was extracted from infected cells. FLAG-tagged and untagged constructs were generated from PCR amplification of CeHV-2 viral DNA. FLAG-tagged CeHV-2 gB, gD, gH, and gL were generated by subcloning the glycoproteins (without the native signal sequence from amino acids 1 to 28 for gB, 1 to 24 for gD, 1 to 37 for gH, and 1 to 24 for gL, respectively) into the pFLAG-myc-CMV-21 expression vector downstream of the FLAG peptide. The signal peptides from the BV and CeHV-2 glycoproteins were predicted by using the SignalP 4.0 server (http://www.cbs.dtu.dk/services/SignalP/). All plasmids made for this study were sequenced by the Northwestern Genomics Core facility. Glycoproteins from BV and CeHV-2 match amino acid sequences published on NCBI with accession numbers NC_004812 and AY714813, respectively.
CELISA.
Cell-based enzyme-linked immunosorbent assay (CELISA) was used to check the cell surface expression of the expression constructs. CHO-K1 cells seeded in 96-well plates were transfected with 60 ng of empty vector or plasmids expressing FLAG-tagged gB, gD, and gH/gL (30 ng DNA for gH and 30 ng DNA for gL) from BV and CeHV-2 and 0.15 μl of Lipofectamine 2000 (Invitrogen), both diluted in Opti-MEM (Invitrogen). The cells were washed once with phosphate-buffered saline (PBS) 24 h after transfection, and CELISA was performed as described previously (29). Briefly, after incubation of the live cells with the FLAG monoclonal antibody (F1084; Sigma), the cells were washed, fixed, and incubated with biotinylated goat anti-mouse IgG (Sigma), followed by streptavidin-horseradish peroxidase (HRP) (GE Healthcare) and an HRP substrate (BioFX).
Western blots.
Western blotting was performed to check the whole-cell lysate expression of glycoproteins from BV and CeHV-2. CHO-K1 cells seeded in 6-well plates were transfected with 1.5 μg of empty vector or a plasmid expressing FLAG-tagged gB, gD, gH, and gL from BV and CeHV-2 and 5 μl of Lipofectamine 2000. After 24 h of incubation, the cells were detached using Versene (0.2 g EDTA/liter in PBS), washed with PBS, and lysed with 200 μl of lysis buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 10 mM NaF, 1 mM Na3VO3, and 1% Nonidet P-40) containing a protease inhibitor mixture (Roche Diagnostics, Indianapolis, IN). Proteins were separated by SDS-PAGE on 4 to 20% gels after boiling for 5 min under reducing conditions. Western blot analyses were performed using rabbit anti-FLAG (Sigma, F7425) at a 1:1,000 dilution for 1 h at room temperature. Anti-rabbit secondary antibody coupled to HRP and ECL Western blotting detection reagents (GE Healthcare) were used.
Cell fusion assay.
The assay was done as previously described (43). CHO-K1 cells were seeded in 6-well plates 1 day before transfection. The CHO-K1 cells (effector cells) were transfected with 400 ng (each) of plasmids expressing T7 RNA polymerase, gB, gD, gH, and gL from HSV-1, FLAG-tagged gB, gD, gH, and gL from BV or CeHV-2, and 5 μl of Lipofectamine 2000. The target CHO-K1 cells were transfected with 400 ng of a plasmid carrying the firefly luciferase gene under the control of the T7 promoter, 1.5 μg of empty vector (pcDNA3), or a plasmid expressing either human PILRα (pQF003), human HVEM (pBEC10), simian HVEM homolog (HVEMs), or human nectin-1 (pBG38) and 5 μl of Lipofectamine 2000. Six h after transfection, the cells were detached with Versene and suspended in 1.5 ml of F12 medium supplemented with 10% FBS. Effector and target cells were mixed in a 1:1 ratio and replated in 96-well plates for 18 h. Luciferase activity was quantitated by a luciferase reporter assay system (Promega) using a Wallac-Victor luminometer (Perkin Elmer).
BV infections, RNA isolation, and quantitative real-time PCR.
Sixty-millimeter dishes of 80%-confluent Vero, CHO-K1, or CHO cells stably expressing human HVEM (38), nectin-1 (16), or PILRα were infected with BV at a multiplicity of infection (MOI) of 5. At 1 h postinfection (p.i.), cells were scraped from the dishes and pelleted in a low-speed centrifuge. After removing the supernatant, RNA was harvested using the mirVana microRNA (miRNA) isolation kit (Ambion) according to the manufacturer's instructions. The infections were repeated three times with all cell types.
Infected cell protein 0 (ICP0), an immediate-early protein of HSV-1, activates viral and cellular gene expression and functions as an E3 ubiquitin ligase that degrades several cellular proteins (18). ICP0 plays a major role in establishing permissive conditions for viral infection and therefore was used to detect BV infection to determine whether the cells were BV infected; a previously developed reverse transcription-quantitative PCR (RT-qPCR) assay to detect expression of the immediate-early gene ICP0 was used (1). Briefly, RNA was subjected to reverse transcription using 500 nM ICP0 reverse transcription primer (5′-AACTGGTGCCCCTACCAC-3′) (IDT), 5× iScript select reaction mix (Bio-Rad), GSP enhancer solution (Bio-Rad), and iScript reverse transcriptase (Bio-Rad) in a total volume of 20 μl. RTs were carried out at 42°C for 60 min and 85°C for 5 min. One microliter of the resulting cDNA was added to 1× iQ SYBR green supermix (Bio-Rad), 500 nM ICP0 forward primer (5′-CAGACGTGCCTCGCGTA-3′) (IDT), and 500 nM ICP0 reverse primer (5′-TCGACAACGCGTACCCG-3′) (IDT) in a total volume of 25 μl. The reactions were carried out in triplicate for each cell type for all three infections. To estimate the number of molecules of ICP0 mRNA in each cell type, standard curves were generated using RNA isolated from uninfected Vero cells with a known amount of ICP0 RNA in vitro transcribed using the mMessage mMachine kit (Ambion) from pMAICP0, which contains a portion of the second exon of the immediate-early gene encoding ICP0 (1). Reverse transcription and qPCR were performed using the iScript Select cDNA synthesis kit (Bio-Rad) and iQ SYBR green supermix (Bio-Rad), respectively, according to the manufacturer's instructions.
Heterotypic cell fusion assay.
A heterotypic cell fusion assay was performed similarly to the cell fusion assay described above. The only difference was that gB, gD, gH, gL, or gH/gL of one virus was replaced by a homologous glycoprotein(s) of another virus during transfection of the CHO-K1 cells (effector cells), and the fusion experiment was done as described above.
RESULTS
Cloning and expression of glycoproteins from BV and CeHV-2.
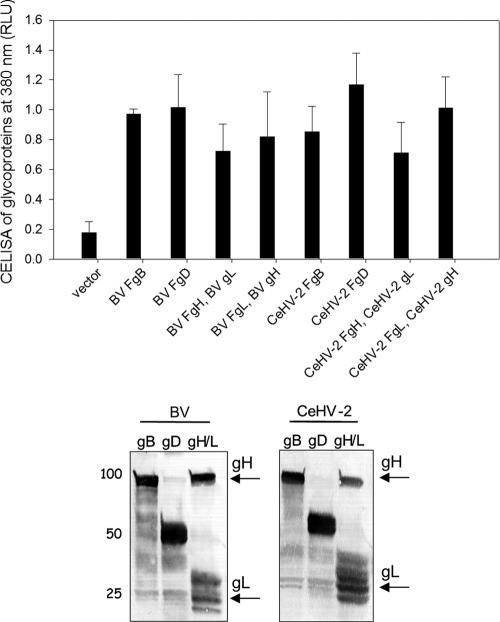
To begin our studies, we first generated FLAG-tagged, gB, gD, gH, and gL and untagged gH and gL for BV and CeHV-2 as shown in Table 1 and described in detail in Materials and Methods. To verify expression of the various constructs shown in Table 1, CHO-K1 cells were transfected as previously done (13) with the FLAG-tagged glycoprotein expression constructs. Since gH requires gL as a chaperone, these two proteins were cotransfected using two different transfections in which only gH or gL was FLAG tagged. For gD and gB, the FLAG-tagged constructs were transfected alone. To monitor cell surface expression, a CELISA was done as previously described (29). For all of the expression constructs, high levels of expression FLAG epitope were readily detected on the cell surface for all transfections (Fig. 1, upper panel). To further confirm expression, Western blotting of total cell lysate was performed (Fig. 1, lower panel). FLAG-tagged gH and gL were transfected together, and FLAG-tagged gD and gB were transfected separately. Each of the glycoproteins was expressed at the expected size
Fig 1.
Expression of gB, gD, and gH/L from BV and CeHV-2 on the cell surface by CELISA and in whole-cell lysates by Western blotting. The upper panel shows the CELISA for cell surface expression of the glycoproteins from BV and CeHV-2. CHO cells were transfected in a 96-well plate with gB, gD, or gH/gL (F indicates a FLAG-tagged glycoprotein) and empty vector. FLAG-tagged gH (or gL) was cotransfected with wild-type (WT) gL (or gH), which was cloned into pCAGGS. The cells were washed and incubated with an anti-FLAG M2 antibody and washed extensively prior to fixation and incubation with a mouse secondary antibody and an HRP detection system. Each bar shows the mean of 3 independent determinations, with the results of actual absorbance at 380 nm. The lower panel shows the Western blot experiments analyzing the expression of glycoproteins. CHO cells expressing wild-type FLAG-tagged gB, gD, and gH/gL were lysed and were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with rabbit anti-FLAG antibody followed by goat anti-rabbit IgG. gB, gD, gH, and gL run at the expected molecular weights.
BV human receptor usage in fusion.
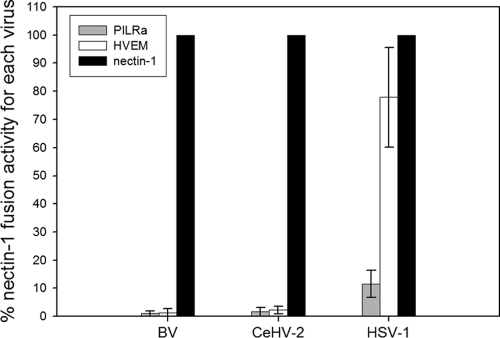
Fusion experiments were performed by transfecting target CHO-K1 cells with a plasmid encoding luciferase under a T7 promoter and either empty vector, PILRα, nectin-1, or HVEM. Effector CHO-K1 cells were transfected with a plasmid encoding T7 RNA polymerase and either gB, gD, gH, and gL from HSV-1, FLAG-tagged gB, gD, gH, and gL from BV, or FLAG-tagged gB, gD, gH, and gL from CeHV-2. Effector and target cells were mixed, and luciferase activity was recorded as a measure of cell-cell fusion. The background luciferase activity recorded when target cells were transfected with empty vector was subtracted from each experiment, and that for each virus fusion with target cells expressing nectin-1 was set at 100% (Fig. 2). For BV and CeHV-2 glycoproteins, fusion with nectin-1-expressing target cells was significantly greater than that observed with either PILRα- or HVEM-expressing target cells. In contrast, levels of fusion for HSV-1 glycoproteins mediated by HVEM and nectin-1 were quite similar, with HVEM showing a modest reduction in fusion activity compared to nectin-1 (78% of nectin-1 levels), which was similar to what has previously been observed (43, 55). Also as previously observed (12), the levels of PILRα-mediated fusion for HSV-1 were much lower than that of nectin-1- or HVEM-mediated fusion. Fusion with PILRα-expressing target cells for both BV and CeHV-2 was slightly above the background level and much lower than that for HSV-1 (1% for BV, 2% for CeHV-2, and 12% for HSV-1) compared to nectin-1 fusion levels. Most interestingly, there was a dramatic difference in levels of fusion for HVEM compared to levels of fusion for nectin-1. Fusion mediated by HVEM was 1% (for BV), 2% for (CeHV-2), or 78% (for HSV-1) of the levels of fusion mediated by nectin-1. These results suggest that nectin-1 is the receptor for BV and CeHV-2, and HVEM and PILRα do not function as entry receptors for these viruses.
Fig 2.
Cell fusion activities of BV, CeHV-2, and HSV-1 mediated by HSV-1 receptors. Target CHO cells were transfected with pCDNA3 (empty vector), HVEM, PILRα, or nectin-1, along with a reporter plasmid expressing luciferase under the control of the T7 promoter. The transfected cells were replated with effector CHO cells transfected with gB, gD, gH, and gL from HSV-1 or FLAG-tagged homologous glycoproteins from BV and CeHV-2, along with a plasmid expressing T7 polymerase. Cell fusion activity mediated by each receptor is presented as a percentage of that for nectin-1(each virus set as 100%) after subtracting the data from the empty vector. Each bar shows the mean and standard deviation of at least three independent determinations. Levels of overall nectin-1 fusion for BV and CeHV2 were approximately 35% of the levels observed for HSV-1 when relative luciferase levels were compared between the different viral fusion assays.
BV human receptor usage in infection.
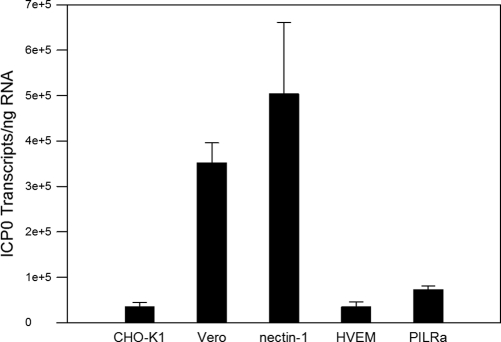
To confirm the cell fusion results, cell lines stably expressing human nectin-1, HVEM, or PILRα were infected with BV. CHO-K1 cell and Vero cells were used as negative and positive controls, respectively. CHO cells are resistant to the entry of HSV and other viruses, such as pseudorabies virus (PRV), hepatitis B virus (HBV), and HIV, because receptors for these viruses are either missing in the CHO genome or lacking expression in the CHO-K1 transcriptome (65), so they are poor target cells in cell fusion assays unless transfected to express appropriate entry/fusion receptors. Since simplexviruses do not form plaques on CHO-K1 cells and in the absence of a recombinant BV carrying an appropriate marker to monitor infection, infection was quantified by measuring the expression of the viral immediate-early gene encoding ICP0. RNA was used to perform RT-qPCR for ICP0 mRNA transcripts. The results showed that after 60 min of infection, the number of ICP0 transcripts in Vero cells was 3.52 × 105 per ng of harvested RNA, while the number of ICP0 transcripts in nectin-1 cells was 5.04 × 105 (139% of the Vero cells) (Fig. 3). However, the number of ICP0 transcripts in HVEM and PILRα was 3.5 × 104 per ng of harvested RNA and 7.3 × 104 per ng of harvested RNA, or 9.8% and 18.8% of the levels in Vero cells, respectively, similar to results for CHO-K1 cells (10% of the Vero cells) (Fig. 3). These results indicate that BV uses human nectin-1 as the functional entry receptor but does not use human HVEM or PILRα as an entry receptor.
Fig 3.
ICP0 transcripts in BV-infected cells. Vero E6, CHO-K1, and CHO cells stably expressing human HVEM, nectin-1 (16), and PILRα were infected with BV at an MOI of 5. RNA was harvested at 1 h p.i., and quantitative real-time PCR was performed to determine the ICP0 transcript number. The data are presented as transcripts of ICP0 per ng of harvested RNA. Each bar shows the mean and standard deviation of results from three independent experiments.
BV and CeHV-2 do not utilize African green monkey HVEM.
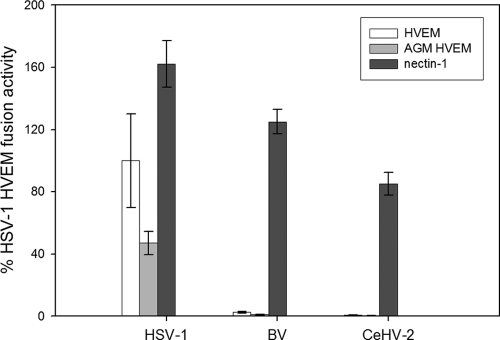
According to the cell fusion assay and infection assay, BV did not utilize human HVEM as an entry receptor. We next chose to determine if BV and CeHV-2 use African green monkey (AGM) HVEM as a receptor. To answer this question, we performed cell-cell fusion by transfecting target CHO K1 cells with a plasmid encoding luciferase under a T7 promoter and either empty vector, nectin-1, AGM HVEM, or human HVEM. Effector CHO-K1 cells were transfected with a plasmid encoding T7 polymerase and either gB, gD, gH, and gL from HSV-1, FLAG-tagged gB, gD, gH, and gL from BV, or FLAG-tagged gB, gD, gH, and gL from CeHV-2. The empty vector background was subtracted from each experiment, and HSV-1 glycoprotein-mediated fusion with human HVEM-expressing target cells was set to 100% (Fig. 4). Nectin-1 was used as a positive control, and it showed similar results, as shown in Fig. 2. HSV-1 can efficiently use simian HVEM (Fig. 4), agreeing with findings of previous studies (14). HSV-1 glycoproteins mediated fusion with AGM HVEM-expressing target cells at about 50% of the level of fusion seen with human HVEM-expressing target cells. However, BV and CeHV-2 glycoproteins mediated fusion with AGM HVEM-expressing target cells at only 1% or 0%, respectively, compared to the level of fusion mediated by HSV-1 glycoproteins using human HVEM. BV and CeHV-2 did not use human HVEM (Fig. 4), similar to results shown in Fig. 2.
Fig 4.
Cell fusion activities of BV, CeHV-2, and HSV-1 mediated by African green monkey (AGM) HVEM. Target CHO cells were transfected with pCDNA3 (empty vector), HVEM, AGM HVEM, or nectin-1, along with a reporter plasmid expressing luciferase under the control of the T7 promoter. The transfected cells were replated with effector CHO cells transfected with gB, gD, gH, and gL from HSV-1 or FLAG-tagged homologous glycoproteins from BV and CeHV-2, along with plasmids expressing T7 polymerase. Cell fusion activity mediated by each receptor is presented as a percentage of that of human HVEM to HSV-1 (set as 100%) after subtracting the data from the empty vector. Each bar shows the mean and standard deviation of at least three independent determinations.
Heterotypic cell fusion assay.
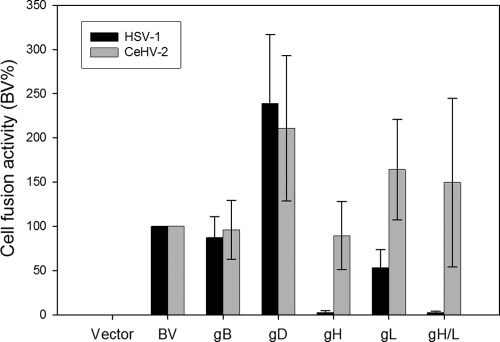
To determine whether gB, gD, gH, and gL could exhibit functional heterotypic interactions, we singularly replaced each virus glycoprotein or both gH/gL with their counterparts in HSV-1, BV, and CeHV-2 and determined the extent to which the heterotypic combinations could mediate fusion with nectin-1-expressing target cells. The empty vector background was subtracted from each experiment, and cell fusion activity was scored as a percentage of fusion when a homotypic set of HSV-1, BV, and CeHV-2 glycoproteins was used (set at 100%). The data are summarized in Table 2, and representative data are shown in Fig. 5.
Table 2.
Heterotypic fusion activities of HSV-1, BV, and CeHV-2 mediated by nectin-1a
| Source of substitution glycoprotein | Relative fusion activity |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HSV-1 glycoprotein set |
BV glycoprotein set |
CeHV-2 glycoprotein set |
|||||||||||||
| gB | gD | gH | gL | gH/L | gB | gD | gH | gL | gH/L | gB | gD | gH | gL | gH/L | |
| HSV-1 | +++ | +++ | +++ | +++ | +++ | +++ | ++++ | − | ++ | − | ++ | ++ | − | + | − |
| BV | ++ | − | − | − | + | +++ | +++ | +++ | +++ | +++ | ++++ | ++ | ++ | ++ | +++ |
| CeHV-2 | +++ | + | − | − | + | +++ | ++++ | +++ | ++++ | ++++ | +++ | +++ | +++ | +++ | +++ |
CHO effector cells were transfected with plasmids expressing T7 RNA polymerase and gB, gD, gH, and gL combinations from HSV-1, BV, or CeHV-2. Glycoproteins from BV and CeHV-2 were FLAG tagged. The target cells were transfected with luciferase under a T7 promoter and either empty vector or nectin-1. Fusion assays were done as described in Materials and Methods. Fusion data were normalized to the fusion activity of each receptor to the whole set of glycoproteins required for fusion (set at 100%) for a specific virus. Glycoproteins (as shown in the column heads) of the viruses were replaced by homologous glycoproteins from other viruses (as shown in the left column). The symbols represent fusion activity relative to that for the whole set of glycoproteins required for fusion for a specific virus: >150% (++++), 80 to 150% (+++), 50 to 80% (++), 10 to 49% (+), or <10% (−).
Fig 5.
Cell fusion activity when BV glycoproteins are replaced by homologous glycoproteins from HSV-1 and CeHV-2. Target CHO cells were transfected with pCDNA3 (empty vector), PILRα, or nectin-1, along with a reporter plasmid expressing luciferase under the control of the T7 promoter. The transfected cells were replated with effector CHO cells transfected with a complete set of BV glycoproteins, or glycoproteins were replaced by gB, gD, gH, gL, or gH/gL from CeHV-2 and HSV-1, along with plasmids expressing T7 polymerase. Relative cell fusion activity of each replacement is presented as a percentage of that of the complete BV glycoprotein set required for fusion of nectin-1. Each bar shows the mean and standard deviation of three independent determinations. The data shown are representative of the complete data set obtained by the glycoprotein substitutions as shown in Table 2.
In general, glycoproteins between BV and CeHV-2 were more interchangeable than BV or CeHV-2 glycoproteins with HSV-1 glycoproteins, most likely due to a higher sequence identity between BV and CeHV-2 (89%, 82%, 82%, and 75% for gB, gD, gH, and gL, respectively, whereas the homology between HSV-1 and BV or CeHV-2 glycoproteins is lower). gB was interchangeable for all three viruses, and substitution of gB only modestly altered fusion activity (Table 2). This may be due to greater sequence homology among the gB genes than for those encoding all of the other glycoproteins (sequence identity between HSV-1 and either BV or CeHV-2 is 84%, and sequence identity between BV and CeHV-2 is 89%). BV gH/CeHV-2 gL and BV gL/CeHV-2 gH was expressed well on the cell surface by CELISA (data not shown), which may explain why exchange of BV and CeHV-2 gH or gL did not abrogate cell fusion activity (Table 2).
Most interestingly, when HSV-1 gH/gL, gH, or gL was expressed with the BV or CeHV-2 glycoprotein set, fusion with nectin-1-expressing cells was significantly reduced (Table 2 and Fig. 5). HSV-1 gH/gL did not work with BV or CeHV-2 glycoproteins. In addition, BV or CeHV-2 gD did not work well with HSV-1 glycoproteins. Since the gBs are interchangeable, this suggests that a homotypic gH/gL-gD interaction is required for fusion (Table 2). When HSV-1 gH or gL was used individually, fusion was also reduced, indicating that both gH and gL may contain important interaction sites for gD since gB was replaceable and functional (Table 2 and Fig. 5). Another possible reason for why gH and gL were not exchangeable among the viruses is that the presence of heterologous gL may lead to misfolding or lack of transport of the gH/gL complex to the cell surface. BV gL/HSV-1 gH and CeHV-2 gH/HSV-1 gL were not expressed on the cell surface, while BV gH/HSV-1 gL and CeHV2 gL/HSV-1 gH were expressed (data not shown), indicating that expression was responsible for the defect in some cases whereas the absence of functional interactions may be responsible in other cases.
DISCUSSION
Despite the genomes of BV and CeHV-2 being completely sequenced (42, 56), little molecular work has been done with these two viruses. Here we report the receptor usage of BV and CeHV-2 using cell-cell fusion for both viruses and virus infection for BV. Both BV and CeHV-2, like HSV-1, can use nectin-1 as a receptor; however, they do not use PILRα as an entry receptor. In addition, neither BV nor CeHV-2 uses HVEM or African green monkey (AGM) (Chlorocebus sabaeus) HVEM as an entry receptor (Fig. 2 to 4). We also investigated the heterotypic fusion activities among HSV-1, BV, and CeHV-2 glycoproteins and found that only the BV and CeHV-2 glycoproteins readily complemented each other, whereas the HSV-1 glycoproteins did not function efficiently with the BV or CeHV-2 glycoproteins (Table 2 and Fig. 5). These findings may provide clues for controlling BV infections in humans by understanding required interactions between the relevant glycoproteins required for virus fusion and entry and developing inhibitors that target these essential interactions.
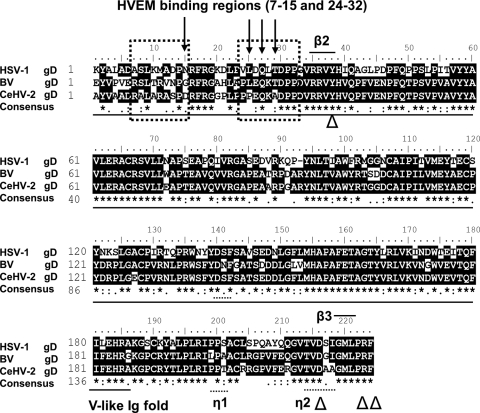
The recently solved structure of gD bound to nectin-1 (10) provides a basis for understanding why nectin-1 functions as an entry receptor for BV whereas HVEM does not. This structure revealed that the nectin-1 binding site on gD differs from the binding site of gD on HVEM (4, 10). Nectin-1 contacts gD exclusively with one β-sheet of the V domain and the intervening loops of nectin-1 (10). Compatible with this structural data, the N-terminal Ig-like V domains of nectin-1 and nectin-2 have been shown to be important for HSV entry (27, 34, 51, 52). Double or triple amino acid substitutions at positions located downstream of the Ig fold in gD (D215, R222, F223) abrogated physical and functional interactions with nectin-1 and nectin-2 (33). Substitutions at position Y38 also reduced physical and functional interactions with nectin-1 but not with HVEM (7). Figure 6 shows the amino sequence alignment of HSV-1, BV, and CeHV-2 gD, indicating that the four important nectin-1 interaction residues (Y38, D215, R222, and F223) are conserved in both BV and CeHV-2 gD, as well as the gD-nectin-1 interface β-strands β2 (residues 35–38) and β3 (residues 219–221 (10), explaining why both BV and CeHV-2 can efficiently use nectin-1 as a fusion receptor.
Fig 6.
Sequence alignment of partial HSV-1 gD, BV gD, and CeHV-2 gD sequences. gDs from K1 to F223 were aligned; the amino acid numbers begin with the first residue in the mature protein after signal sequence cleavage. Underlined portions are the V-like Ig fold domain, arrow-marked residues are important for the association of gD with HVEM, and “△”-marked residues are important for the association of gD with nectin-1. Dotted boxes are gD-HVEM interfaces. Short black bars show the gD–nectin-1 interfaces (gD β2, residues 35 to 38; and β3, residues 219 to 221), gD helical turns (η1, residues 199 to 201; and η2, residues 214 to 217). “*” means that the residues in that column are identical in all sequences in the alignment. “:” means that conserved substitutions have been observed. “.” means that semiconserved substitutions are observed.
Amino acid substitutions, insertions, and deletions of the N terminus of gD abrogated physical and functional interactions of either HSV-1 gD or HSV-2 gD with HVEM but did not alter interactions with nectin-1 (22, 67). The gD-HVEM crystal structure indicated that all of the HVEM contact residues are within an N-terminal hairpin loop (residues 7 to 15 and residues 24 to 32) of gD (4). Mutation of gD at Q27R (Rid-2 virus), Q27P (Rid-1 virus), and L25P (US10 virus) or three mutations, L25P, Q27R, and T230I (Ang virus), results in the loss of HVEM functioning as a receptor for HSV-1 (4, 38, 64). The HSV-1 gD Q27P mutation was shown to eliminate HVEM receptor usage, and an L25P mutation reduced HVEM usage (67). Structure-based mutagenesis demonstrated that three regions at the gD-HVEM binding interface contribute to function: an intermolecular β-sheet involving gD residues 27 to 29, the central region of the interface that includes HVEM-Y23, gD-M11, and gD-L25, and the region encompassing HVEM CRD2 and gD-N15 (4, 8, 9).
According to our results, both BV and CeHV-2 cannot utilize human HVEM as a fusion receptor (Fig. 2 to 4). Sequence alignments showed that the lack of HVEM usage is likely due to sequence differences between HSV-1 gD, BV gD, and CeHV-2 gD within the first 32 gD amino acids (Fig. 6). BV gD and CeHV-2 gD contain R11, while HSV-1 gD contains M11; BV gD contains G15 and CeHV-2 gD contains D15, which is different from N15 at HSV-1 gD; CeHV-2 gD P25 and A29 are also different from sequence of HSV-1 gD and BV gD. All these changes occur within gD residues that contact HVEM, and the changes may contribute to the lack of HVEM usage by BV and CeHV-2. When BV gD was replaced by HSV-1 gD and cotransfected with BV gB and gH/gL, cell fusion activity mediated by HVEM was restored to about 50% of the fusion activity of HSV-1 gD with HVEM (Table 2 and data not shown), confirming the interaction of HSV-1 gD and HVEM.
The amino sequences of human HVEM and AGM HVEM share 83% identity, whereas AGM and macacine HVEM share 96% identity. Interestingly, cysteine-rich domain 1 (CRD1) and CRD2 from both AGM HVEM and macacine HVEM are 100% identical. Changes exist within CRD3; AGM HVEM encodes residues N144 and S161, whereas Macacine HVEM encodes T144 and F161. All of the gD contacts in HVEM occur within CRD1 and CRD2 (4), and therefore we expect that BV does not use macacine HVEM during the course of natural infection.
Association of the HSV-1 gB receptor PILRα with gB depends on the O-glycosylation sites on gB, and gB residues T53 and T480 are part of the interaction site (59). Mutation of T53 and T480 alone did not abrogate the binding of gB to PILRα. Mutation of T53 affected the association with PILRα more than that of T480, and concurrent mutation of both residues totally abrogated the association of HSV-1 gB with PILRα (59). Sequence alignments of HSV-1, BV, and CeHV-2 gB indicate that only the threonine residue corresponding to HSV-1 gB T480 is conserved in BV and CeHV-2 (BV gB T485 and CeHV-2 gB T483, respectively). The amino acid corresponding to T53 in BV is S43, and that in CeHV-2 is D39. However, it is very likely that other glycosylation sites on BV gB and CeHV-2 gB function together with T485 or T483 to facilitate PILRα association, because HSV-1 gB has multiple O-glycosylation sites (59). These differences may account for why BV and CeHV-2 do not efficiently use PILRα as a receptor.
Dramatic differences in pathogenesis are seen in HSV-1 in the human host compared to its pathogenesis in primates and vice versa. There is continuing concern that BV represents a potential threat to humans because of potential spread of this virus from pet monkeys or monkeys at primate centers to the human host. Whereas BV can be deadly to humans, similarly, HSV-1 infection of marmosets can be deadly (19, 23, 41, 45). The pathogenesis of these viruses in humans and monkeys is likely multifactorial, but the results of our current studies suggest that receptor usage may in part be responsible for the dramatic differences in pathogenesis these virus cause during zoonotic transmission. Nectin-1 is a pan-alphaherpesvirus entry receptor that mediates entry of HSV (6, 16, 36, 37, 48), bovine herpesvirus 1 (BHV-1) (5), and porcine pseudorabies virus (PRV) (25, 44). Nectin-1 is commonly used by both human HSV-1 and the simian viruses, potentially due to interspecies transfer of parts of the viral genomes. In contrast, the two species have adapted to use different receptors as well, which may explain their altered virulence, tropism, or pathogenesis. Recent studies with HSV may provide clues to how differential receptor usage may alter pathogenesis in infected hosts. Studies using murine models of HSV infection have demonstrated receptor preferences of HVEM and nectin-1 when HSV-1 or HSV-2 was investigated experimentally using vaginal, direct intracranial, or eye infection (24, 26, 54). Infection of the vaginal epithelium with HSV-2 showed that nectin-1 was the primary receptor for vaginal infection and necessary, while nectin-1 is not the sole receptor capable of enabling spread of HSV infection from the vaginal epithelium to the peripheral nervous system (PNS) and central nervous system (CNS) (54). However, expression of nectin-1 is necessary for HSV-2 infection via the intracranial route and for encephalitis, while HVEM played a less important role and was largely irrelevant (26). In the ocular model of HSV-1 infection, both HVEM and nectin-1 must be present for maximal HSV-1 infection of the cornea, and the findings suggested that receptor requirements for HSV vary depending on the route of inoculation and/or serotype (24).
Finally, engagement of HVEM by gD has been shown to modestly alter the immune response following vaginal infection of mice with a strain of HSV-2 expressing a gD mutant unable to engage HVEM (66). In these studies, there was a transient increase in mucosal chemokine and interleukin 6 (IL-6) levels compared to findings for infection with HSV-2 expressing wild-type gD (66), indicating HVEM gD interaction may alter early innate events in the murine immune response to infection. Future studies will investigate if the inability of BV gD to utilize HVEM is important for the pathogenesis of BV. Understanding human and simian receptor usage for BV and HSV may provide clues to understanding the pathogenesis of these viruses, as well as related viruses, in their natural host and in cross-species infections.
ACKNOWLEDGMENTS
We thank Gus Kousoulas for providing us with the AGM HVEM expression plasmid. We thank N. Susmarski for timely and excellent technical assistance and members of the Longnecker laboratory for their help in these studies, particularly Sarah Connolly and Cindy Rowe for reading the manuscript. Sequencing services were performed at the Northwestern University Genomics Core Facility.
R.L. is the Dan and Bertha Spear Research Professor in Microbiology-Immunology. This work was supported by NIH grants CA021776 and AI067048 to R.L. and P51 RR013986 and R21 RR026287 to A.G. and was carried out in facilities constructed with funding from grant C06 RR012087. M.A. is supported by grant 5T32AI007271.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Amen MA, Griffiths A. 2011. Identification and expression analysis of herpes B virus-encoded small RNAs. J. Virol. 85:7296–7311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arii J, et al. 2010. Non-muscle myosin IIA is a functional entry receptor for herpes simplex virus-1. Nature 467:859–862 [DOI] [PubMed] [Google Scholar]
- 3. Atwal JK, et al. 2008. PirB is a functional receptor for myelin inhibitors of axonal regeneration. Science 322:967–970 [DOI] [PubMed] [Google Scholar]
- 4. Carfi A, et al. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169–179 [DOI] [PubMed] [Google Scholar]
- 5. Chase CCL, Lohff C, Letchworth GJ. 1993. Resistance and susceptibility of bovine cells expressing herpesviral glycoprotein D homologs to herpesviral infections. Virology 194:365–369 [DOI] [PubMed] [Google Scholar]
- 5a. Chosewood LC, et al. (ed). 2009. Biosafety in microbiological and biomedical laboratories, 5th ed. HHS publication no. (CDC)21-1112. CDC, Atlanta, GA [Google Scholar]
- 6. Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. 1998. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 72:9992–10002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Connolly SA, et al. 2005. Potential nectin-1 binding site on herpes simplex virus glycoprotein D. J. Virol. 79:1282–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Connolly SA, et al. 2003. Structure-based mutagenesis of herpes simplex virus glycoprotein D defines three critical regions at the gD-HveA/HVEM binding interface. J. Virol. 77:8127–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Connolly SA, et al. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J. Virol. 76:10894–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Di Giovine P, et al. 2011. Structure of herpes simplex virus glycoprotein d bound to the human receptor nectin-1. PLoS Pathog. 7:e1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eberle R, Hilliard J. 1995. The simian herpesviruses. Infect. Agents Dis. 4:55–70 [PubMed] [Google Scholar]
- 12. Fan Q, Lin E, Satoh T, Arase H, Spear PG. 2009. Differential effects on cell fusion activity of mutations in herpes simplex virus 1 glycoprotein B (gB) dependent on whether a gD receptor or a gB receptor is overexpressed. J. Virol. 83:7384–7390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fan Q, Longnecker R. 2010. The Ig-like v-type domain of paired Ig-like type 2 receptor alpha is critical for herpes simplex virus type 1-mediated membrane fusion. J. Virol. 84:8664–8672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Foster TP, Chouljenko VN, Kousoulas KG. 1999. Functional characterization of the HveA homolog specified by African green monkey kidney cells with a herpes simplex virus expressing the green fluorescence protein. Virology 258:365–374 [DOI] [PubMed] [Google Scholar]
- 15. Fournier N, et al. 2000. FDF03, a novel inhibitory receptor of the immunoglobulin superfamily, is expressed by human dendritic and myeloid cells. J. Immunol. 165:1197–1209 [DOI] [PubMed] [Google Scholar]
- 16. Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618–1620 [DOI] [PubMed] [Google Scholar]
- 17. Golomb E, et al. 2004. Identification and characterization of nonmuscle myosin II-C, a new member of the myosin II family. J. Biol. Chem. 279:2800–2808 [DOI] [PubMed] [Google Scholar]
- 18. Hagglund R, Roizman B. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huemer HP, Larcher C, Czedik-Eysenberg T, Nowotny N, Reifinger M. 2002. Fatal infection of a pet monkey with human herpesvirus. Emerg. Infect. Dis. 8:639–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huff JL, Barry PA. 2003. B-virus (Cercopithecine herpesvirus 1) infection in humans and macaques: potential for zoonotic disease. Emerg. Infect. Dis. 9:246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jainkittivong A, Langlais RP. 1998. Herpes B virus infection. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 85:399–403 [DOI] [PubMed] [Google Scholar]
- 22. Jogger CR, Montgomery RI, Spear PG. 2004. Effects of linker-insertion mutations in herpes simplex virus 1 gD on glycoprotein-induced fusion with cells expressing HVEM or nectin-1. Virology 318:318–326 [DOI] [PubMed] [Google Scholar]
- 23. Juan-Salles C, et al. 1997. Spontaneous herpes simplex virus infection in common marmosets (Callithrix jacchus). J. Vet. Diagn. Invest. 9:341–345 [DOI] [PubMed] [Google Scholar]
- 24. Karaba AH, Kopp SJ, Longnecker R. 2011. Herpesvirus entry mediator and nectin-1 mediate herpes simplex virus 1 infection of the murine cornea. J. Virol. 85:10041–10047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karger A, Mettenleiter TC. 1993. Glycoproteins gIII and gp50 play dominant roles in the biphasic attachment of pseudorabies virus. Virology 194:654–664 [DOI] [PubMed] [Google Scholar]
- 26. Kopp SJ, et al. 2009. Infection of neurons and encephalitis after intracranial inoculation of herpes simplex virus requires the entry receptor nectin-1. Proc. Natl. Acad. Sci. U. S. A. 106:17916–17920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krummenacher C, et al. 1999. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J. Virol. 73:8127–8137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kubagawa H, et al. 1999. Biochemical nature and cellular distribution of the paired immunoglobulin-like receptors, PIR-A and PIR-B. J. Exp. Med. 189:309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin E, Spear PG. 2007. Random linker-insertion mutagenesis to identify functional domains of herpes simplex virus type 1 glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 104:13140–13145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu BP, Fournier A, GrandPre T, Strittmatter SM. 2002. Myelin-associated glycoprotein as a functional ligand for the Nogo-66 receptor. Science 297:1190–1193 [DOI] [PubMed] [Google Scholar]
- 31. Lopez M, et al. 2000. Nectin2α (PRR2α or HveB) and nectin2δ are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J. Virol. 74:1267–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Malherbe H, Harwin R. 1958. Neurotropic virus in African monkeys. Lancet ii:530 [Google Scholar]
- 33. Manoj S, Jogger CR, Myscofski D, Yoon M, Spear PG. 2004. Mutations in herpes simplex virus glycoprotein D that prevent cell entry via nectins and alter cell tropism. Proc. Natl. Acad. Sci. U. S. A. 101:12414–12421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martinez WM, Spear PG. 2002. Amino acid substitutions in the V domain of nectin-1 (HveC) that impair entry activity for herpes simplex viruses 1 and 2 but not for pseudorabies virus or bovine herpesvirus 1. J. Virol. 76:7255–7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McGee AW, Yang Y, Fischer QS, Daw NW, Strittmatter SM. 2005. Experience-driven plasticity of visual cortex limited by myelin and Nogo receptor. Science 309:2222–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Menotti L, Avitabile E, Dubreuil P, Lopez M, Campadelli-Fiume G. 2001. Comparison of murine and human nectin-1 binding to herpes simplex virus glyc0protein D (gD) reveals a weak interaction of murine nectin-1 to gD and a gD-dependent pathway of entry. Virology 282:256–266 [DOI] [PubMed] [Google Scholar]
- 37. Menotti L, et al. 2000. The murine homolog of human Nectin1δ serves as a species nonspecific mediator for entry of human and animal αherpesviruses in a pathway independent of a detectable binding to gD. Proc. Natl. Acad. Sci. U. S. A. 97:4867–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Montgomery RI, Warner MS, Lum BJ, Spear PG. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427–436 [DOI] [PubMed] [Google Scholar]
- 39. Mousseau DD, Banville D, L'Abbe D, Bouchard P, Shen S-H. 2000. PILRα, a novel immunoreceptor tyrosine-based inhibitory motif-bearing protein, recruits SHP-1 upon tyrosine phosphorylation and is paired with the truncated counterpart PILRβ. J. Biol. Chem. 275:4467–4474 [DOI] [PubMed] [Google Scholar]
- 40. Munitz A, McBride ML, Bernstein JS, Rothenberg ME. 2008. A dual activation and inhibition role for the paired immunoglobulin-like receptor B in eosinophils. Blood 111:5694–5703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murphy BL, Maynard JE, Krushak DH, Berquist KR. 1972. Microbial flora of imported marmosets: viruses and enteric bacteria. Lab. Anim. Sci. 22:339–343 [PubMed] [Google Scholar]
- 42. Perelygina L, et al. 2003. Complete sequence and comparative analysis of the genome of herpes B virus (Cercopithecine herpesvirus 1) from a rhesus monkey. J. Virol. 77:6167–6177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pertel P, Fridberg A, Parish ML, Spear PG. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313–324 [DOI] [PubMed] [Google Scholar]
- 44. Petrovskis EA, Meyer AL, Post LE. 1988. Reduced yield of infectious pseudorabies virus and herpes simplex virus from cell lines producing viral glycoprotein gp50. J. Virol. 62:2196–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Potkay S. 1992. Diseases of the Callitrichidae: a review. J. Med. Primatol. 21:189–236 [PubMed] [Google Scholar]
- 46. Satoh T, et al. 2008. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132:935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shiratori I, Ogasawara K, Saito T, Lanier LL, Arase H. 2004. Activation of natural killer cells and dendritic cells upon recognition of a novel CD99-like ligand by paired immunoglobulin-like type 2 receptor. J. Exp. Med. 199:525–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shukla D, Dal Canto M, Rowe CL, Spear PG. 2000. Striking similarity of murine nectin-1α to human nectin-1α (HveC) in sequence and activity as a gD receptor for alphaherpesvirus entry. J. Virol. 74:11773–11781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shukla D, et al. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13–22 [DOI] [PubMed] [Google Scholar]
- 50. Shukla D, Spear PG. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Invest. 108:503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Struyf F, Martinez WM, Spear PG. 2002. Mutations in the N-terminal domains of nectin-1 and nectin-2 reveal differences in requirements for entry of various alphaherpesviruses and for nectin-nectin interactions. J. Virol. 76:12940–12950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Struyf F, Plate AE, Spear PG. 2005. Deletion of the second immunoglobulin-like domain of nectin-1 alters its intracellular processing and localization and ability to mediate entry of herpes simplex virus. J. Virol. 79:3841–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Takai Y, Miyoshi J, Ikeda W, Ogita H. 2008. Nectins and nectin-like moleecules: roles in contact inhibition of cell movement and proliferation. Nat. Rev. Mol. Cell Biol. 9:603–615 [DOI] [PubMed] [Google Scholar]
- 54. Taylor JM, et al. 2007. Alternative entry receptors for herpes simplex virus and their roles in disease. Cell Host Microbe 2:19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Terry-Allison T, Montgomery RI, Warner MS, Geraghty RJ, Spear PG. 2001. Contributions of gD receptors and glycosaminoglycan sulfation to cell fusion mediated by herpes simplex virus 1. Virus Res. 74:39–45 [DOI] [PubMed] [Google Scholar]
- 56. Tyler SD, Peters GA, Severini A. 2005. Complete genome sequence of cercopithecine herpesvirus 2 (SA8) and comparison with other simplexviruses. Virology 331:429–440 [DOI] [PubMed] [Google Scholar]
- 57. Uehara T, et al. 2001. Inhibition of IgE-mediated mast cell activation by the paired Ig-like receptor PIR-B. J. Clin. Invest. 108:1041–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10:778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang J, et al. 2009. Binding of herpes simplex virus glycoprotein B (gB) to paired immunoglobulin-like type 2 receptor alpha depends on specific sialylated O-linked glycans on gB. J. Virol. 83:13042–13045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang KC, et al. 2002. Oligodendrocyte-myelin glycoprotein is a Nogo receptor ligand that inhibits neurite outgrowth. Nature 417:941–944 [DOI] [PubMed] [Google Scholar]
- 61. Ware CF. 2008. Targeting lymphocyte activation through the lymphotoxin and LIGHT pathways. Immunol. Rev. 223:186–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Warner MS, et al. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2 and pseudorabies virus. Virology 246:179–189 [DOI] [PubMed] [Google Scholar]
- 63. Weigler BJ. 1992. Biology of B virus in macaque and human hosts: a review. Clin. Infect. Dis. 14:555–567 [DOI] [PubMed] [Google Scholar]
- 64. Whitbeck JC, et al. 1997. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J. Virol. 71:6083–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xu X, et al. 2011. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat. Biotechnol. 29:735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yoon M, et al. 2011. Functional interaction between herpes simplex virus type 2 gD and HVEM transiently dampens local chemokine production after murine mucosal infection. PLoS One 6:e16122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yoon M, Zago A, Shukla D, Spear PG. 2003. Mutations in the N termini of herpes simplex virus type 1 and 2 gDs alter functional interactions with the entry/fusion receptors HVEM, nectin-2 and 3-O-sulfated heparan sulfate but not with nectin-1. J. Virol. 77:9221–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]