Abstract
Fibroblasts can be reprogrammed into induced pluripotent stem cells (iPSC) by ectopic expression of key transcription factors. Current methods for the generation of integration-free iPSC are limited by the low efficiency of iPSC generation and by challenges in reprogramming methodology. Recombinant adeno-associated virus (rAAV) is a potent gene delivery vehicle capable of efficient transduction of transgenic DNA into cells. rAAV stays mainly as an episome in nondividing cells, and the extent of integration is still poorly defined for various replicating cells. In this study, we aimed to induce iPSC from mouse and human fibroblasts by using rAAV vector-mediated transient delivery of reprogramming factors. We succeeded in deriving induced pluripotent stem cells from mouse but not human fibroblasts. Unexpectedly, the rAAV vector-mediated reprogramming led to frequent genomic integration of vector sequences during the reprogramming process, independent of the amount of virus used, and to persistent expression of reprogramming factors in generated iPSC clones. It thus appears that rAAV vectors are not compatible with the derivation of integration-free iPSC.
INTRODUCTION
Mouse and human somatic cells can be reprogrammed into induced pluripotent stem cells (iPSC) by ectopic expression of retrovirally delivered key transcription factors (30). Although retroviruses are effectively silenced in stem cells, they integrate randomly into genomic DNA, and there is a potential risk of virally induced tumor formation. To date, several techniques have been used for generation of nonintegrative iPSC, such as the use of plasmids, proteins, Sendai viruses, and modified RNAs (7, 23, 32, 35). However, many of the nonintegrative methods still have severe limitations, such as the challenges in generation and purification of proteins and Sendai viruses (7, 35), the need for repeated administration of synthetic mRNA (32), and the low reprogramming efficiency of plasmid- and protein-based methods (23, 35). Therefore, there is a need for efficient nonintegrative methods of reprogramming.
Adeno-associated virus (AAV) is a small nonpathogenic parvovirus with a 4.7-kb single-stranded linear genome (26). The recombinant AAV (rAAV) genome does not carry rep and cap genes, which are supplied in trans from a helper plasmid during generation of viral particles in host cells (3). AAVs are potent gene delivery vehicles capable of transducing both dividing and nondividing cells (4). Transduction of postmitotic cells, such as skeletal muscle, leads mainly to formation of episomal monomeric and concatemeric circles or linear episomes, which assimilate into chromatin with a typical nucleosomal pattern (21, 25). Previously, AAV was shown to be able to integrate at a specific site, AAVS1, on human chromosome 19, although this requires the product of the AAV-carried Rep gene (29), which is unavailable in recombinant virions. In proliferating cells, nonintegrated viral genomes are unstable and are lost soon upon proliferation of the transduced cells (21). Since reprogramming to pluripotency is accompanied by extensive cell proliferation, we hypothesized that rAAV-mediated delivery of reprogramming factors could be beneficial in terms of producing vector-free iPSC (7, 23, 28, 32, 33).
Our aim was to study the reprogramming of mouse and human fibroblasts by using rAAVs encoding the established reprogramming factors OCT4, SOX2, KLF4, and c-Myc. We were able to generate iPSC from mouse but not human fibroblasts. In the course of this study, we found that all generated iPSC colonies contained genomic integration of the transgene sequences.
MATERIALS AND METHODS
Cell culture.
All cell lines were cultured in an incubator at 37°C and 5% CO2. Human foreskin fibroblasts (HFFs; ATCC line CRL-2429), mouse embryonic fibroblasts (MEFs), and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) containing 10% fetal bovine serum (FBS; Promocell), 2 mM GlutaMAX (Gibco), and 100 μg/ml penicillin-streptomycin (Sigma). Mouse iPSC were cultured in mES medium (KnockOut high-glucose DMEM at 4,500 mg/liter [Gibco] supplemented with 1 mM sodium pyruvate [Invitrogen], 15% FBS [Sigma], 2 mM GlutaMAX [Gibco], 0.10 mM nonessential amino acids [Gibco], 0.1 mM β-mercaptoethanol, and leukemia inhibitory factor [LIF]).
Vector construction.
To generate rAAV particles carrying the transgene under the control of the cytomegalovirus (CMV) promoter, we used previously described pSubCMV-WPRE vectors (24). For rAAV carrying the transgene under the control of the CMV early enhancer/chicken beta actin (CAG) promoter, we used the pSubCAG-WPRE plasmid, which is a derivative of pSubCMV-WPRE in which the CMV promoter is replaced with the CAG promoter from pDRIVE-CAG (Invivogen). Human OCT4, SOX2, and KLF4 genes and mouse c-Myc were cut out from pMXs vectors, used routinely in our laboratory for iPSC inductions, by standard cloning procedures (13). Fragments were blunted and cloned into the aforementioned rAAV plasmids, which were opened with PmlI.
rAAV production.
rAAVs were produced as described by Zolotukhin et al. (36). Briefly, for each virus, 23 × 106 293T cells in total were plated into three 15-cm dishes and transfected with gene-carrying plasmids, constructed on the basis of the aforementioned pSubCMV-WPRE and pSubCAG-WPRE plasmids, and with a serotype-determining helper plasmid(s) (pDG [for rAAV2] [10] or the combination of pBS-E2A-VA-E4 and p5e18-VD2/9 [for rAAV9] [8, 15]), using JetPEI (Polyplus Transfection) according to the manufacturer's instructions. Cells were collected 2 days later and resuspended in lysis buffer (150 mM NaCl, 50 mM Tris, pH 8.5). Cells were lysed with three freeze-thaw cycles, treated with DNase, and centrifuged. Viruses were purified from supernatant by iodixanol gradient ultracentrifugation using Beckman quick-seal tubes and a 70.1TI rotor. Viruses were then divided into aliquots and stored at −80°C. Virus concentrations were determined as the number of viral genomes per volume by quantitative PCR (qPCR) using WPRE-specific primers (see Table S3 in the supplemental material). Briefly, rAAV preparations were diluted 1:10,000 with deionized water, and aliquots were loaded directly into DyNAmo HS SYBR Green qPCR (Finnzymes) master mix, diluted with deionized water, as specified by the manufacturer. A calibration curve was built in parallel with known amounts of the gene-carrying plasmid pSubCMV-WPRE, expressed as the number of single-stranded molecules per reaction volume.
Determining the transduction efficiency of rAAV.
HFFs and MEFs were plated on 24-well plates at a density of 1 × 104 cells/well. The next day, cells were transduced with rAAV2 encoding enhanced green fluorescent protein (EGFP) under the control of the CMV promoter, with viral dilutions of 1 × 102 to 1 × 105 viral genomes per cell. Cells were incubated in the virus-containing medium for 48 h and then fixed with 4% paraformaldehyde (PFA). Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) (Vectashield; Vector). Transduction efficiency was determined as the percentage of GFP-positive cells in the total number of cells. Four to eight random frames were counted per condition.
Generation of iPSC from human foreskin fibroblasts.
HFFs were plated on 6-well plates at a density of 1 × 105 cells/well. The next day, equal amounts of rAAVs containing the OCT4, SOX2, and KLF4 genes and c-Myc expressed under the control of the CMV or CAG promoter were added to the cells, which were transduced 1 to 4 times every second day (see Table S1 in the supplemental material). On day 4, cells were split 1:2 on Matrigel or mitomycin C-inactivated MEF-coated 6-well plates. Medium was changed to MEF-conditioned hES medium (KnockOut DMEM [Gibco] supplemented with 20% KO serum replacement [Gibco], 1% GlutaMAX [Gibco], 0.0915 mM β-mercaptoethanol, 1% nonessential amino acids [Gibco], and 6 ng/ml basic fibroblast growth factor [bFGF; Sigma]) or hES medium. Colonies were picked manually and plated on mitomycin C-inactivated MEF- or Matrigel-coated 24-well plates, depending on the induction conditions. The following small-molecule compounds were tested for their efficiency in enhancing reprogramming efficiency: PD0325901, 1 μM; SB431542, 2 μM; valproic acid (VPA), 2 mM; and sodium butyrate (NaB), 0.25 mM.
Generation of iPSC from mouse embryonic fibroblasts.
Mouse (ICR background; outbred) embryonic fibroblasts were plated on gelatin-coated 6-well plates at a density of 1 × 105 cells/well. The cells were transduced using equal amounts of rAAVs encoding the reprogramming factors OCT4, SOX2, KLF4, and c-Myc (see Table S2 in the supplemental material). Two days after the first transduction, the culture medium was changed to mES medium. Cells were transduced 1 to 3 times every second day and split on gelatin-coated plates on day 4 (Fig. 1C; see Table S2). Medium was changed every other day. Colonies were picked manually onto 24-well plates coated with mitomycin C-inactivated MEFs at a density of 15 × 103 cells/cm2 in mES medium. For passaging, murine induced pluripotent stem cells (miPSC) were dissociated to single cells by use of TrypLE Select (Gibco) and were plated onto new mitomycin C-inactivated MEF-coated plates. The efficiency of reprogramming was determined as the percentage of embryonic stem (ES) cell-like colonies in the total number of fibroblasts (1 × 105).
Fig 1.
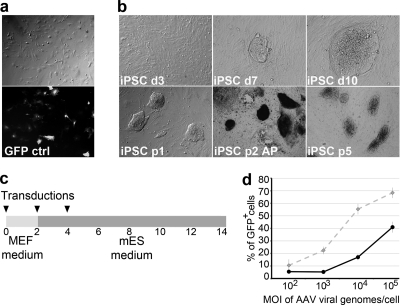
iPSC induction using AAVs. (a) MEFs were transduced efficiently with rAAV2, as seen by GFP expression. (b) Morphology of MEFs 3 days (d3) after rAAV-mediated transduction of reprogramming factors at an MOI of 2 × 104 viral genomes/cell/factor. The first iPSC colonies appeared at day 7 (d7) and were picked from day 10 (d10). iPSC retained embryonic stem cell morphology after passaging (p1, p2, and p5) and stained positive for alkaline phosphatase (AP). (c) Schematic representation of iPSC induction using rAAVs. Fibroblasts were transduced one, two, or three times (black arrowheads) with rAAVs at a range of MOIs. (d) Effects of various MOIs of rAAV-GFP viral genomes per cell (x axis) on number of GFP-positive MEFs (black solid line) and HFFs (gray dashed line) (percentage of total GFP-positive cells; y axis).
Western blotting.
293T cells were seeded into 12-well plates (1 × 105 cells/well). Cells were transduced with rAAVs at 109 to 1010 viral genomes/well and grown for 5 days. The cells were then lysed in cell culture lysis buffer (Promega) with Complete protease inhibitor mix (Roche). Samples were run in SDS-PAGE gels (4 to 20% ID231s IDGel precast gels) and transferred onto nitrocellulose membranes (Hybond-C Extra). Membranes were blocked with 5% milk in Tris-buffered saline plus Tween 20 (TBST) and exposed to antibodies against OCT4 (1:1,000) (sc-9081; Santa Cruz), SOX2 (1:2,500) (Ab5603; Millipore), and KLF4 (1:1,000) (ab34814; Abcam). Blots were probed with horseradish peroxidase (HRP)-conjugated anti-mouse, anti-rabbit, and anti-goat IgG secondary antibodies and visualized using a Bio-Rad Immun-Star WesternC kit for signal detection.
Immunocytochemistry.
For immunocytochemistry, cells were fixed in 4% PFA for 15 min at room temperature and then washed with phosphate-buffered saline (PBS). For nuclear staining, cells were permeabilized using 0.2% Triton X-100 (Sigma) for 15 min at room temperature. Cells were then treated with Ultra V block (Thermo) for 10 min. Immunohistochemical analysis of embryoid bodies (EBs) was done from paraffin sections. To reveal antigenic sites, the rehydrated sections were treated in a microwave in 1 mM EDTA, pH 8, for 3 min and cooled at room temperature for 20 min. Primary antibodies were against SSEA1 (1:50) (MAB4301; Millipore), Nanog (1:100) (sc-30331; Santa Cruz), OCT4 (1:200) (sc-8628; Santa Cruz), SOX2 (1:200) (ab5603; Millipore), KLF4 (1:100) (ab34814; Abcam), c-Myc (1:25) (c-33 sc-42; Santa Cruz), desmin (1:50) (sc-14026; Santa Cruz), FOXA2 (1:500) (sc-9187; Santa Cruz), and TUJ-1 (1:5,000) (Beta Cell Biology Consortium). Secondary antibodies were Alexa Fluor 488-conjugated donkey anti-goat (1:500) (A11055; Invitrogen), Alexa Fluor 488-conjugated donkey anti-mouse (1:500) (A21202; Invitrogen), Alexa Fluor 488-conjugated donkey anti-rabbit (1:500) (A21206; Invitrogen), and horseradish peroxidase (1:200) antibodies. DAPI (Vectashield; Vector) was used for nuclear staining.
RT-PCR.
Total RNA was extracted from cells by use of a Nucleospin RNA II kit (Macherey-Nagel). DNase treatment was performed separately using RQ1 DNase (Promega) in the presence of RNase inhibitor (Protector; Roche). RNA was purified with a Nucleospin RNA cleanup kit (Macherey-Nagel). The amount of RNA was determined using a Nanodrop spectrophotometer (Thermo Scientific). Two micrograms of total RNA was used for reverse transcription (RT) with Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega), random hexamers, and oligo(dT) according to the manufacturer's instructions. One microgram of cDNA was used for RT-PCR. PCR was run for 35 cycles with an annealing temperature of 60°C. Primers used for PCR are listed in Table S3 in the supplemental material.
Southern blotting.
Ten micrograms of genomic DNA purified from AAV miPSC by use of a GenElute mammalian genomic DNA miniprep kit (Sigma-Aldrich) was digested with NcoI (Promega). Samples were run in a 1% agarose gel and blotted onto positively charged nylon membranes (Hybond-N+; Amersham). Transgenic sequences were detected with [α-32P]dCTP-labeled probes by use of a Prime-a-Gene kit (Promega). The left inverted terminal repeat (ITR) and part of the promoter from the pSubCAG-WPRE plasmid digested with NcoI and PvuII were used as a template for probe synthesis.
EB assay.
AAV miPSC were trypsinized to single cells and plated for 30 min to allow feeders to attach. iPSC were then re-collected and plated on nonadherent plates. Medium was changed to mES medium without LIF and was refreshed every other day. EBs were allowed to grow for 14 days and fixed with 4% PFA for immunohistochemical analysis.
RESULTS
Optimization of AAV reprogramming.
Since the serotype of AAV determines the target cell specificity of the virus, two serotypes (AAV2 and AAV9) were tested for their transduction potential. In a control experiment, mouse and human fibroblasts were transduced with viral vectors encoding EGFP under the control of the CMV promoter, using a multiplicity of infection (MOI) of approximately 105 viral genomes/cell, and EGFP expression was monitored by fluorescence microscopy. Both mouse and human fibroblasts were transduced efficiently with AAV2 vectors, while AAV9 had a poor transduction efficiency (Fig. 1a). Further experiments were therefore carried out with AAV2.
To determine the transduction efficiency as a function of MOI, mouse and human fibroblasts were transduced with AAV2-EGFP at different dilutions, with the MOI ranging from 1 × 102 to 1 × 105 viral genomes/cell, and the percentage of GFP-positive cells was counted. At the highest MOI, 68% of HFFs and 41% of MEFs were GFP positive (Fig. 1d).
Next, we cloned reprogramming factors (OCT4, SOX2, KLF4, and c-Myc) from pMXs vectors, used routinely in our laboratory, into an AAV2 vector employing either the CMV or CAG promoter (13, 24). The expression of reprogramming factors was confirmed by RT-PCR, immunocytochemistry, and Western blotting (see Fig. S1 in the supplemental material). To determine the optimal MOI of AAV required for iPSC induction, human and mouse fibroblasts were treated with a series of viral dilutions. Since reprogramming is associated with rapid cell proliferation, we tested multiple protocols for serial AAV transductions to sustain sufficient transgene expression for reprogramming (see Tables S1 and S2).
For human fibroblasts, rAAV vector transduction at an MOI of 2 × 104 viral genomes/cell for each factor was found to be optimal for the expression of the transgenes without compromising cellular viability. When cells were transduced with 1 × 105 viral genomes/cell for each factor, they did not proliferate properly, which was reminiscent of cell cycle arrest. For mouse fibroblasts, MOIs of up to 2 × 105 viral genomes/cell for each factor could be used without a prominent effect on cell viability.
Reprogramming of mouse and human fibroblasts.
rAAV-mediated transduction of reprogramming factors into human foreskin fibroblasts resulted in the formation of colonies that appeared 10 to 12 days after induction. Most of them proceeded to form non-iPSC-like colonies, whereas smaller colonies with more promising morphology stopped growing at some point during the process (see Fig. S2 in the supplemental material). In order to further analyze these colonies, the most promising ones were picked at different stages of induction. However, in eight separate iPSC induction experiments (see Table S1), none of the picked colonies sustained iPSC-like morphology upon further propagation. The addition of small-molecule inhibitors that have been described to increase induction efficiency (12, 17, 18) also did not result in the generation of stable human iPSC clones. RT-PCR analysis of the propagated colonies showed that these retained the expression of at least some of the reprogramming factors (see Fig. S2).
To reprogram mouse fibroblasts, we used the protocol shown in Fig. 1c (also see Table S2 in the supplemental material). In four separate experiments, the cells were transduced with rAAVs expressing reprogramming factors at MOIs of 2 × 104 and 2 × 105 viral genomes/cell for each factor. We observed the first colonies with characteristic ES cell-like morphology and growth properties 7 days after transduction (Fig. 1b). The iPSC induction efficiency was determined as the percentage of ES cell-like colonies in the total number of fibroblasts and was found to be 0.001 to 0.006% when the factors were expressed under the control of the CMV promoter and 0.003 to 0.09% when the CAG promoter was used. We did not detect iPSC-like colonies when lower MOIs were used (2 × 102 or 2 × 103 viral genomes/cell per factor; n = 2).
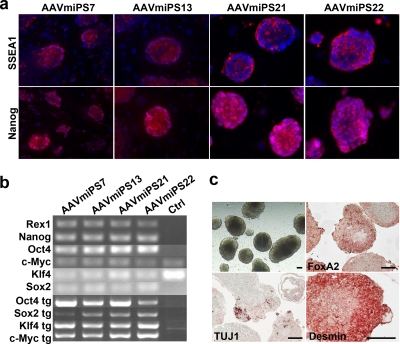
Four mouse iPSC clones, i.e., AAVmiPS7, AAVmiPS13, AAVmiPS21, and AAVmiPS22, were passaged and characterized further for pluripotency. All of the clones maintained ES cell-like morphology and proliferation for at least seven passages (Fig. 1b). All iPSC clones expressed the cell surface marker SSEA1, which is characteristic of mouse embryonic stem cells, and the nuclear transcription factor Nanog (Fig. 2a). Furthermore, the iPSC clones expressed the pluripotency markers Rex1 and Nanog and endogenous Oct4, c-Myc, Klf4, and Sox2 mRNAs, as detected by RT-PCR. However, all clones also expressed the transgenic reprogramming factor RNAs (Fig. 2b).
Fig 2.
Characterization of mouse iPSC clones. (a) Mouse iPSC clones express the cell surface marker SSEA1 (red) and stain positive for the transcription factor Nanog (red). The nucleus was stained blue (DAPI), and the images were merged with the SSEA1 and Nanog images. (b) RT-PCR analysis of mouse iPSC clones for the pluripotent markers Rex1, Nanog, Oct4, c-Myc, Klf4, and Sox2 and the transgenes Oct4 tg, Sox2 tg, Klf4 tg, and c-Myc tg. Ctrl, MEFs. (c) EBs under bright-field microscopy on day 14 and immunostaining of EB sections for FoxA2, TUJ-1, and desmin. Bar, 100 μm.
The pluripotent differentiation potential of the rAAV-induced iPSC clones was determined by EB assay. Cells positive for markers of all three embryonic germ layers, namely, βIII-tubulin (ectoderm), FOXA2 (endoderm), and desmin (mesoderm), were found in sections of 14-day EBs (Fig. 2c).
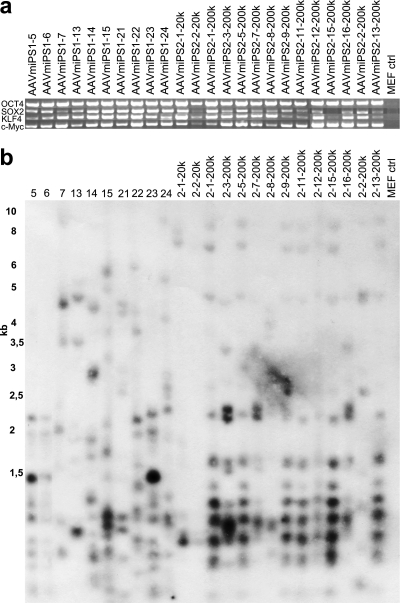
Since all of the previously analyzed clones expressed the transgenic reprogramming factors, 24 AAV-induced miPSC clones were further analyzed for the presence of the integrated transgene sequences in their genomes. Of the 24 clones, 12 (10 + 2) were from 2 separate inductions at an MOI of 2 × 104 and 12 were from single inductions at an MOI of 2 × 105. PCR amplification from purified genomic DNA samples from AAV-induced miPSC indicated that sequences of the reprogramming factors persisted in all iPSC clones (Fig. 3a). To exclude the possibility of contaminating episomal DNA in the genomic DNA preparations, integrations were validated by Southern blot analysis. All 24 clones analyzed showed multiple integrations of rAAV vectors in their genomes (Fig. 3b). Many of the iPSC clones also shared identical integration patterns indicative of a subclonal origin of the cell lines, resulting either from integration of vectors prior to splitting of the cells or from different clones being picked from satellite colonies.
Fig 3.
Integration of rAAVs into genomic DNA. (a) PCR amplification of different mouse iPSC clones to detect genomically integrated rAAV vectors. (b) Genomic integrations further confirmed by Southern blotting. Genomic DNA was digested with an enzyme cutting approximately 500 bp downstream from the 3′ end of AAV and detected using a probe recognizing the left ITR and part of the CAG promoter. The first 12 clones, from clone 5 to clone 2-2-20k, were induced at an MOI of 2 × 104, and the rest of the clones were induced at an MOI of 2 × 105.
DISCUSSION
In the present study, we compared the reprogramming efficiencies of rAAV2-delivered OCT4, KLF4, SOX2, and c-Myc transcription factors in mouse and human fibroblasts. We were able to obtain several iPSC clones with the mouse cells, but all human clones proved to be unstable. Human fibroblasts in general show a lower reprogramming efficiency and slower reprogramming kinetics than mouse fibroblasts (30, 34), which can partly explain the difficulty in inducing human iPSC clones with rAAV. It has been reported that mouse pluripotent stem cells normally exist in a so-called naïve pluripotent state, whereas human pluripotent stem cells are in a primed state representing developmentally more advanced cells (22). The transition of primed pluripotent cells to a naïve state has been shown to be inducible by overexpression of reprogramming-related transcription factors (2, 22). Since all of the rAAV-induced clones retained persistent expression of the reprogramming factors, it is possible that transgene expression does not allow human cells to stabilize properly to a primed pluripotent state. Apparently, the situation is different for mouse cells, where transgene expression may help in achieving and maintaining the naïve state.
Since reprogramming is accompanied by active cell proliferation, we hypothesized that integration of rAAV vectors into the host genome would not occur and that transgene-free iPSC clones could be generated (20, 31). However, we found that all stable iPSC clones that were studied had the viral vectors integrated into the host genome. Since rAAV integration can be dependent on the MOI, lowering the viral load could provide the possibility of avoiding viral integration. However, the lowest effective MOI (2 × 104 viral genomes/cell for each of the 4 factors) was always associated with genomic integration, and we were not able to generate any miPSC at lower MOIs. Since vector integration seemed to happen at an unexpectedly high rate, it is possible that the process of reprogramming either favored events where integration happened or increased the probability of vector integration during reprogramming.
Pluripotent reprogramming has recently been shown to be associated with genomic instability, resulting in the generation of chromosomal abnormalities such as deletions, insertions, and amplifications (9, 13, 16). These can appear as a result of reparation of DNA double-strand breaks (DSBs). The processing of rAAV genomes has been shown to occur in nuclear foci overlapping with DSB repair proteins of the MRE11-RAD50-NBS1 (MRN) complex (5). MRN complex proteins have also been implied in the processing of self-complementary recombinant adeno-associated virus (scAAV) genomes into a circular form (6), and MRE11 and NBS1 have been shown to bind directly to the ITR sequences of AAV (27). Integration of rAAV genomes has been associated with an increased frequency of DSBs (19). AAV integration is also found often in the vicinity of DNA palindromes, which can constitute fragile genomic sites susceptible to breakage (14). This suggests that DNA breakage prior to or during iPSC induction might facilitate AAV integration. In support of this view, rAAV was shown to efficiently transduce male germ line stem cells, which undergo meiotic divisions and are accompanied by a frequent occurrence of DSBs, thus resulting in stable transgene transmission through generations (1, 11). In addition, AAV genomes have been shown to integrate preferentially at actively transcribed genes in neonatal and adult mouse liver, and these integration events are frequently associated with chromosomal deletions (20).
In conclusion, we have demonstrated that rAAV-based vectors are efficient gene delivery vehicles for the reprogramming of mouse but not human fibroblasts into iPSC. Unexpectedly, rAAV vectors appear to stably integrate into host genomes of the reprogrammed cells. Furthermore, in contrast to retrovirally reprogrammed iPSC, the AAV-induced iPSC did not show silencing of the transgenes during further propagation. Our results do not exclude the possibility of inducing integration-free iPSC with rAAV vectors, but they imply that the probability of such events may be too low for the use of rAAV to be practical for the purpose. Because of these reasons, the rAAV constructs tested herein are not compatible with the derivation of integration-free iPSC.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the AAV Gene Transfer and Cell Therapy Core Facility of the University of Helsinki and Biocenter Finland for help with virus production. We thank Timo Tuuri for a critical reading of the manuscript.
This study was supported by grants from the Academy of Finland, The Sigrid Jusélius Foundation, Biocentrum Finland, and the ESTOOLS Consortium under the Sixth Research Framework Program of the European Union.
Footnotes
Published ahead of print 1 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Bickel JS, et al. 2010. Structural maintenance of chromosomes (SMC) proteins promote homolog-independent recombination repair in meiosis crucial for germ cell genomic stability. PLoS Genet. 6:e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buecker C, et al. 2010. A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell 6:535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buning H, Perabo L, Coutelle O, Quadt-Humme S, Hallek M. 2008. Recent developments in adeno-associated virus vector technology. J. Gene Med. 10:717–733 [DOI] [PubMed] [Google Scholar]
- 4. Carter P, Samulski R. 2000. Adeno-associated viral vectors as gene delivery vehicles. Int. J. Mol. Med. 6:17–27 [DOI] [PubMed] [Google Scholar]
- 5. Cervelli T, et al. 2008. Processing of recombinant AAV genomes occurs in specific nuclear structures that overlap with foci of DNA-damage-response proteins. J. Cell Sci. 121:349–357 [DOI] [PubMed] [Google Scholar]
- 6. Choi VW, McCarty DM, Samulski RJ. 2006. Host cell DNA repair pathways in adeno-associated viral genome processing. J. Virol. 80:10346–10356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. 2009. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B 85:348–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao G-P, et al. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 99:11854–11859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gore A, et al. 2011. Somatic coding mutations in human induced pluripotent stem cells. Nature 471:63–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grimm D, Kern A, Rittner K, Kleinschmidt JA. 1998. Novel tools for production and purification of recombinant adenoassociated virus vectors Hum. Gene Ther. 9:2745–2760 [DOI] [PubMed] [Google Scholar]
- 11. Honaramooz A, et al. 2008. Adeno-associated virus (AAV)-mediated transduction of male germ line stem cells results in transgene transmission after germ cell transplantation. FASEB J. 22:374–382 [DOI] [PubMed] [Google Scholar]
- 12. Huangfu D, et al. 2008. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 26:795–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hussein SM, et al. 2011. Copy number variation and selection during reprogramming to pluripotency. Nature 471:58–62 [DOI] [PubMed] [Google Scholar]
- 14. Inagaki K, et al. 2007. DNA palindromes with a modest arm length of greater, similar 20 base pairs are a significant target for recombinant adeno-associated virus vector integration in the liver, muscles, and heart in mice. J. Virol. 81:11290–11303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jalkanen J, et al. 2003. Adeno-associated virus-mediated gene transfer of a secreted decoy human macrophage scavenger receptor reduces atherosclerotic lesion formation in LDL receptor knockout mice. Gene Ther. 8:903–910 [DOI] [PubMed] [Google Scholar]
- 16. Laurent LC, et al. 2011. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 8:106–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin T, et al. 2009. A chemical platform for improved induction of human iPSCs. Nat. Methods 6:805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mali P, et al. 2010. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells 28:713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miller DG, Petek LM, Russell DW. 2004. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat. Genet. 36:767–773 [DOI] [PubMed] [Google Scholar]
- 20. Nakai H, et al. 2003. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat. Genet. 34:297–302 [DOI] [PubMed] [Google Scholar]
- 21. Nakai H, et al. 2000. Recruitment of single-stranded recombinant adeno-associated virus vector genomes and intermolecular recombination are responsible for stable transduction of liver in vivo. J. Virol. 74:9451–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nichols J, Smith A. 2009. Naive and primed pluripotent states. Cell Stem Cell 4:487–492 [DOI] [PubMed] [Google Scholar]
- 23. Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. 2008. Generation of mouse induced pluripotent stem cells without viral vectors. Science 322:949–953 [DOI] [PubMed] [Google Scholar]
- 24. Paterna JC, Moccetti T, Mura Feldon J, Büeler H. 2000. Influence of promoter and WHV post-transcriptional regulatory element on AAV-mediated transgene expression in the rat brain. Gene Ther. 7:1304–1311 [DOI] [PubMed] [Google Scholar]
- 25. Penaud-Budloo M, et al. 2008. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. J. Virol. 82:7875–7885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Russell DW, Kay MA. 1999. Adeno-associated virus vectors and hematology. Blood 94:864–874 [PMC free article] [PubMed] [Google Scholar]
- 27. Schwartz RA, et al. 2007. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J. Virol. 81:12936–12945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. 2008. Induced pluripotent stem cells generated without viral integration. Science 322:945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Surosky RT, et al. 1997. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J. Virol. 71:7951–7959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676 [DOI] [PubMed] [Google Scholar]
- 31. van Os R, et al. 1999. Recombinant adeno-associated virus-based vectors provide short-term rather than long-term transduction of primitive hematopoietic stem cells. Stem Cells 17:117–120 [DOI] [PubMed] [Google Scholar]
- 32. Warren L, et al. 2010. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7:618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu J, et al. 2009. Human induced pluripotent stem cells free of vector and transgene sequences. Science 324:797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu J, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920 [DOI] [PubMed] [Google Scholar]
- 35. Zhou H, et al. 2009. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4:381–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zolotukhin S, et al. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6:973–985 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.