Abstract
Herpes simplex virus 1 (HSV-1) and HSV-2 are medically significant pathogens. The development of an effective HSV vaccine remains a global public health priority. HSV-1 and HSV-2 immunodominant “asymptomatic” antigens (ID-A-Ags), which are strongly recognized by B and T cells from seropositive healthy asymptomatic individuals, may be critical to be included in an effective immunotherapeutic HSV vaccine. In contrast, immunodominant “symptomatic” antigens (ID-S-Ags) may exacerbate herpetic disease and therefore must be excluded from any HSV vaccine. In the present study, proteome microarrays of 88 HSV-1 and 84 HSV-2 open reading frames(ORFs) (ORFomes) were constructed and probed with sera from 32 HSV-1-, 6 HSV-2-, and 5 HSV-1/HSV-2-seropositive individuals and 47 seronegative healthy individuals (negative controls). The proteins detected in both HSV-1 and HSV-2 proteome microarrays were further classified according to their recognition by sera from HSV-seropositive clinically defined symptomatic (n = 10) and asymptomatic (n = 10) individuals. We found that (i) serum antibodies recognized an average of 6 ORFs per seropositive individual; (ii) the antibody responses to HSV antigens were diverse among HSV-1- and HSV-2-seropositive individuals; (iii) panels of 21 and 30 immunodominant antigens (ID-Ags) were identified from the HSV-1 and HSV-2 ORFomes, respectively, as being highly and frequently recognized by serum antibodies from seropositive individuals; and (iv) interestingly, four HSV-1 and HSV-2 cross-reactive asymptomatic ID-A-Ags, US4, US11, UL30, and UL42, were strongly and frequently recognized by sera from 10 of 10 asymptomatic patients but not by sera from 10 of 10 symptomatic patients (P < 0.001). In contrast, sera from symptomatic patients preferentially recognized the US10 ID-S-Ag (P < 0.001). We have identified previously unreported immunodominant HSV antigens, among which were 4 ID-A-Ags and 1 ID-S-Ag. These newly identified ID-A-Ags could lead to the development of an efficient “asymptomatic” vaccine against ocular, orofacial, and genital herpes.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) and HSV-2 are infectious pathogens that cause serious diseases at every stage of life, from fatal disseminated disease in newborns to cold sores, genital ulcerations, eye disease, and fatal encephalitis in adults (14, 15, 18, 82, 83). HSV-1 infects 60% of the U.S. population, who develop painful recurrent orolabial infections, causing a significant cumulative health care burden (37). For example, infection of the brain and eyes can lead to irreversible brain damage and blindness (37). Over 400,000 adults in the United States have a history of recurrent ocular disease capable of causing a loss of vision (62, 63, 65, 67, 68, 83). Virtually all herpetic orolabial disease is caused by HSV-1 (42). HSV-1 infection is responsible for approximately 50% of clinical first episodes of genital herpes in the United States. The geographic distribution of HSV-1 is worldwide, with infection occurring in both developed and underdeveloped countries. The virus is transmitted from infected to susceptible individuals during close personal contact only. There is no seasonal variation in the incidence of infection. HSV-1 infection is rarely fatal and establishes latency in the trigeminal ganglia after primary infection. Over one-third of the world's population has recurrent HSV-1 infections, and hence, the probability of transmitting HSV-1 is during the episodes of productive infection and not during latent infection. As such, recurrent herpes labialis is the largest reservoir of HSV-1 infections in the community. Recurrent genital herpes infection (primarily by HSV-2) also leads to an immunopathological response that develops into genital ulcerations and scarring (8, 61). The global prevalences of HSV-2-seropositive individuals of 15 years of age and older are estimated to be at least 45 million within the United States (24, 84) and well over 530 million worldwide, with a greater frequency of infection in women (53).
The shedding of reactivated HSV-1 is estimated to occur at rates of 3 to 28% in adults who harbor latent HSV-1 in their sensory neurons (44, 78–80). However, the vast majority of these individuals do not experience recurrent herpetic disease and are designated asymptomatic patients (32, 52, 80). In contrast, for some individuals (symptomatic patients), the reactivation of latent virus leads to the induction of ineffective or “symptomatic” HSV-specific CD4+ and CD8+ T cells (25, 32, 80). While some people have frequent recurrences of herpes disease (i.e., symptomatic patients, with 1 to 5 episodes of recurrent disease/year), others have less frequent recurrent disease to no history of recurrent disease (i.e., asymptomatic patients, with 0 to 1 episodes of recurrent disease/year). Interestingly, the difference between the symptomatic and asymptomatic groups is not a result of how often the latent herpesvirus reactivates, as both groups shed the virus at similar rates (75, 80). Instead, the difference is very likely related to variations in the number and nature of HSV antigens (Ags) that are targeted. In animal models, HSV antigens have been reported (i) to be protective against the disease (7, 28–30, 69, 73) and (ii) to cause/exacerbate the disease (11, 27, 31, 70). However, the number and nature of “protective” (i.e., asymptomatic) and “pathogenic” (i.e., “symptomatic”) antigens remain largely to be determined (18). Regardless of the number and nature of these antigens, it is logical that an efficient vaccine must include immunodominant asymptomatic antigens (ID-A-Ags) but not immunodominant symptomatic antigens (ID-S-Ags), to avoid inducing or exacerbating herpes disease. While an enzyme-linked immunosorbent assay (ELISA) is often used to identify one or several antigens targeted by antibodies (Abs), new promising antibody-profiling technologies, such as protein microarrays, are emerging and can now be used to simultaneously identify hundreds, or even thousands, of pathogen-derived protein Ags (1, 2, 9).
In the present study, proteome microarrays of 88 HSV-1 and 84 HSV-2 open reading frames (ORFs) expressed in Escherichia coli-based in vitro transcription-translation reactions (ORFomes) were constructed and probed with sera from 32 HSV-1-, 6 HSV-2-, and 5 HSV-1/HSV-2-seropositive individuals and 47 seronegative healthy controls. Twenty-one and 30 immunodominant antigens (ID-Ags) were identified from the HSV-1 and HSV-2 ORFomes, respectively. Interestingly, using sera from asymptomatic versus symptomatic individuals, we identified four HSV-1 and HSV-2 cross-reactive ID-A-Ags that are highly and frequently recognized by serum antibodies from HSV-1-infected and HSV-2-infected asymptomatic, but not symptomatic, individuals. The identification of HSV-1 and HSV-2 ID-A-Ags should provide new insights into immune mechanisms that correlate with protection and may lead to the development of an effective immunotherapeutic vaccine against ocular, orofacial, and genital herpes.
MATERIALS AND METHODS
Study population.
From August 2003 to August 2011, we screened 345 HSV-1- and/or HSV-2-seropositive individuals. Among these, a cohort of 43 immunocompetent individuals, with an age range of 18 to 63 years (median, 31 years), who were seropositive or seronegative for HSV-1 and/or HSV-2 were enrolled in the present study. Table 1 shows the characteristics of this study population with respect to sex, age, HSV serology, and HSV disease. Thirty-two patients were HSV-1 seropositive and HSV-2 seronegative, among which 22 patients were healthy and asymptomatic (no history of recurrent HSV disease). The other 10 patients were defined as HSV-1/2-seropositive symptomatic individuals who suffered frequent and severe recurrent ocular and/or orofacial lesions, with two patients having had well-characterized herpes stromal keratitis (HSK). Six patients were HSV-2 seropositive and HSV-1 seronegative, 5 of whom were healthy and asymptomatic (no history of recurrent HSV disease). One patient was defined as an HSV-2-seropositive symptomatic individual who suffered frequent and severe recurrent genital lesions. Five individuals tested positive for both HSV-1 and HSV-2. Control individuals (n = 47) were seronegative for both HSV-1 and HSV-2 and had no history of ocular HSK, genital lesions, or orofacial herpes disease. Fifty-one individuals were Caucasian, 39 were non-Caucasian (African, Asian, Hispanic, and others), 51 were females, and 39 were males. All patients were negative for HIV and hepatitis B virus (HBV) and had no history of immunodeficiency. All subjects were enrolled at the University of California—Irvine (UCI) under an institutional review board-approved protocol (protocol no. 2009-6963). All subjects provided written informed consent.
Table 1.
Demographics and clinical features of the study populationa
| Subject-level characteristic | Value for all subjects (n = 90) |
|---|---|
| No. (%) of subjects of gender | |
| Female | 51 (56) |
| Male | 39 (44) |
| No. (%) of subjects of race | |
| Caucasian | 51 (57) |
| Non-Caucasian | 39 (43) |
| Median age (yr) (range) | 31 (18–63) |
| No. (%) of subjects with HSV status of: | |
| HSV-1 seropositive | 32 (35) |
| HSV-2 seropositive | 06 (07) |
| HSV-1 and HSV-2 seropositive | 05 (06) |
| HSV seronegative | 47 (52) |
| No. (%) of subjects with herpes disease status of: | |
| Seropositive symptomatic | 10 (11) |
| Seropositive asymptomatic | 33 (37) |
Sera from a total of 90 individuals attending the UCI Medical Center were used to probe the chips displaying the HSV-1 and HSV-2 protein microarrays. These 90 individuals were comprised of 32 HSV-1-, 6 HSV-2-, and 5 HSV-1/HSV-2-seropositive and 47 HSV-1- and HSV-2-seronegative individuals. Sera from all 90 individuals were serotyped by a using commercial gG1 and gG2 ELISA (FocuSelect 1 and 2 IgG). The seronegative individuals were used as negative controls and were used to establish baseline responses for each antigen.
HSV-1 and HSV-2 seropositivity screening.
Sera that had been banked at −80°C were coded and supplied for serological analysis without patient identifiers or clinical information. In addition to probing against protein arrays, the sera were assayed by FDA-approved commercial HSV-1 and HSV-2 ELISAs (FocuSelect 1 and 2) according to the manufacturer's instructions (Focus Diagnostics, Cypress, CA) (23). The sensitivity and specificity of these ELISAs were 91.2% (HSV-1) to 96.1% (HSV-2) and 92.3% (HSV-1) to 97% (HSV-2), respectively. Although this assay generally gives a clear-cut result, in some instances, the stereotyping was also validated by Western blotting, as previously described (72).
Construction of HSV-1 and HSV-2 proteome microarrays.
The construction of HSV-1 and HSV-2 proteome microarrays was described in detail in a previous report (39); proteome microarrays were fabricated, as described previously (19, 20, 54), by the PCR amplification of coding sequences in genomic DNA, followed by the insertion of amplicons into a T7 expression vector by homologous recombination and expression in coupled in vitro transcriptions-translations (IVTT) prior to printing onto microarrays. Gene sequences for PCR primer design were obtained from the NCBI (accession no. NC001806 and NC001798 for HSV-1 strain 17 and HSV-2 strain HG52, respectively). The gene nomenclature used is that published in the Oral Pathogen Genome Sequence Databases (ORALGEN) at the Los Alamos National Laboratory (http://www.oralgen.lanl.gov/). HSV-1 strain 17 DNA was supplied as 5 overlapping genomic fragments cloned into cosmids. HSV-2 strain HG52 DNA was supplied as virion-extracted DNA, and primers used for PCR amplification contained 20-bp nucleotides specific for each gene, with an extension of 20 bp complementary to ends of the linear pXT7 vector at the 5′ ends (19, 20, 54). The genome of herpes simplex viruses are CG rich (68% for HSV-1 and 70% for HSV-2). For PCR, genes were amplified by using AccuPrine GC-rich DNA polymerase (catalog no. 12337-016; Invitrogen) or 2× Phusion high-fidelity PCR master mix with GC buffer (catalog no. F-532S; Finnzymes/Thermo Scientific), with the addition of dimethyl sulfoxide (DMSO) (final concentration, 2%) and 8 ng/μl bovine serum albumin (BSA), using touchdown PCR with cycling conditions of an initial denaturation step at 98°C for 1 min followed by 20 cycles of 98°C for 10 s, 68°C for 20 s with a decremental temperature of 0.5°C/cycle, and 72°C for 30 s/kb, followed by 20 cycles of 98°C for 10 s, 58°C for 20 s, and 72°C for 30 s/kb. In vivo homologous recombination takes place between the PCR product and the pXT7 vector in competent DH5α cells. The recombinant plasmids were isolated from this culture by using a QIAprep 96 Turbo kit (Qiagen). Cloned genes were sequenced, and it was verified that the correct sequence was inserted.
For array fabrication, purified minipreparations of DNA were expressed in the E. coli-based in vitro transcription-translation expression system (RTS-100; Roche). Ten-microliter reaction mixtures were set up in sealed 384-well plates and incubated for 16 h at 24°C in a platform shaker at 300 rpm. A protease inhibitor cocktail (Complete; Roche) and Tween 20 at a final concentration of 0.05% were then added prior to printing. The rapid translation system (RTS) reactions were printed in singlicate without further purification onto 8-pad nitrocellulose-coated Fast slides (Whatman) by using a Gene Machine OmniGrid Accent microarray printer (Digilabs Inc.) in a 1-by-4 subarray format. Each subarray included multiple negative-control spots comprising “mock” RTS reaction mixtures lacking a DNA template. Each subarray also included positive-control spots of 4 serial dilutions of mouse, rat, and human whole immunoglobulin G (IgG) and 2 serial dilutions of human IgM and mouse IgM. Together, these positive and negative controls were used to normalize the data from different arrays (see below). Also included were 4 serial dilutions of purified recombinant Epstein-Barr virus nuclear antigen 1 (EBNA-1; DevaTal Inc., Hamilton, NJ), which is recognized by the majority of humans and which serves as a useful guide for serum quality.
To monitor the protein expression in each spot, we used antibodies against the N-terminal poly-His (clone His-1; Sigma) and the C-terminal hemagglutinin (HA) (clone 3F10; Roche) tags engineered into each protein. Arrays were first blocked for 30 min in protein array blocking buffer (Whatman) at room temperature (RT) and then probed for 1 h with anti-tag antibodies diluted 1/1,000 in blocking buffer. The slides were then washed six times in Tris-buffered saline (TBS) containing 0.05% (vol/vol) Tween 20 (T-TBS) and incubated with biotinylated secondary antibodies (Jackson ImmunoResearch). After washing the slides six times in T-TBS, bound antibodies were detected by incubation with streptavidin-conjugated SureLight P-3 (Columbia Biosciences). The slides were then washed three times each in T-TBS followed by TBS and dipped in distilled water prior to air drying by brief centrifugation. Slides were scanned with a Perkin-Elmer ScanArray confocal laser scanner, and data were acquired by using ScanArrayExpress software.
For probing with human sera, samples were diluted 1/200 in protein array blocking buffer supplemented with E. coli lysate (Antigen Discovery Inc.) at a final concentration of 10 mg/ml protein to block anti-E. coli antibodies and incubated at 37°C for 30 min with constant mixing. Meanwhile, the arrays were incubated in protein array blocking buffer for 30 min and probed with the pretreated sera overnight at 4°C with gentle rocking. The slides were then washed six times in T-TBS and incubated with biotinylated anti-human IgG(H+L) (Jackson ImmunoResearch) diluted 1/400 in protein array blocking buffer. After the slides were washed three times each with T-TBS and TBS, bound antibodies were visualized as described above.
ELISA.
Anti-HSV IgG responses was measured in serum samples by a solid-phase ELISA on microtiter plates, as we previously described (6, 64). Plates were coated overnight at 4°C with 50 μl of a 5-μg/ml solution of a protein (i.e., RTS) per well in 0.1 M carbonate-bicarbonate buffer (pH 9.6) per well. The plates were blocked with 1% BSA in phosphate-buffered saline (PBS) (pH 7.3). Serial 2-fold dilutions of sera in PBS–0.05% Tween 20–1% BSA (final volume, 50 μl) were added to the plates, which were incubated at 37°C for 1 h and then washed. Fifty microliters of a 1:2,000 dilution of peroxidase-conjugated mouse anti-human IgG (heavy and light chains) was then added to each well, and the plates were incubated for 1 h at 37°C. The unbound conjugate was removed by washing, and 50 μl of 0.04% o-phenylenediamine–hydrogen peroxidase in citrate-phosphate buffer was added to detect the bound enzyme. The reaction was stopped after 10 min by the addition of 25 μl of 2 M sulfuric acid per well, and the absorbance (A450) was determined with an automatic plate reader. Titers of anti-RTS antibodies are expressed as optical densities at 450 nm (OD450) as an ELISA ratio calculated as follows: OD450 of postimmune sera divided by OD450 of preimmune sera.
Array data analysis and statistical treatment.
Raw array data were collected as the mean pixel signal intensity data for each spot. To stabilize the variance of the raw data, a variant of the log transformation (asinh) was used (22, 33), and negative- and positive-control spots (the “no-DNA” and IgG spots, respectively) were used to normalize the data by use of the “VSN” package in R from the Bioconductor suite (http://Bioconductor.org/). Reactive antigens were defined as positive when the normalized log signal intensity was above the mean plus 2 standard deviations (SD) of the average no-DNA control spots. We then calculated P values for the log-normalized data by comparing signals between groups of donors by using a Bayes-regularized t test adapted from Cyber-T for use with protein arrays (22, 35). To account for multiple test conditions, we calculated P value adjustments by the Benjamini-Hochberg method (33, 43). Positive antigens were classified as type specific or cross-reactive according to significance (P values of <0.05 and ≥0.05, respectively). Receiver operator characteristic (ROC) analyses were performed with log-transformed/normalized array data for single antigens by testing signals for each donor as a threshold cutoff to discriminate HSV-1 and HSV-2 infections. Sensitivity and specificity values were calculated for each cutoff, and the data were plotted to give an ROC curve. The area under the curve (AUC) was used as a relative measure of each antigen's ability to discriminate between HSV-1 and -2 infections. For frequency-of-recognition (FR) analyses, a cutoff was defined for each antigen on the array by using the log-transformed/normalized data and was set as the average signal plus 3 SD of the seronegative population (as defined by a commercial ELISA). The numbers of individuals above the cutoff in each of the seropositive groups were determined and expressed as a percentage.
RESULTS
Construction and verification of HSV-1 and HSV-2 proteome microarrays.
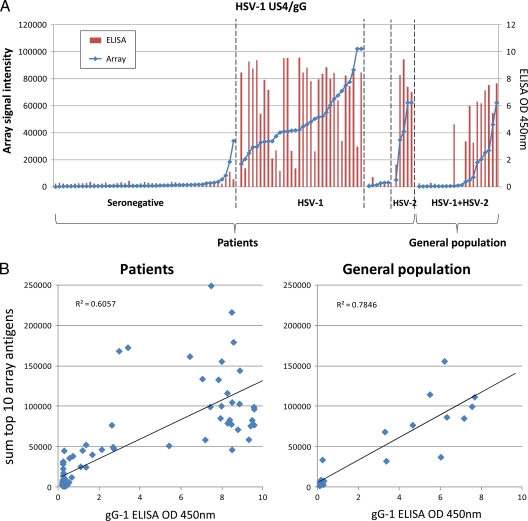
The construction of HSV-1 and HSV-2 proteome microarrays was described in detail in a previous report (39). Briefly, HSV-1 strain 17 template DNA was supplied as 5 overlapping genomic fragments cloned into cosmids (17). HSV-2 strain 333 DNA was prepared from virion-extracted DNA. Open reading frames were amplified and cloned into the T7 vector pXi, as described previously (19, 54). Recombinant plasmids were verified by “QC-PCR” (in which the correct size of the insert was verified by PCR) and/or by sequencing. Plasmids were expressed in vitro and printed onto nitrocellulose glass slides, and protein expression was verified by using monoclonal antibodies to terminal polyhistidine and hemagglutinin epitope tags, as described previously (19). Seroreactivity was defined as positive if average triplicate signals were above a threshold set as the average of control spots (IVTT reaction mixtures lacking a template plasmid) plus 2 standard deviations (SD). A total of 99% of HSV-1 and 97% of HSV-2 proteins were reactive with one or both of the antitag antibodies, and the remaining 1% and 3% were negative for both. To ascertain the sensitivity and specificity of our proteomic microarray, we compared the screening of either a panel of HSV ORFs or the HSV US4 ORF by the array to an ELISA. Figure 1A shows that the proteomic array is highly sensitive and specific compared to the ELISA. A strong correlation exists between the array and ELISA results when tested against a single antigen (i.e., US4/glycoprotein G [gG]) or against a panel of HSV antigens (Fig. 1B).
Fig 1.
Validation of the microarray. (A) The protein microarray signals for HSV-1 US4/gG were compared to OD450 readings by a FocuSelect ELISA using sera from 90 patients and 21 randomly selected general population controls, The patients were grouped according to serodiagnosis based on the commercial ELISA results and ranked within each group by the signal to HSV-1 US4/gG on the array. (B) Linear regression analysis between the sum of the top 10 HSV-1 antigens recognized on the array by HSV-1-seropositive individuals and ODs for patient sera (left) and population controls (right) determined by a FocuSelect ELISA.
The following paragraphs describe the identification of immunodominant antigens from HSV-1 and HSV-2 using the proteome-wide microarray. Details on the potential use of the identified antigens for diagnostic purposes at a point of care (POC) were discussed in a previous report (39).
Broad and diverse immunodominant HSV antigens targeted by serum antibodies from HSV-1- and HSV-2-seropositive individuals.
Protein microarray chips displaying either HSV-1 or HSV-2 ORFomes were used to identify immunodominant protein antigens (ID-Ags) that are recognized by serum antibodies from individuals infected with HSV-1, HSV-2, or both HSV-1 and HSV-2. A total of 90 individuals were enrolled, and their serum antibodies were used to probe the chips displaying the HSV-1 and HSV-2 protein microarrays. These individuals included 32 HSV-1-, 6 HSV-2-, and 5 HSV-1/HSV-2-seropositive individuals that were serotyped by use of commercial gG-1 and gG-2 ELISAs (FocuSelect). FocuSelect is an ELISA that is based on the recombinant glycoprotein G from HSV-1 (gG1) and HSV-2 (gG2). A control group, comprising 47 HSV-1- and HSV-2-seronegative individuals, was also enrolled to establish baseline responses for each antigen. All individuals were hepatitis and HIV negative. Table 1 shows demographics and clinical features of this study population with respect to gender, age, HSV serology, and HSV disease. At the time of blood drawing, none of the 43 HSV-1- and/or HSV-2-seropositive individuals had ongoing acute ocular, orofacial, or genital herpes disease. Each serum sample was probed against the microarrays in a single application. A serum sample was defined as being seropositive for a particular antigen if the signal was >C plus 2 SD (where C is the average signal of 16 control spots of IVTT reaction mixtures lacking template DNA). Reactive antigens were ranked by the normalized signal intensity and by Benjamini-Hochberg-corrected P values after comparing the seropositive and seronegative donors by using Bayesian t tests, as we described previously (39). The intensity of antibody responses, specific to the products of the HSV-1 and HSV-2 ORFomes, was determined for each individual and ranked as follows: strong responses, with an average signal intensity (ASI) of >20,000; medium responses, with an ASI of between >10,000 and <20,000; low responses, with an ASI of between >5,000 and <10,000; and no responses, with an ASI of <5,000. Positive antibody responses were compared with negative-control wells without Ag.
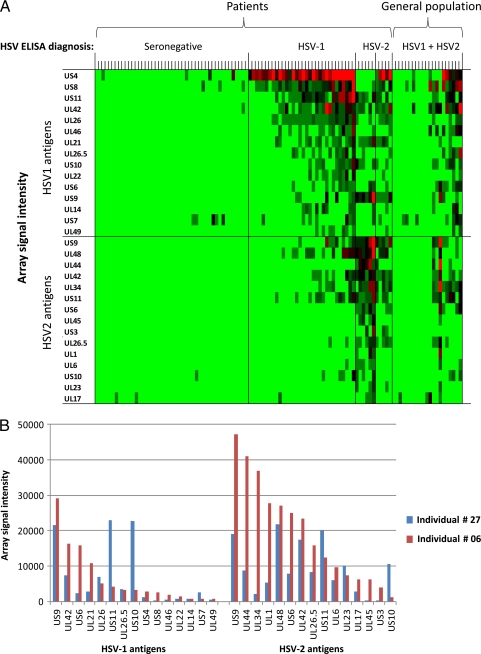
Figure 2A shows a heat map that provides an overview of the reactivities of serum antibodies against the top 15 seroreactive antigens from the HSV-1 and HSV-2 ORFomes. An antigen was defined as being seroreactive if the average signal intensity of one or more of the 5 donor groups was above a cutoff defined as C plus 2 SD. By this criterion, 26 different HSV-1 antigens and 30 different HSV-2 antigens were seroreactive. As expected, many ID-Ags belonged to the envelope proteins, but many tegument proteins were also reactive to serum antibodies. The heat map shows that the seronegative samples were minimally reactive against few HSV-1 or HSV-2 ORFs. The sera from a single seropositive individual targeted an average of 6 ORFs, with a maximum of 11 ORFs. These data were analyzed for type-specific and type-common antigens in our previous report (39). Figure 2B shows representative examples of array signal intensities of the antibody responses of two HSV-2-seropositive individuals. Overall, the reactivity of donor 6 against the individual HSV-2 antigens was stronger than the corresponding signal of donor 27.
Fig 2.
Intensities of antibody responses from HSV-1- and HSV-2-seropositive individuals against HSV-1 and HSV-2 antigens. (A) Heat map overview of broadly targeted HSV-1 and HSV-2 immunodominant antigens determined by the intensities of antibody responses. Columns correspond to sera used to probe the array, and rows are arrayed antigens. Sera were serotyped by using FocuSelect 1 and 2 IgG ELISAs (Focus Diagnostics), as shown at the top, and were used as the reference for sample categorizations. The patient sera were thus classified into seronegative (n = 47), HSV-1-seropositive-only (n = 32), HSV-2-seropositive-only (n = 6), and HSV-1- and HSV-2-seropositive (n = 5) groups. For comparison, sera from the general population were probed (n = 21). Only those antigens that were reactive against sera from the HSV-1- or HSV-2-seropositive populations are shown. An antigen was defined as reactive when the average signal intensity for a donor population was more than the mean plus 2 SD of the control spots consisting of IVTT reaction mixtures lacking a DNA template (C plus 2 SD). The HSV-1 antigens are ranked by descending average signal intensity of the HSV-1-seropositive population, and the HSV-2 antigens are similarly ranked by the HSV-2-positive population. In each case, only the top 15 antigens are shown. The sera are also ranked from left to right within each group by the increasing sum of the signals. The heat map was generated from log-normalized data that were retransformed to approximate raw values, and the signal was converted into a color (red, high; green, low). (B) Representative data for serum antibodies from two HSV-2-seropositive individuals recognizing protein microarrays derived from HSV-2 ORFs. IgG antibodies from seropositive individuals 6 and 27 reacted strongly to 7 and 11 ORFs, respectively.
Altogether, these results showed that (i) serum antibodies from HSV-1- and/or HSV-2-seropositive individuals frequently recognized many HSV antigens with a high to medium intensity compared to those from seronegative individuals; (ii) while, as expected, the envelope proteins appeared to be highly recognized by serum antibodies, many tegument proteins were also reactive to serum antibodies; (iii) interestingly, serum antibodies from a single HSV-seropositive individual appeared to broadly target HSV antigens, and serum antibodies recognized an average of 6 ORFs per seropositive individual; and (iv) the antibody responses to HSV antigens were diverse among HSV-1- and HSV-2-seropositive individuals. Whether the diversity in the antibody responses among HSV-1- and HSV-2-seropositive individuals is associated with the frequency of viral reactivation or with symptomatic and asymptomatic statuses warrants further investigation.
Immunodominant HSV-1 and HSV-2 antigens confirmed by the frequency of antibody responses from seropositive individuals.
To ascertain the immunodominance of the above-identified HSV-1 and HSV-2 ID-Ags, the frequency of antibody responses specific to each HSV-1 and HSV-2 ORFome was determined. The frequency of antibody responses was ranked as follows: frequent responses, with >75% average responders (ARs); medium frequency, with between <75% and >50% ARs; low frequency, with between <50% and >10% ARs; and no responses, with <10% ARs. An antigen was designated immunodominant when it was recognized by serum antibodies with a high to medium frequency from seropositive individuals with a high to medium intensity. The intensity was ranked as described above (i.e., strong responses, with an ASI of >20,000; medium responses, with an ASI of between >10,000 and <20,000 ASI; low responses, with an ASI of between >5,000 and <10,000; and no responses, with an ASI of <5,000). The various signals for a particular antigen probed with different sera (i.e., reading left to right across the heat map in Fig. 2) are directly correlated with antibody titers, since the concentration of a given protein was constant between different arrays. Thus, to identify a positive antigen, we derived P values by comparing seropositive and seronegative individuals using Bayesian t tests.
Based on these criteria, we confirmed that a panel of 21 ID-Ags from the HSV-1 ORFome was frequently recognized by serum antibodies from 32 HSV-1-seropositive individuals: US11, US4, US8, UL30, UL42, US11, UL26, UL46, US10, UL26.5, UL22, UL44, UL14, US7, UL10, UL39, UL49, UL1, UL25, UL21, and UL51, in descending order of average signal intensity. Another panel of 30 ID-Ags from the HSV-2 ORFome was recognized by serum antibodies from 6 HSV-2-seropositive individuals, US9, UL44, UL42, UL34, US11, US6, UL26.5, UL45, RL2 (ICP0), UL1, UL6, UL23, UL26, UL17, UL32, UL18, UL3, US8, UL51, UL28, UL14, UL10, UL49, UL41, UL7, UL50, US7, UL54, UL27, and UL5, in descending order of frequency. Some of the antigens were overlapping between the HSV-1 and HSV-2 ID-Ags. We also confirmed that the human antibody response to HSV-1 and HSV-2 antigens is remarkably broad, targeting an average of 6 ORFs per individual.
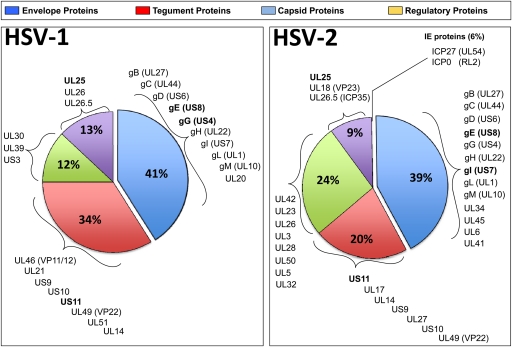
Although, as expected, the surface glycoproteins were among the most recognized antigens, sera from HSV-1/HSV-2-seropositive individuals also frequently recognized many tegument and regulatory proteins. The pie chart in Fig. 3 summaries the proportions of glycoproteins, teguments proteins, regulatory proteins, and capsid proteins, derived from either HSV-1 or HSV-2, that were recognized by serum antibodies from HSV-1- and/or HSV-2-seropositive individuals. The detection of a higher number of HSV-2 ID-Ags than HSV-1 ID-Ags might be attributed to (i) the few HSV-2-seropositive individuals enrolled in this study, compared to the relatively higher number of HSV-1-seropositive individuals (i.e., 6 versus 32 individuals), and (ii) the frequency of HSV-1 and HSV-2 reactivation.
Fig 3.
Pie chart summarizing the proportions of HSV-1 and HSV-2 envelope glycoproteins, tegument proteins, capsid proteins, and regulatory proteins recognized by serum antibodies from HSV-1- and/or HSV-2-seropositive individuals. Note that HSV-2 US4/gG was not present on the array.
Next, we hypothesized that the screening of HSV-1 and HSV-2 proteomes with sera from symptomatic versus asymptomatic individuals would uncover “symptomatic” and “asymptomatic” ID-Ags.
Definition of symptomatic and asymptomatic individuals.
A “black-and-white” definition of symptomatic and asymptomatic HSV patients is difficult, mainly because the spectrum of ocular, orofacial, and genital disease is broad, complex, and multifaceted (18). For simplicity, we concentrated on extreme disease situations, with the following definitions. Symptomatic individuals were defined as seropositive individuals with known HSV-1 and/or HSV-2 infection, as determined by a FocuSelect test and by physician examination or self-report, with 1 or more recurrent episodes of ocular, orofacial, and/or genital herpes per year for the past 2 years. Symptoms that typically define genital herpes include a burning feeling or pain in the genital area, dysuria, itching, tingling, sores, genital lesions, and vaginal discharge. Symptoms that typically define orofacial herpes are an eruption of painful sores in and around the oral cavity. This includes lips (cold sores), cheeks, nose, chin, roof of the mouth, tongue, and gums (51). Symptoms that typically define ocular herpes include a clinically well-documented history of recurrent HSV-1 ocular disease (HSK), such as herpetic lid lesions, herpetic conjunctivitis, dendritic or geographic keratitis, stromal keratitis, decreased corneal sensation, and iritis consistent with HSK. Asymptomatic individuals were defined as seropositive individuals with known HSV-1 and/or HSV-2 infection based on physician examination or self-report but who never had symptoms typical of any herpes disease. Seronegative individuals were defined as healthy individuals who were seronegative for both HSV-1 and HSV-2 infection and had no history of genital, orofacial, or ocular herpes disease.
Proteomic analysis of serum from HSV-1- and HSV-2-seropositive asymptomatic versus symptomatic individuals reveals potential symptomatic and asymptomatic antigens.
Sera from clinically defined symptomatic and asymptomatic HSV-seropositive individuals were used to screen HSV-1 and HSV-2 immunodominant antigens. The serum proteomic analysis was performed in a blinded study. Sera from symptomatic (n = 10) and asymptomatic (n = 10) individuals were collected from patients attending the UCI Institute for Clinical and Translational Science (ICTS) and clinics and were confirmed to be seropositive by a Focus HerpeSelect test, a single-antigen-based ELISA using recombinant gG antigens from HSV-1 and HSV-2. None of the sera were from donors with acute herpetic disease, thereby ruling out the parameter for ongoing acute HSV-1 and HSV-2 infections. Negative sera (n = 10) were obtained from seronegative individuals and were also confirmed to be negative by the FocuSelect test. To rank an antigen as seroreactive, both the frequency and the intensity of antibody responses, specific to the antigens of each HSV-1 and HSV-2 ORFome, were determined by using the same criteria described above. An antigen was designated asymptomatic (ID-A-Ag) when it was recognized with a high to medium frequency by serum antibodies from seropositive asymptomatic individuals with a high to medium intensity. Inversely, an antigen was designated as an immunodominant symptomatic antigen (ID-S-Ag) when it is was recognized with a high to medium frequency by serum antibodies from seropositive symptomatic individuals with a high to medium intensity.
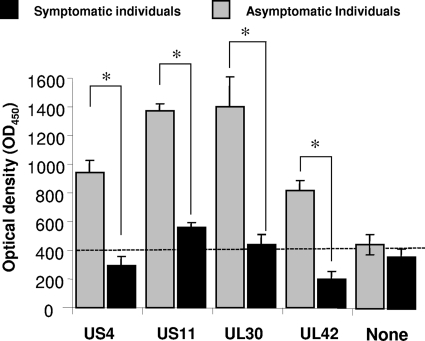
As shown in Table 2, four HSV ID-A-Ags (US4, US11, US10, and UL42) were frequently recognized by serum antibodies from 10 out of 10 asymptomatic patients but not by serum antibodies from 10 symptomatic patients (P < 0.001 by an analysis of variance [ANOVA] posttest). In contrast, serum antibodies from symptomatic patients preferentially recognized the US10 antigen (P < 0.001). The high intensity of recognition of the four HSV ID-A-Ags by serum antibodies from HSV-1- and HSV-2-seropositive asymptomatic individuals was confirmed by an ELISA. Figure 4 shows the average intensities of recognition of RTS expressing US4, US11, UL30, and UL42 proteins by HSV-seropositive symptomatic (n = 10) and asymptomatic (n = 10) patients, confirmed by an ELISA. The P value was calculated by using the ANOVA two-tailed test. Note the difference in serum antibody responses induced by US11 and UL30 in asymptomatic versus symptomatic patients. The antibody recognitions of four antigens (US4, US11, UL30, and UL42) were different by >2-fold between symptomatic and asymptomatic serum samples. Elevated antibody responses were confirmed by immunohistochemistry and immunoblotting (not shown). Altogether, these data identified, for the first time, novel ID-A-Ags that are recognized specifically by antibodies from symptomatic versus asymptomatic individuals.
Table 2.
Frequencies of antibody responses in symptomatic versus asymptomatic individualsa
| HSV-1 antigen | Description | % frequency of response in subjects |
P value | ||
|---|---|---|---|---|---|
| Seronegative (n = 47) | Symptomatic (n = 10) | Asymptomatic (n = 10) | |||
| UL26.5 | Capsid scaffold protein, ICP35, VP22a | 0.0 | 80.0 | 87.5 | >0.05 |
| UL46 | Tegument protein, VP11/12 | 2.1 | 90.0 | 81.3 | >0.05 |
| UL30 | DNA-directed polymerase homolog | 2.1 | 10.0 | 71.9 | >0.05 |
| UL26 | Serine protease, VP40 | 4.3 | 78.0 | 78.1 | >0.05 |
| US6 | Virion gD | 4.3 | 90.0 | 80 | <0.001 |
| US4 | Virion gG | 2.1 | 10.0 | 80 | <0.001 |
| US11 | RNA-binding tegument protein | 2.1 | 65.0 | 65.6 | >0.05 |
| UL39 | Ribonucleotide reductase subunit homolog | 2.1 | 60.0 | 62.5 | >0.05 |
| US8 | gE | 2.1 | 60.0 | 62.5 | >0.05 |
| US11 | RNA-binding tegument protein | 4.3 | 4.0 | 56.3 | <0.05 |
| US10 | Tegument protein | 2.1 | 60.0 | 10 | <0.005 |
| UL42 | DNA polymerase accessory protein | 2.1 | 10.0 | 60 | <0.005 |
| UL10 | Virion gM | 2.1 | 50.0 | 50.0 | >0.05 |
| UL51 | Tegument protein | 2.1 | 40.0 | 46.9 | >0.05 |
| UL22 | Virion gH | 4.3 | 40.0 | 46.9 | >0.05 |
| UL44 | Virion gC | 4.3 | 40.0 | 43.8 | >0.05 |
| UL7 | Regulatory protein | 0.0 | 40.0 | 40.6 | >0.05 |
| US9 | Tegument protein | 0.0 | 40.0 | 40.6 | >0.05 |
Shown are frequencies of recognition of HSV-1 immunodominant antigens in seropositive symptomatic and asymptomatic individuals measured by ELISA. Percentages shown are those for 10 asymptomatic and 10 symptomatic individuals reactive for individual HSV-1 and HSV-2 antigens, using a cutoff defined by the mean plus 3 SD of the seronegative population. Patient groups were defined as described in Table 1. Antigens that discriminate HSV-1- and HSV-2-seropositive asymptomatic and symptomatic individuals (i.e., P < 0.05 when the two populations are compared by t tests) are indicated in boldface type. ORF product descriptions were obtained from the Uniprot database (http://www.uniprot.org/) and the Oralgen database (http://www.oralgen.lanl.gov/), with additional annotation from recent mass spectrometry data (42).
Fig 4.
Intensities of antibody responses against four immunodominant HSV-1 antigens from symptomatic and asymptomatic patients. The average intensity of recognition of RTS expressing US4, US11, UL30, and UL42 proteins by serum antibodies from HSV-seropositive symptomatic (n = 10) and asymptomatic patients (n = 10) was determined by an ELISA. The optical density (OD) at 450 nm determined by an ELISA is shown for each serum sample. The P value was calculated by t tests. Note the difference in serum antibody responses induced by US11 and UL30 in asymptomatic versus symptomatic patients. The antibody recognition of four antigens (US4, US11, UL30, and UL42) was different by >2-fold between serum samples from symptomatic and asymptomatic subjects.
Interestingly, the surface gG encoded by US4 appeared to be the only ID-A-Ag recognized by serum antibodies from asymptomatic individuals (i.e., gG with an unknown function) (Table 2). The other three ID-A-Ags, US11, UL30, and UL42, are either tegument or regulatory proteins (Table 2). US11 is a tegument-associated phosphoprotein involved in RNA binding and posttranscriptional regulation. Both UL30 and UL42 are polymerase accessory proteins involved in DNA replication. The only detected ID-S-Ag, US10, is a capsid/tegument-associated phosphoprotein with an unknown function.
DISCUSSION
HSV-1 and HSV-2 are medically significant viral pathogens. The failure to develop an HSV vaccine despite decades of effort might be attributed to the historic focus on two glycoproteins, gB and gD, as potential vaccine targets. In this study, panels of 21 and 30 ID-Ags were identified from the HSV-1 and HSV-2 ORFomes, respectively, as being highly and frequently recognized by serum antibodies from seropositive individuals. Interestingly, the antibody responses to HSV antigens were broad and diverse among HSV-1- and HSV-2-seropositive individuals, suggesting that a panel of antigens rather than one antigen in isolation may induce the natural protective immunity observed for asymptomatic individuals. This implies that an efficient HSV vaccine should be multivalent and targeted at a select panel of key ID-A-Ags.
To advance HSV vaccine development, we have constructed protein microarrays representing the entire HSV-1 and HSV-2 proteomes and have probed these arrays with serum antibodies from asymptomatic individuals with “natural protection” versus symptomatic individuals with “no protection.” We have identified a set of four ID-A-Ags from HSV-1 and HSV-2 that was strongly recognized by antibodies from asymptomatic individuals but not by antibodies from symptomatic individuals. These HSV-1 and HSV-2 ID-A-Ags are considered potentially “protective” antigens and therefore can be included in a therapeutic vaccine against herpes disease. In contrast, one ID-S-Ag, US10, was strongly recognized by antibodies from symptomatic individuals with frequent recurrent herpetic disease but not by antibodies from asymptomatic individuals. This ID-S-Ag recognized by antibodies from symptomatic individuals is considered potentially “pathogenic” and must be excluded from any HSV vaccine formulation. Our results do not imply that the identified ID-A-Ags and ID-S-Ags are the only ones. Many of our ORFs are recognized by sera from both symptomatic and asymptomatic individuals, many of which have yet to be characterized as either symptomatic or asymptomatic Ags using a larger population of seropositive symptomatic and asymptomatic individuals.
While many people have frequent recurrences of herpes disease (i.e., 1 or more episodes/year), others have no history of recurrent disease (i.e., less than 1 episode/year). A complete comparison of the B- and T-cell responses in symptomatic and asymptomatic patients is lacking yet essential in order to develop efficient immunotherapeutic vaccine strategies. The primary goal of this study was to identify human asymptomatic antigens on HSV-1 and HSV-2 ORFomes (i.e., ID-A-Ags) to assist in the building of a profile of “surrogate markers” of protection and of potential “protective” HSV ID-A-Ags to be incorporated into an effective therapeutic vaccine. This also involves the identification and elimination of human symptomatic antigens (i.e., ID-S-Ags) that may be used as surrogate markers of pathogenicity. Regardless of the mechanism by which ID-S-Ags induce “pathogenic” disease, it is logical to exclude those ID-S-Ags from vaccines, since they may be harmful by exacerbating recurrent herpetic disease (be that ocular, orofacial, or genital). We emphasize that our results do not imply that the currently identified symptomatic antigen US10 is the only one present in HSV-1 and HSV-2. It is likely that other antigens, including glycoprotein K (gK) or UL53 (57, 58), exist and may be identified once the library of antigenic ORFs and the panel of symptomatic and asymptomatic sera are expanded.
Over two-thirds of the global population (∼4.66 billion people) is currently infected with HSV-1 and/or HSV-2 (24, 53, 84). The majority of those infected (∼90%) will never have active ocular, orofacial, or genital herpes and will remain asymptomatic (18). Only a minority are symptomatic and often experience recurrent ocular, orofacial, and/or genital herpes disease (18). As a result, many HSV-seropositive individuals remain undiagnosed because of uncharacteristic clinical presentations or the lack of noticeable lesions (53). Currently, there is no simple point-of-care (POC) in-clinic test for the classification of symptomatic versus asymptomatic patients. In addition, there is no reliable, cost-effective, and sensitive POC diagnostic test to discriminate between HSV-1 and HSV-2 infections. The only currently available commercial test, the FocuSelect test, a single-antigen ELISA that is based on the recombinant gG antigens from HSV-1 (gG1) and HSV-2 (gG2), remains an expensive and not-practicable diagnostic test for developing countries in regions where HSV is endemic, such as the sub-Saharan countries. The ID-Ag identified in this study could obviously be used to develop a reliable, cost-effective, and sensitive POC diagnostic test that would discriminate between HSV-1 and HSV-2 infection, as discussed in our previous report (39).
Considerable homology exists between the HSV-1 and HSV-2 genomes (42). These homologous sequences are distributed over the entire genomic map, and most of the ORFs specified by one viral type are antigenically related to ORFs of the other viral type. This results in considerable cross-reactivity between the HSV-1 and HSV-2 glycoproteins, although unique antigenic determinants exist for each virus. Eleven glycoproteins of HSV have been identified (gB, gC, gD, gE, gG, gH, gI, gJ, gK, gL, and gM), with a 12th being predicted (gN). gD is the most potent inducer of neutralizing antibodies and appears to be related to viral entry into a cell, and gB is also required for infectivity. Antigenic specificity is provided by gG, with the resulting antibody response allowing for a distinction between HSV-1 (gG1) and HSV-2 (gG2).
Importantly, whether some of the identified antigens can be used as surrogate markers to identify potential symptomatic and asymptomatic individuals warrants further investigation. The monitoring of herpes disease and immunity has become more difficult for individuals showing low-level antibody responses. Commonly available techniques for HSV diagnostics and seroepidemiology have limited sensitivity during early infection and remain costly. Thus, the second aim will be to develop a sensitive tool to assess a patient's level of exposure to HSV with a very small amount of serum, saliva, or tears or with vaginal swabs. As a good start, we were able to test serum antibodies of 32 HSV-1- and 6 HSV-2-seropositive individuals from Southern California regions only with different levels of herpes infections. A multiplex diagnostic assay based on US4, US11, UL30, and UL42 ID-A-Ags would therefore be rapid, sensitive, and reproducible for a small volume of serum. It would provide a useful tool to evaluate antibody responses to multiple Ags in large populations where antibody titers might be low, as is the case of the symptomatic individuals tested in our study. This could replace the current diagnostic FocuSelect test, which is costly, especially for developing countries, such as countries in the sub-Saharan region, where herpes disease is endemic. This present technology would be a rapid, comprehensive, and high-throughput serodiagnostic test that would discriminate between HSV-1 and HSV-2 infections. These results were discussed our previous report (39).
The limitation of E. coli-based expression systems is the lack of posttranslational modification machinery. Therefore, antibodies that recognize proteins based on the posttranslational modification of the target antigen, such as phosphorylation, glycosylation, or lipidation, will not be identified with this method. However, it was shown previously by using microarrays that in the case of vaccinia virus, the known glycosylated antigens were recognized by sera from immunized humans and animals (21). Since none of the proteins on the array were posttranslationally modified, this finding implies that at least a portion of the natural polyclonal response to these proteins is directed against epitopes, or domains, that have not been posttranslationally modified. Similar limitations may occur with conformational B-cell epitopes and those formed by disulfide bonds or the multimerization of proteins. Epitopes requiring disulfide bonds for Ab recognition can either be seen or not depending on whether they are expressed in vitro under oxidizing conditions and whether the protein is correctly folded. Regardless of this limitation, if the long-term goal is to produce a vaccine with a recombinant protein, the use of an antigen that induces protection as a result of posttranslational modifications, or conformational epitopes, may represent a shortcoming for large-scale manufacturing. Another limitation of the type of microarray used in the current study is that antibody reactivity toward the poly-His or the HA tag cannot be used to quantify the amount of antigen present in each individual spot. The binding of the antibodies to the poly-His and HA tags appears to be affected by many factors, which may include the availability of the tags for binding due to folding. We therefore cannot compare quantitatively the antibody responses between different antigens. We can, however, quantitatively monitor the antibody response to a particular antigen, since the amount of protein per spot for each antigen is the same from array to array.
It should be pointed out that there are limitations of protein microarray technologies, including the difficulty in purifying proteins in combination with high-throughput gene expression systems (54). On one hand, because of the complexity of protein folding and posttranslational modifications, proteins are often difficult to re-create on a microarray platform. On the other hand, standard criteria for array production and data normalization with noise models, variance estimation, and differential expression analysis techniques have become powerful tools for the interpretation of results. Based on this progress, successful attempts have been made using protein microarrays for profiling the antibody responses to numerous infectious pathogens (3, 21, 40, 54, 66).
Homologous sequences exist between the HSV-1 and HSV-2 genomes and are distributed over the entire genomic map (42). Most of the ORFs specified by one viral type are antigenically related to ORFs of the other viral type. This indicates that considerable cross-reactivity does exist between the HSV-1 and HSV-2 antigens, although unique antigenic determinants might be present for each virus, as we recently described (39). Therefore, it is unlikely that the HSV-1 US4 antigen does not cross-react with its HSV-2 US4 ortholog. If the US4 asymptomatic antigen is absent in the HSV-2 ORF, we will not be able to determine whether the antibodies induced in HSV-1-seropositive individuals specifically recognize HSV-2 US4. However, sera from HSV-2-seropositive individuals did not recognize HSV-1 US4. Our approach appears be very specific, given the appropriate antigen, as in the case of HSV-1 US4. The specificity of US4 is already well established, and it was our intention to discover additional useful antigens. Not obtaining the HSV-2 US4 was unfortunate, but it does not automatically rule out others of the 80+ HSV-2 antigens that are potentially type specific.
Considering the wealth of data addressing the role of protective versus pathogenic immune effectors in animal models, it is surprising how there are few reports that have explored the immunological basis of symptomatic and asymptomatic HSV infections in humans. The identification of these immune mechanisms by which asymptomatic patients control herpes disease and symptomatic patients do not, or at least the antigens involved, is critical for the advancement of HSV vaccine development. The symptomatic (i.e., pathogenic) and asymptomatic (i.e., protective) antigens identified in this study may be key to an understanding of the mechanisms of pathogenicity versus protection in HSV-infected humans. Multiple and complex mechanisms might be in play, including (i) differences in precursor frequencies, proliferative capacities, and functional properties of symptomatic versus asymptomatic epitope-specific B cells; (ii) the possibility that asymptomatic antigens might trigger neutralizing protective antibodies, while symptomatic antigens might trigger nonneutralizing pathogenic antibodies; (iii) the possibility that symptomatic antigens may direct antibody responses away from those that are best suited to neutralize the viral infection with a minimal pathogenic reaction; and (iv) finally, the possibility that B-cell cross-reactivity with antigens and epitopes from other viral pathogens, within or outside the herpesvirus family, may also play a role in protective immunity versus damaging immunopathology (41, 81).
T cells, mainly of the CD4+ subsets, rather than antibodies appear to mediate herpetic disease (26, 74, 77). Until our recent studies with gB and gD (13), no symptomatic or asymptomatic HSV T-cell epitopes had been identified. The symptomatic and asymptomatic B-cell antigens identified in this study would certainly help speed up the identification of more symptomatic and asymptomatic T-cell antigens by serodiagnosing more symptomatic and asymptomatic individuals. In addition, many HSV and non-HSV antigens tend to contain both B- and T-cell epitopes (36, 59, 60). It was suggested previously that the most immunodominant B-cell antigens in HSV may be those of envelope proteins, such as the envelope glycoproteins UL27/gB and US6/gD (10). However, many CD4+ and CD8+ T-cell epitopes have been recently identified, by our laboratory and others, for gB and gD (5, 13, 16, 45, 47, 48, 50, 55, 56, 71). Tegument proteins such as those encoded by HSV-1/2 UL39, UL41, UL46/VP11/12, UL47/VP13/14, UL48/VP16, and UL49 and immediate-early (IE) proteins such as RL2/ICP0 and RS1/ICP4 have also been identified as major targets for effector T cells (12, 34, 37, 46, 49). Of these, the arrays detected strong antibody signals against US6/gD (from both HSV-1 and -2) and UL48 of HSV-2, both of which are also CD4+ target antigens. The overlap between the targets of antibody and CD4+ cells, rather than CD8+ cells, is also consistent with recent findings for vaccinia virus antigens (36, 59, 60). Further investigations of T-cell responses to ID-A-Ags versus ID-S-Ags may be needed to break new ground in our understanding of the protective versus pathogenic immune mechanisms against ocular, orofacial, and genital herpes. We should, however, emphasize that T- and B-cell epitopes have frequently been observed in other systems to cluster within a limited region of Ags (6, 38, 76). We are currently using a genomic approach to detect HSV-specific T-cell dominance in symptomatic versus asymptomatic individuals.
Whether the ID-A-Ags identified in this study are truly protective and whether the ID-S-Ags are pathogenic remain to be determined. A logical extension of this screening study is to construct and test their protective efficacy in animal models of HSV infection (mice, guinea pigs, and rabbits) using vaccine formulations that will exclusively incorporate the ID-A-Ags (but exclude the symptomatic antigens). We expect that the next immunogenicity and protective preclinical studies that use the four identified ID-A-Ags (US4, US11, UL30, and UL42) individually and in combination will confirm their protective function. Inversely, we also expect that the ID-S-Ag (UL10) identified in this study will exacerbate herpes disease following the vaccination of latently infected animals. Results from these studies will be the subject of future reports.
In conclusion, the present study (i) validates the use of a high-throughput microarray analysis for the characterization of the Ab response to HSV-1 and HSV-2 antigens; (ii) shows that the antibody responses to HSV-1 and HSV-2 antigens among seropositive individuals were broad and diverse; (iii) reports 21 and 30 immunodominant HSV-1 and HSV-2 antigens, respectively; and (iv) characterizes four previously unreported ID-A-Ags that, considering the recent unsuccessful clinical HSV vaccine (4), are potentially useful to be included in a novel asymptomatic vaccine against HSV.
ACKNOWLEDGMENTS
This work was supported by NIH Public Health Service grants EY14900 and EY019896 (L.B.M.), R44AI058365 (D.H.D.), and U01AI078213 (P.L.F.); the Discovery Eye Foundation; the Henry L. Guenther Foundation; and a Research To Prevent Blindness challenge grant.
We thank Steven L. Wechsler and Dale Carpenter for providing the HSV-2 DNA.
Footnotes
Published ahead of print 8 February 2012
REFERENCES
- 1. Anderson KS, et al. 2008. Application of protein microarrays for multiplexed detection of antibodies to tumor antigens in breast cancer. J. Proteome Res. 7:1490–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson KS, et al. 2011. Protein microarray signature of autoantibody biomarkers for the early detection of breast cancer. J. Proteome Res. 10:85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbour AG, et al. 2008. A genome-wide proteome array reveals a limited set of immunogens in natural infections of humans and white-footed mice with Borrelia burgdorferi. Infect. Immun. 76:3374–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Belshe PB, et al. 2012. Efficacy results of a trial of a herpes simplex vaccine. N. Engl. J. Med. 366:34–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. BenMohamed L, et al. 2003. Identification of novel immunodominant CD4+ Th1-type T-cell peptide epitopes from herpes simplex virus glycoprotein D that confer protective immunity. J. Virol. 77:9463–9473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. BenMohamed L, et al. 1997. Lipopeptide immunization without adjuvant induces potent and long-lasting B, T helper, and cytotoxic T lymphocyte responses against a malaria liver stage antigen in mice and chimpanzees. Eur. J. Immunol. 27:1242–1253 [DOI] [PubMed] [Google Scholar]
- 7. Bernstein DI, Spruance SL, Arora SS, Schroeder JL, Meng TC. 2005. Evaluation of imiquimod 5% cream to modify the natural history of herpes labialis: a pilot study. Clin. Infect. Dis. 41:808–814 [DOI] [PubMed] [Google Scholar]
- 8. Bettahi I, Zhang X, Afifi RE, BenMohamed L. 2006. Protective immunity to genital herpes simplex virus type 1 and type 2 provided by self-adjuvanting lipopeptides that drive dendritic cell maturation and elicit a polarized Th1 immune response. Viral Immunol. 19:220–236 [DOI] [PubMed] [Google Scholar]
- 9. Burbelo PD, Ching KH, Bush ER, Han BL, Iadarola MJ. 2010. Antibody-profiling technologies for studying humoral responses to infectious agents. Expert Rev. Vaccines 9:567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burbelo PD, et al. 2009. Serological diagnosis of human herpes simplex virus type 1 and 2 infections by luciferase immunoprecipitation system assay. Clin. Vaccine Immunol. 16:366–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cantin E, Tanamachi B, Openshaw H, Mann J, Clarke K. 1999. Gamma interferon (IFN-gamma) receptor null-mutant mice are more susceptible to herpes simplex virus type 1 infection than IFN-gamma ligand null-mutant mice. J. Virol. 73:5196–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carmack MA, et al. 1996. T cell recognition and cytokine production elicited by common and type-specific glycoproteins of herpes simplex virus type 1 and type 2. J. Infect. Dis. 174:899–906 [DOI] [PubMed] [Google Scholar]
- 13. Chentoufi AA, et al. 2008. Asymptomatic human CD4+ cytotoxic T-cell epitopes identified from herpes simplex virus glycoprotein B. J. Virol. 82:11792–11802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chentoufi AA, et al. 2010. A novel HLA (HLA-A*0201) transgenic rabbit model for preclinical evaluation of human CD8+ T cell epitope-based vaccines against ocular herpes. J. Immunol. 184:2561–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chentoufi AA, et al. 2010. Nasolacrimal duct closure modulates ocular mucosal and systemic CD4(+) T-cell responses induced following topical ocular or intranasal immunization. Clin. Vaccine Immunol. 17:342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chentoufi AA, et al. 2008. HLA-A*0201-restricted CD8+ cytotoxic T lymphocyte epitopes identified from herpes simplex virus glycoprotein D. J. Immunol. 180:426–437 [DOI] [PubMed] [Google Scholar]
- 17. Cunningham C, Davison AJ. 1993. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology 197:116–124 [DOI] [PubMed] [Google Scholar]
- 18. Dasgupta G, Nesburn AB, Wechsler SL, BenMohamed L. 2010. Developing an asymptomatic mucosal herpes vaccine: the present and the future. Future Microbiol. 5:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Davies DH, et al. 2005. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl. Acad. Sci. U. S. A. 102:547–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davies DH, et al. 2005. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J. Virol. 79:11724–11733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davies DH, et al. 2008. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus Ankara is comparable to that of Dryvax. J. Virol. 82:652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Durbin BP, Hardin JS, Hawkins DM, Rocke DM. 2002. A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics 18(Suppl 1):S105–S110 [DOI] [PubMed] [Google Scholar]
- 23. Field PR, Ho DW, Irving WL, Isaacs D, Cunningham AL. 1993. The reliability of serological tests for the diagnosis of genital herpes: a critique. Pathology 25:175–179 [DOI] [PubMed] [Google Scholar]
- 24. Fleming DT, et al. 1997. Herpes simplex virus type 2 in the United States, 1976 to 1994. N. Engl. J. Med. 337:1105–1111 [DOI] [PubMed] [Google Scholar]
- 25. Freeman ML, Sheridan BS, Bonneau RH, Hendricks RL. 2007. Psychological stress compromises CD8+ T cell control of latent herpes simplex virus type 1 infections. J. Immunol. 179:322–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gangappa S, Deshpande SP, Rouse BT. 1999. Bystander activation of CD4(+) T cells can represent an exclusive means of immunopathology in a virus infection. Eur. J. Immunol. 29:3674–3682 [DOI] [PubMed] [Google Scholar]
- 27. Geiger KD, et al. 1997. Interferon-gamma protects against herpes simplex virus type 1-mediated neuronal death. Virology 238:189–197 [DOI] [PubMed] [Google Scholar]
- 28. Ghiasi H, Cai S, Slanina S, Nesburn AB, Wechsler SL. 1995. Vaccination of mice with herpes simplex virus type 1 glycoprotein D DNA produces low levels of protection against lethal HSV-1 challenge. Antiviral Res. 28:147–157 [DOI] [PubMed] [Google Scholar]
- 29. Ghiasi H, Kaiwar R, Nesburn AB, Wechsler SL. 1992. Baculovirus-expressed glycoprotein G of herpes simplex virus type 1 partially protects vaccinated mice against lethal HSV-1 challenge. Virology 190:233–239 [DOI] [PubMed] [Google Scholar]
- 30. Ghiasi H, Perng GC, Cai S, Nesburn AB, Wechsler SL. 1996. The UL3 open reading frame of herpes simplex virus type 1 codes for a phosphoprotein. Virus Res. 44:137–142 [DOI] [PubMed] [Google Scholar]
- 31. Ghiasi H, Slanina S, Nesburn AB, Wechsler SL. 1994. Characterization of baculovirus-expressed herpes simplex virus type 1 glycoprotein K. J. Virol. 68:2347–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Herpetic Eye Disease Study Group 1998. Acyclovir for the prevention of recurrent herpes simplex virus eye disease. N. Engl. J. Med. 339:300–306 [DOI] [PubMed] [Google Scholar]
- 33. Hochberg Y, Benjamini Y. 1990. More powerful procedures for multiple significance testing. Stat. Med. 9:811–818 [DOI] [PubMed] [Google Scholar]
- 34. Hosken N, et al. 2006. Diversity of the CD8+ T-cell response to herpes simplex virus type 2 proteins among persons with genital herpes. J. Virol. 80:5509–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl 1):S96–S104 [DOI] [PubMed] [Google Scholar]
- 36. Jing L, et al. 2008. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J. Virol. 82:7120–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jing L, et al. 2012. Cross-presentation and genome-wide screening reveal candidate T cells antigens for a herpes simplex virus type 1 vaccine. J. Clin. Invest. 122:654–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kabilan L, et al. 1988. T-cell epitopes in Pf155/RESA, a major candidate for a Plasmodium falciparum malaria vaccine. Proc. Natl. Acad. Sci. U. S. A. 85:5659–5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kalantari-Dehaghi M, et al. 2012. Discovery of potential diagnostic and vaccine antigens in herpes simplex virus 1 and 2 by proteome-wide antibody profiling. J. Virol. 86:4328–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kalantari-Dehaghi M, et al. 2011. New targets of pemphigus vulgaris antibodies identified by protein array technology. Exp. Dermatol. 20:154–156 [DOI] [PubMed] [Google Scholar]
- 41. Kim SK, et al. 2005. Private specificities of CD8 T cell responses control patterns of heterologous immunity. J. Exp. Med. 201:523–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kimberlin DW, et al. 2011. Oral acyclovir suppression and neurodevelopment after neonatal herpes. N. Engl. J. Med. 365:1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klipper-Aurbach Y, et al. 1995. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med. Hypotheses 45:486–490 [DOI] [PubMed] [Google Scholar]
- 44. Knaup B, Schunemann S, Wolff MH. 2000. Subclinical reactivation of herpes simplex virus type 1 in the oral cavity. Oral Microbiol. Immunol. 15:281–283 [DOI] [PubMed] [Google Scholar]
- 45. Koelle DM. 2003. Expression cloning for the discovery of viral antigens and epitopes recognized by T cells. Methods 29:213–226 [DOI] [PubMed] [Google Scholar]
- 46. Koelle DM, Abbo H, Peck A, Ziegweid K, Corey L. 1994. Direct recovery of herpes simplex virus (HSV)-specific T lymphocyte clones from recurrent genital HSV-2 lesions. J. Infect. Dis. 169:956–961 [DOI] [PubMed] [Google Scholar]
- 47. Koelle DM, et al. 2001. CD8 CTL from genital herpes simplex lesions: recognition of viral tegument and immediate early proteins and lysis of infected cutaneous cells. J. Immunol. 166:4049–4058 [DOI] [PubMed] [Google Scholar]
- 48. Koelle DM, Corey L. 2003. Recent progress in herpes simplex virus immunobiology and vaccine research. Clin. Microbiol. Rev. 16:96–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Koelle DM, et al. 1994. Antigenic specificities of human CD4+ T-cell clones recovered from recurrent genital herpes simplex virus type 2 lesions. J. Virol. 68:2803–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koelle DM, et al. 2003. Immunodominance among herpes simplex virus-specific CD8 T cells expressing a tissue-specific homing receptor. Proc. Natl. Acad. Sci. U. S. A. 100:12899–12904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kriesel JD, et al. 2011. C21orf91 genotypes correlate with herpes simplex labialis (cold sore) frequency: description of a cold sore susceptibility gene. J. Infect. Dis. 204:1654–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leigh JF, Acharya N, Cevallos V, Margolis TP. 2008. Does asymptomatic shedding of herpes simplex virus on the ocular surface lead to false-positive diagnostic PCR results? Br. J. Ophthalmol. 92:435–436 [DOI] [PubMed] [Google Scholar]
- 53. Looker KJ, Garnett GP, Schmid GP. 2008. An estimate of the global prevalence and incidence of herpes simplex virus type 2 infection. Bull. World Health Organ. 86:805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Luevano M, et al. 2010. High-throughput profiling of the humoral immune responses against thirteen human papillomavirus types by proteome microarrays. Virology 405:31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mikloska Z, Cunningham AL. 1998. Herpes simplex virus type 1 glycoproteins gB, gC and gD are major targets for CD4 T-lymphocyte cytotoxicity in HLA-DR expressing human epidermal keratinocytes. J. Gen. Virol. 79(Pt 2):353–361 [DOI] [PubMed] [Google Scholar]
- 56. Mikloska Z, Kesson AM, Penfold ME, Cunningham AL. 1996. Herpes simplex virus protein targets for CD4 and CD8 lymphocyte cytotoxicity in cultured epidermal keratinocytes treated with interferon-gamma. J. Infect. Dis. 173:7–17 [DOI] [PubMed] [Google Scholar]
- 57. Mott KR, et al. 2009. The role of a glycoprotein K (gK) CD8+ T-cell epitope of herpes simplex virus on virus replication and pathogenicity. Invest. Ophthalmol. Vis. Sci. 50:2903–2912 [DOI] [PubMed] [Google Scholar]
- 58. Mott KR, et al. 2007. Role of anti-glycoproteins D (anti-gD) and K (anti-gK) IgGs in pathology of herpes stromal keratitis in humans. Invest. Ophthalmol. Vis. Sci. 48:2185–2193 [DOI] [PubMed] [Google Scholar]
- 59. Moutaftsi M, et al. 2007. Vaccinia virus-specific CD4+ T cell responses target a set of antigens largely distinct from those targeted by CD8+ T cell responses. J. Immunol. 178:6814–6820 [DOI] [PubMed] [Google Scholar]
- 60. Moutaftsi M, et al. 2010. Uncovering the interplay between CD8, CD4 and antibody responses to complex pathogens. Future Microbiol. 5:221–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nesburn AB, et al. 2006. Topical/mucosal delivery of sub-unit vaccines that stimulate the ocular mucosal immune system. Ocul. Surf. 4:178–187 [DOI] [PubMed] [Google Scholar]
- 62. Nesburn AB, Burke RL, Ghiasi H, Slanina SM, Wechsler SL. 1998. Therapeutic periocular vaccination with a subunit vaccine induces higher levels of herpes simplex virus-specific tear secretory immunoglobulin A than systemic vaccination and provides protection against recurrent spontaneous ocular shedding of virus in latently infected rabbits. Virology 252:200–209 [DOI] [PubMed] [Google Scholar]
- 63. Nesburn AB, Burke RL, Ghiasi H, Slanina SM, Wechsler SL. 1998. A therapeutic vaccine that reduces recurrent herpes simplex virus type 1 corneal disease. Invest. Ophthalmol. Vis. Sci. 39:1163–1170 [PubMed] [Google Scholar]
- 64. Nesburn AB, et al. 2005. Local and systemic B cell and Th1 responses induced following ocular mucosal delivery of multiple epitopes of herpes simplex virus type 1 glycoprotein D together with cytosine-phosphate-guanine adjuvant. Vaccine 23:873–883 [DOI] [PubMed] [Google Scholar]
- 65. Nesburn AB, et al. 1998. Local periocular vaccination protects against eye disease more effectively than systemic vaccination following primary ocular herpes simplex virus infection in rabbits. J. Virol. 72:7715–7721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nnedu ON, et al. 2011. Humoral immune responses to Plasmodium falciparum among HIV-1 infected Kenyan adults. Proteomics Clin. Appl. 5:613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. O'Farrell N. 1999. Increasing prevalence of genital herpes in developing countries: implications for heterosexual HIV transmission and STI control programmes. Sex. Transm. Infect. 75:377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. O'Farrell N, Tovey SJ. 1994. High cumulative incidence of genital herpes amongst HIV-1 seropositive heterosexuals in south London. Int. J. STD AIDS 5:415–418 [DOI] [PubMed] [Google Scholar]
- 69. Perng GC, et al. 1994. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J. Virol. 68:8045–8055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Perng GC, et al. 1999. Herpes simplex virus type 1 serum neutralizing antibody titers increase during latency in rabbits latently infected with latency-associated transcript (LAT)-positive but not LAT-negative viruses. J. Virol. 73:9669–9672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Posavad CM, et al. 2003. T cell immunity to herpes simplex viruses in seronegative subjects: silent infection or acquired immunity? J. Immunol. 170:4380–4388 [DOI] [PubMed] [Google Scholar]
- 72. Schmid DS, et al. 1999. Limits in reliability of glycoprotein G-based type-specific serologic assays for herpes simplex virus types 1 and 2. J. Clin. Microbiol. 37:376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tengvall S, O'Hagan D, Harandi AM. 2008. Rectal immunization generates protective immunity in the female genital tract against herpes simplex virus type 2 infection: relative importance of myeloid differentiation factor 88. Antiviral Res. 78:202–214 [DOI] [PubMed] [Google Scholar]
- 74. Thomas J, Rouse BT. 1998. Immunopathology of herpetic stromal keratitis: discordance in CD4+ T cell function between euthymic host and reconstituted SCID recipients. J. Immunol. 160:3965–3970 [PubMed] [Google Scholar]
- 75. Tronstein E, et al. 2011. Genital shedding of herpes simplex virus among symptomatic and asymptomatic persons with HSV-2 infection. JAMA 305:1441–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Troye-Blomberg M, et al. 1988. T cell reactivity of defined peptides from a major Plasmodium falciparum vaccine candidate: the Pf155/RESA antigen. Immunol. Lett. 19:229–233 [DOI] [PubMed] [Google Scholar]
- 77. Verjans GM, et al. 1998. Identification and characterization of herpes simplex virus-specific CD4+ T cells in corneas of herpetic stromal keratitis patients. J. Infect. Dis. 177:484–488 [DOI] [PubMed] [Google Scholar]
- 78. Wald A, et al. 1997. Frequent genital herpes simplex virus 2 shedding in immunocompetent women. Effect of acyclovir treatment. J. Clin. Invest. 99:1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wald A, et al. 2002. Genital shedding of herpes simplex virus among men. J. Infect. Dis. 186(Suppl 1):S34–S39 [DOI] [PubMed] [Google Scholar]
- 80. Wald A, et al. 2000. Reactivation of genital herpes simplex virus type 2 infection in asymptomatic seropositive persons. N. Engl. J. Med. 342:844–850 [DOI] [PubMed] [Google Scholar]
- 81. Welsh RM, Selin LK. 2002. No one is naive: the significance of heterologous T-cell immunity. Nat. Rev. Immunol. 2:417–426 [DOI] [PubMed] [Google Scholar]
- 82. Whitley RJ, Miller RL. 2001. Immunologic approach to herpes simplex virus. Viral Immunol. 14:111–118 [DOI] [PubMed] [Google Scholar]
- 83. Whitley RJ, Roizman B. 2001. Herpes simplex virus infections. Lancet 357:1513–1518 [DOI] [PubMed] [Google Scholar]
- 84. Xu F, et al. 2006. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. JAMA 296:964–973 [DOI] [PubMed] [Google Scholar]