Abstract
Nonstructural proteins 7 and 8 of severe acute respiratory syndrome coronavirus (SARS-CoV) have previously been shown by X-ray crystallography to form an 8:8 hexadecamer. In addition, it has been demonstrated that N-terminally His6-tagged SARS-CoV Nsp8 is a primase able to synthesize RNA oligonucleotides with a length of up to 6 nucleotides. We present here the 2.6-Å crystal structure of the feline coronavirus (FCoV) Nsp7:Nsp8 complex, which is a 2:1 heterotrimer containing two copies of the α-helical Nsp7 with conformational differences between them, and one copy of Nsp8 that consists of an α/β domain and a long-α-helix domain. The same stoichiometry is found for the Nsp7:Nsp8 complex in solution, as demonstrated by chemical cross-linking, size exclusion chromatography, and small-angle X-ray scattering. Furthermore, we show that FCoV Nsp8, like its SARS-CoV counterpart, is able to synthesize short oligoribonucleotides of up to 6 nucleotides in length when carrying an N-terminal His6 tag. Remarkably, the same protein harboring the sequence GPLG instead of the His6 tag at its N terminus exhibits a substantially increased, primer-independent RNA polymerase activity. Upon addition of Nsp7, the RNA polymerase activity is further enhanced so that RNA up to template length (67 nucleotides) can be synthesized. Further, we show that the unprocessed intermediate polyprotein Nsp7-10 of human coronavirus (HCoV) 229E is also capable of synthesizing oligoribonucleotides up to a chain length of six. These results indicate that in case of FCoV as well as of HCoV 229E, the formation of a hexadecameric Nsp7:Nsp8 complex is not necessary for RNA polymerase activity. Further, the FCoV Nsp7:Nsp8 complex functions as a noncanonical RNA polymerase capable of synthesizing RNA of up to template length.
INTRODUCTION
Coronaviruses are enveloped, positive-strand RNA viruses with extraordinarily large genomes comprising up to 31,000 nucleotides. Among the RNA viruses, the transcription of coronavirus RNA is unique. First, the large size of the genome requires unusual enzymatic activities, such as an exoribonuclease and an endoribonuclease activity (11), in order to maintain genetic stability. Second, the synthesis of a nested set of subgenomic mRNAs by the discontinuous transcription strategy used by coronaviruses demands a huge and complicated replication/transcription complex (RTC) (22).
In addition to (as-yet-unidentified) host cell factors, the RTC is composed of virus-encoded nonstructural proteins (Nsps), which are synthesized as part of the polyproteins pp1a and pp1ab. These polyproteins are translation products of open reading frame 1 (Orf1); the synthesis of pp1ab requires a (−1) ribosomal frameshift after translation of about 2/3 of Orf1 (30). Through the proteolytic activities of cysteine proteases contained within Nsp3 and Nsp5, the polyproteins are processed into intermediate and mature nonstructural proteins, ultimately generating up to 15 or 16 Nsps. Many of these have well-characterized functions within the RTC; for example, Nsp12 is an RNA-dependent RNA polymerase (RdRp), Nsp13 is a helicase, and Nsp14 harbors a methyltransferase that, along with the Nsp16:Nsp10 complex, is responsible for RNA capping (4, 5, 11).
In several cases, possible functions of Nsps were derived from their three-dimensional structures as determined by X-ray crystallography (11). Thus, a crystal structure of the Nsp7:Nsp8 (Nsp7+8) complex of severe acute respiratory syndrome coronavirus (SARS-CoV) revealed a hollow, cylindrical hexadecamer composed of eight copies of Nsp8 and eight copies of Nsp7 (31). The channel with a diameter of about 30 Å at the center of this complex suggested that it could encircle double-stranded RNA (dsRNA), in agreement with the observation that it was lined by positively charged amino acid residues. In addition, a heterodimeric complex formed by the Nsp8 C-terminal half and Nsp7 of SARS-CoV was also reported recently (16). The authors postulated that the truncated complex might be incorporated into the Nsp7+8 supercomplex, thereby interfering with its activity. Furthermore, Imbert et al. described a sequence-specific oligonucleotide-synthesizing activity for Nsp8 of SARS-CoV (12). In that study, the recombinant Nsp8 carrying an N-terminal His6 tag was capable of recognizing a specific short sequence [5′-(G/U)CC-3′] in the single-stranded RNA coronavirus genome as a template for the synthesis of short oligonucleotides (<6 residues) in an Mn2+-dependent manner but with a relatively low fidelity. Consequently, Imbert et al. (12) proposed that oligonucleotides synthesized this way might be utilized as primers by the primer-dependent RdRp, Nsp12 (28). According to this proposition, Nsp8 would be the primase of SARS-CoV and Nsp12 its main RdRp.
However, many questions regarding the roles of Nsp7 and Nsp8 in coronavirus RNA synthesis remain. First, the function of Nsp7 in the primase activity is poorly characterized thus far. At present, Nsp7 is only regarded as “mortar” stabilizing the hexadecameric Nsp7+8 complex structure (31), without having significant influence on the synthesis of RNA primers (12). Second, since Nsp8 alone and the Nsp7+8 complex showed equal primase activity in the study by Imbert et al. (12), we still do not know the stoichiometry between the two components in vivo, or whether various stoichiometries may exist at different transcriptional stages (12). Third, even though the observed Nsp7:Nsp8 hexadecamer presents a positive electrostatic channel suitable for dsRNA binding (31), the mode of action at the molecular level of this noncanonical primase is not understood. Therefore, it is important to investigate Nsp7+8 of coronaviruses other than SARS-CoV, to determine whether the hexadecamer occurs universally in all coronaviruses and to hopefully shed light on the molecular mechanism of RNA synthesis by Nsp8 and its complex with Nsp7.
This study was motivated by dynamic light-scattering experiments, indicating that in solution, the particle diameter of Nsp8 from the alphacoronaviruses HCoV-229E (human coronavirus 229E) and FCoV (feline coronavirus) was decreasing upon the addition of Nsp7. This observation suggested that a form of the Nsp7+8 complex other than the hexadecamer described for the corresponding proteins of the betacoronavirus SARS-CoV was possibly formed (19). We report here that the HCoV-229E Nsp7-10 polyprotein, which is an intermediate of pp1a and pp1ab processing and unlikely to form the same hexadecamer as the SARS-CoV Nsp7+8 complex, exhibits primase activity. Furthermore, we show that in case of FCoV both Nsp7 and Nsp8 are dimeric in solution, but the Nsp7+8 complex is a 2:1 heterotrimer rather than a hexadecamer as described for SARS-CoV. The same stoichiometry was found in solution and in the crystal. In addition to describing the crystal structure of the complex at 2.6-Å resolution, we show here that FCoV Nsp8 and the Nsp7+8 complex exhibit de novo RNA synthesis activities that result in much longer RNA molecules than previously reported for SARS-CoV (12). This suggests that Nsp8 is not only a primase but also provides a primer-independent polymerase function in coronavirus replication and transcription.
MATERIALS AND METHODS
Construct design, cloning, expression, and purification.
Plasmid pET11d-229E710 encoding C-terminally His6-tagged HCoV-229E Nsp7-10 polyprotein was kindly provided by John Ziebuhr (Queen's University, Belfast, United Kingdom). Constructs coding for C-terminally His6-tagged Nsp7 and N-terminally His6-tagged Nsp8 (corresponding to residues Ser3423-Gln3505 and Ser3506-Gln3700 of pp1ab, FCoV strain FIPV WSU-79/1146), were kindly provided by Bruno Coutard (AFMB, University de la Méditerranée, Marseille, France). To produce glutathione S-transferase (GST)-tagged FCoV Nsp7 and Nsp8, DNA sequences encoding Nsp7 and Nsp8 were amplified by PCR using Pfu DNA polymerase (Invitrogen) and primers encoding the 5′ and 3′ ends (for Nsp7 forward, 5′-CGCGGATCCAAACTTACAGAGATGAAGTGT-3′; for Nsp7 reverse, 5′-CCGCTCGAGTTACTGTAAAATAGTGGTGTTCTCA-3′; for Nsp8 forward, 5′-CGCGGATCCGTAGCATCTGCTTATGCTGCTT-3′; for Nsp8 reverse, 5′-CCGCTCGAGTTACTGCAACTTTGTGGTTCTC-3′), and the PCR products were then cloned into the expression vector pGEX-6p-1 using BamHI and XhoI sites. Both sequence-verified clones were transformed into the Escherichia coli strain BL21-Gold (DE3). The culture was grown at 37°C in 2×YT medium with vigorous shaking until the optical density at 600 nm reached 0.8 (Nsp8) or 1.3 (Nsp7). Expression was induced by adding 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubation at 25°C for 16 h. The selenomethionyl (SeMet) derivative of Nsp8 was produced by using the methionine-biosynthesis pathway inhibition method (7). Cells were harvested by centrifugation and lysed by sonication in buffer containing 50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 5 mM dithiothreitol (DTT), and EDTA-free cocktail protease inhibitor (Roche; one tablet per 50 ml of buffer). The lysate was cleared by ultracentrifugation at 30,000 rpm for 90 min at 4°C, and the soluble fraction was applied onto a pre-equilibrated 5-ml GSTrap FF column (GE Healthcare). The column was washed with 20 column volumes of lysis buffer and 10 column volumes of PreScission cleavage buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, and 1 mM DTT), followed by loading the PreScission protease (GE Healthcare) onto the column. Cleavage was performed at 4°C overnight, and the eluted samples were further purified by size-exclusion chromatography (SEC; HiLoad 16/60 Superdex 75; GE Healthcare) equilibrated with 10 mM Tris-HCl (pH 7.5)–200 mM NaCl. Nsp8 was then mixed with 2-fold molar excess of Nsp7 and passed through the Superdex 75 column in the same buffer. Fractions of the Nsp7+8 complex were concentrated to 15 mg/ml and used for either crystallization or biochemical assays. The FCoV Nsp7 and Nsp8 produced as described here contain four additional residues (GPLG) at their N termini.
Recombinant production of the C-terminally His6-tagged HCoV-229E polyprotein Nsp7-10 and of N-terminally His6-tagged FCoV Nsp8 followed the same procedure as for the GST-tagged FCoV Nsp8. For purification, the cell pellets were first resuspended in buffer A (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 20 mM imidazole). The sample was then centrifuged at 30,000 rpm for 90 min at 4°C. The supernatant was loaded onto a HisTrap HP column (GE Healthcare). After being washed with buffer A, the protein was eluted with buffer B (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 500 mM imidazole) and further purified by SEC using the procedure described above. The HCoV-229E polyprotein Nsp7-10 and the N-terminally His6-tagged FCoV Nsp8 were concentrated to 5 and 15 mg/ml, respectively.
Crystallization and diffraction-data collection.
Crystallization screening was performed using a Phoenix robot (Dunn Labortechnik) with the sitting-drop, vapor-diffusion method at 12°C and drops containing 0.3 μl of protein solution plus 0.3 μl of reservoir solution. The following commercial screens were used: SaltRx, PEG/Ion, Index, Crystal Screen, and PEGRx (all from Hampton Research). Tiny crystals appeared in a drop containing 2.4 M (NH4)2HPO4 and 0.1 M Tris-HCl (pH 8.5). Optimized crystals suitable for diffraction were grown in drops containing 1.5 μl of protein solution and 1.5 μl of reservoir solution [2.24 M (NH4)2HPO4, 2 mM tris(2-carboxyethyl)phosphine (TCEP), 0.1 M Tris-HCl (pH 8.7)] at 4°C. Prior to being cryo-cooled in liquid nitrogen, the crystals were soaked in reservoir solution supplemented with 20% glycerol. Diffraction data were collected at 100 K using beamline BL14.2, BESSY (Berlin, Germany), which is equipped with an MX225 charge-coupled device detector (Rayonics). Indexing, integration, and scaling of data was carried out with XDS (via the interface program ixds, developed at BESSY) (14). The program XPREP (Bruker) was used to further analyze and prepare data sets for structure solution and refinement. The statistics of the data sets are summarized in Table 1.
Table 1.
Data collection and refinement statistics
| Statisticsa | Nsp7/SeMet-Nsp8b |
|
|---|---|---|
| Crystal 1 | Crystal 2c | |
| Data collection | ||
| X-ray source | BL14.2, BESSY (Berlin, Germany) | |
| Wavelength (Å) | 0.9809 | 0.97974/0.97990 |
| Space group | C2221 | |
| Unit cell dimensions (Å) | a = 121.45; b = 160.30; c = 102.66 | a = 120.78; b = 161.43; c = 103.25 |
| Resolution range (Å) | 33.45–2.60 (2.70–2.60) | 33.55–3.24 (3.34–3.24) |
| No. of unique reflections | 31,155 | 16,370/16,291 |
| Completeness (%) | 99.8 (100) | 99.8 (99.9)/99.7 (99.3) |
| Mean I/σ(I) | 20.38 (3.33) | 24.74 (4.92)/23.52 (3.62) |
| Multiplicity | 7.23 (7.41) | 14.48 (14.99)/14.60 (14.90) |
| Rint (%) | 5.77 (62.0) | 7.81 (48.4)/8.42 (84.1) |
| Rsigma (%) | 3.1 (30.0) | 2.84 (19.9)/2.81 (27.08) |
| Solvent content (%) | 61.42 | |
| Vm (Å3/Da) | 3.19 | |
| Refinement | ||
| Resolution range | 33.45-2.60 | |
| Rwork (%) | 20.51 | |
| Rfree (%) | 22.74 | |
| Modeled residues | A: −2–73, 79–192; B: 2–83; C: 0–81; D: −2–191; E: 2–82; F: 0–77 | |
| No. of protein atoms | 5,498 | |
| No. of water molecules | 77 | |
| RMSD in bond lengths (Å) | 0.010 | |
| RMSD in bond angles (°) | 1.29 | |
| Ramachandran plot (%) | ||
| Mostly favored | 97.3 | |
| Additionally allowed | 2.4 | |
| Outliers | 0.3 | |
Rint = Σ||Fo|2 − <|Fo|2>| |/Σ|Fo|2. |Fo|2 values are the intensities of the reflections in the unmerged data. <|Fo|2> is the mean intensity for symmetry equivalents, including Friedel pairs. Rsigma = Σ[σ (|Fo|2)]/Σ[|Fo|2]. The |Fo|2 values are the intensities of the reflections in the unique data. R = Σ||Fo| − |Fc||/Σ|Fo|. The |Fo| values are the amplitudes of the structure factors. Rwork is the R value for reflections used in refinement, whereas Rfree is the R value for 5% of the reflections which are selected in thin shells and are not included in the refinement.
Values in parentheses represent the highest-resolution shell.
Values are presented as “peak/inflection” where applicable.
Structure solution, refinement, analysis, and presentation.
The crystal structure of the FCoV Nsp7+8 complex was determined by using the multi-wavelength anomalous diffraction (MAD) method. The selenium substructure was determined using SHELXD (24). Phases were initially calculated to 3.5 Å and then extended to 2.6 Å and improved via density modification using SHELXE (24). Initially, the electron density was interpreted automatically using SHELXE (24) and ARP/wARP (10). Thereafter, model refinement and manual rebuilding were performed alternately. For refinement, Refmac5 (17) was used initially and autoBUSTER (2) was used subsequently, whereas model building was carried out using Coot (8). Molecular figures were prepared by using PyMol (Schrödinger).
Glutaraldehyde cross-linking.
Chemical cross-linking was performed for Nsp7, Nsp8, and the Nsp7+8 complex. All samples were diluted to 2 mg/ml in 10 mM HEPES (pH 7.5)–200 mM NaCl–5 mM DTT. An aliquot of 25% (vol/vol) glutaraldehyde was diluted and added to the protein samples to give a final concentration of 0.01% (for Nsp7 and Nsp8) or 0.05% (for the Nsp7+8 complex). The reaction was performed at 4°C for 10 min, followed by quenching by the addition of 1.0 M Tris-HCl pH 8.0 (0.5% [vol/vol]). Subsequently, an equal volume of loading buffer was added to the reaction mixture, and the samples were heated at 95°C for 10 min, followed by SDS-PAGE (15% acrylamide gels).
SEC.
The solution states of Nsp7, Nsp8, and the Nsp7+8 complex were analyzed by size-exclusion chromatography (SEC). A 1-ml portion of protein solution at a concentration of 15 mg/ml was loaded onto a HiLoad 16/60 Superdex 75 column (GE Healthcare) and then eluted with buffer (10 mM Tris-HCl [pH 7.5], 200 mM NaCl, 5 mM DTT), with detection of the absorbance at 280 nm. The linear relationship between the gel-phase distribution coefficient (Kav) and the average molecular mass (MM) was fitted by Kav = −0.49265 log MM + 2.51301.
SAXS.
Small-angle X-ray scattering (SAXS) measurements for Nsp7 and the Nsp7+8 complex were carried out using the EMBL BioSAXS beamline X33 (20, 21) at the DORIS storage ring, DESY (Hamburg, Germany). A photon-counting PILATUS detector with 1 million pixels was used to record the scattered X-rays at a wavelength of 1.54 Å. The sample-to-detector distance was 2.7 m, yielding a maximum recordable momentum transfer (s = 4π sinθ/λ) of 0.6 Å−1. A concentration series was analyzed for each sample at 20°C (1.0, 2.0, 4.0, 8.0, and 16.0 mg/ml for Nsp7+8 and 1.2, 2.4, 4.7, and 9.4 mg/ml for Nsp7). For either sample, no concentration dependence of the scattering profile was observed, and the SAXS data from the most concentrated sample were used for further analysis. Standard SAXS data reduction and analysis were carried out using PRIMUS (15). The forward scattering I(0) and radius of gyration Rg were evaluated using the Guinier approximation (9), assuming that at very small angles (s < 1.3/Rg), the intensity is represented as I(s) = I(0) exp[−(sRg)2/3]; the Guinier plots were linear, suggesting monodisperse solutions. The molecular mass of the solutes was evaluated by comparison of the forward scattering I(0) to that from a reference solution of bovine serum albumin (molecular mass, 66 kDa; 4.5 mg/ml in 50 mM HEPES [pH 7.5]). The maximum size of the protein molecule (Dmax) and the pair distance distribution function p(r) were calculated using the program GNOM (26). The program CRYSOL (27) was used to calculate the X-ray scattering profiles with the high-resolution model from the crystal structure (for Nsp7 alone, the corresponding part of the crystal structure of Nsp7+8 was used).
RdRp activity assay.
An RNA oligonucleotide representing stem-loop 1 of the 3′ untranslated region (3′UTR) of the FCoV strain FIPV WSU-79/1146 genome (5′-GGCAACCCGAUGUCUAAAACUUGUCUUUCCGAGGAAUUACGGGUCAUCGCGCUGCCUACUCUUGUAC-3′, 67 nucleotides [purchased from Riboxx]) was used as a template in the de novo polymerase reaction. The standard reaction mixture (20 μl) contained 25 mM Tris-HCl (pH 8.0), 10 mM KCl, 4 mM MgCl2, 10 mM DTT, 1 μCi of [α-32P]GTP, 20 μM (each) ATP, CTP, and UTP, and 2 μM RNA templates, along with a 5 μM concentration of purified protein. The reaction was carried out by incubation at 30°C for 1 h and terminated by the addition of EDTA. The products were separated by using denaturing PAGE (17.5% acrylamide, 7 M urea) in TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA [pH 8.0]). Dried gels were exposed to phosphorimaging screens and scanned with a PhosphorImager (Typhoon; GE Healthcare).
Protein structure accession number.
Coordinates and structure factors for the crystal structure of the FCoV Nsp7+8 complex have been deposited in the Protein Data Bank (PDB) with accession code 3UB0.
RESULTS
Overall architecture of the Nsp7+8 complex.
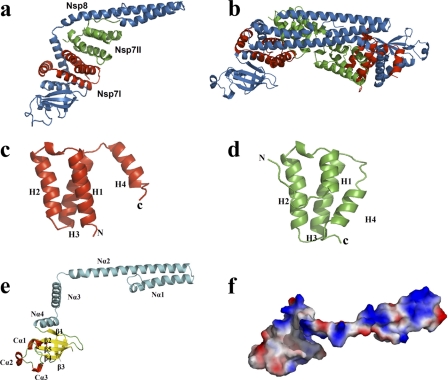
The 2.6-Å FCoV Nsp7+8 complex structure reveals a heterotrimer consisting of two copies of Nsp7 and one copy of Nsp8 (Fig. 1a). The shape of the complex resembles a pistol (Nsp8) equipped with a magazine (2×Nsp7). In the asymmetric unit, two copies of this heterotrimer dock onto each other through interaction between the two Nsp8 molecules, forming a dimer of the 2:1 Nsp7+8 trimer, i.e., a heterohexamer (Fig. 1b).
Fig 1.
Three-dimensional structure of the FCoV Nsp7+8 complex and its constituents. (a and b) Overall structure of the Nsp7+8 heterotrimeric (a) and heterohexameric (b) complex. Nsp8, Nsp7I, and Nsp7II are colored in sky blue, red, and green, respectively. (c and d) Diagram illustration of FCoV Nsp7I (c) and Nsp7II (d). (e) Diagram illustration of FCoV Nsp8. (f) Electrostatic-potential surface of FCoV Nsp8. Red and blue surfaces represent negative and positive potentials (−10 kBT to 10 kBT), respectively. The electrostatic surface potentials were calculated using the Adaptive Poisson-Boltzmann Solver (APBS) within the PyMOL APBS tools. Secondary structures are indicated in Fig. 3.
Structure of Nsp7 in the complex.
Two different conformations of the Nsp7 monomer are observed within the heterotrimer (Fig. 1c and d). Both Nsp7 molecules are composed of four α-helices forming an antiparallel helix bundle. Helix H2 (residues 26 to 41) and helix H3 (residues 45 to 62), as well as the loops preceding and following them are structurally conserved between the two molecules, with a root-mean-square difference (RMSD) of 2.0 Å (for the 44 Cα atoms of segment 21-64). In contrast, helix H1 (residues 3 to 20 for Nsp7I, residues 11 to 20 for Nsp7II) and helix H4 (residues 65 to 77 for Nsp7I, residues 69 to 78 for Nsp7II) are quite different in length (H1 and H4) and in orientation (H4). As seen in Fig. 2a and Fig. 3a, helix H1 of Nsp7I comprises 18 residues, while helix H1 of Nsp7II contains only 10 residues. Besides, in Nsp7I, helix H4 is far apart from helix H1 (the distance between Val12 [Cγ1] and Leu71 [Cδ2] is 12.31 Å), but in Nsp7II, these two helices are much closer to one another (the distance between the two atoms indicated above is only 4.33 Å). The reason for this conformational difference is the insertion of Nsp8 helix Nα3 in between Nsp7 H1 and H4 in molecule Nsp7I but not in Nsp7II (see below). This difference between the two Nsp7 conformations is found identically in both copies of the Nsp7+8 heterotrimer in the asymmetric unit. The RMSD values between the two Nsp7I (for 78 Cα atoms) and the two Nsp7II (for 81 Cα atoms) are 0.37 and 0.34 Å, respectively.
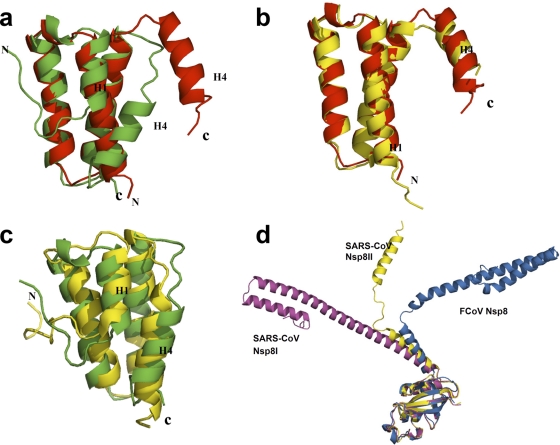
Fig 2.
Structural comparison of FCoV Nsp7I and Nsp7II, and of FCoV Nsp7 and Nsp8 with the corresponding proteins of SARS-CoV. (a) Superimposition of FCoV Nsp7I (red) with FCoV Nsp7II (green), with RMSD values (for 44 Cα atoms) of 2.0 Å. (b) Superimposition of FCoV Nsp7I (red) with SARS-CoV Nsp7 (yellow) in complex with Nsp8. The RMSD (for 74 Cα positions) is 0.97 Å. (c) Superimposition of FCoV Nsp7II (green) with the SARS-CoV Nsp7 (yellow) solution structure at pH 6.5. The RMSD (for 73 Cα positions) is 2.38 Å. (d) C-terminal domain superimposition of FCoV Nsp8 (sky blue) with SARS-CoV Nsp8I (magenta) and Nsp8II (yellow), with RMSD values (for 79 Cα atoms, residues 113 to 191) of 1.9 Å and 1.1 Å, respectively.
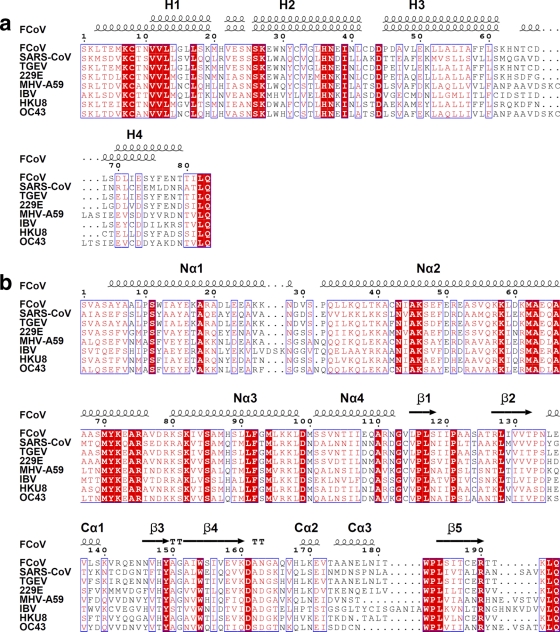
Fig 3.
Sequence alignment of FCoV Nsp7 (a) and Nsp8 (b) with homologues from other coronaviruses. The alignments were achieved by using CLUSTAL W2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/), and colored figures were generated by ESPript2.2 (http://espript.ibcp.fr/ESPript/ESPript/). Residues labeled in red are conserved to >70%, and residues boxed in red are completely identical. Secondary structures of FCoV Nsp7 (above, Nsp7II; below, Nsp7I) and Nsp8 are indicated at the top of the alignments. All of the sequences aligned here are from the National Center for Biotechnology Information (NCBI) database. Abbreviations and accession numbers: feline infectious peritonitis virus (FCoV), DQ010921.1; SARS-CoV, NP_828865 (Nsp7) and NP_828866 (Nsp8); transmissible gastroenteritis coronavirus (TGEV), NP_840005.1 (Nsp7) and NP_840006.1 (Nsp8); human coronavirus 229E (229E), NP_835348.1 (Nsp7) and NP_835349.1 (Nsp8); mouse hepatitis virus strain A59 (MHV-A59), NP_740612.1 (Nsp7) and NP_740613.1 (Nsp8); infectious bronchitis virus (IBV), NP_740625.1 (Nsp7) and NP_740626.1 (Nsp7); bat coronavirus HKU8, YP_001718610.1; and human coronavirus OC43 (OC43), NP_937947.
Interestingly, when we compared our structures for Nsp7 within the heterotrimeric complex to the three structures of SARS-CoV Nsp7 with coordinates available in the PDB (13, 18, 31), we noticed that the structure of FCoV Nsp7I is almost the same as that of SARS-CoV Nsp7 in complex with Nsp8 (Fig. 2b). The RMSD for 74 Cα atoms between the two molecules is 0.97 Å. On the other hand, FCoV Nsp7II is structurally similar to the isolated SARS-CoV Nsp7 in solution, as determined by nuclear magnetic resonance (NMR) spectroscopy at pH 6.5 (13) (Fig. 2c), with an RMSD (for 73 Cα atoms) of 2.38 Å.
Structure of Nsp8 in the complex.
SARS-CoV Nsp8 possesses two different conformations in the hexadecameric complex with Nsp7 (31). In our FCoV Nsp7+8 complex, only one conformation of Nsp8 is observed in the two crystallographically independent copies of the heterotrimer (Fig. 1e). Similar to the “golf club” structure of SARS-CoV Nsp8 (31), the Nsp8 of FCoV has a pistol-like structure composed of an N-terminal shaft domain and a C-terminal “grip” domain (residues 5 to 112 and residues 113 to 191, respectively). The N-terminal shaft domain contains four helices (Nα1, residues 5 to 28; Nα2, residues 32 to 76; Nα3, residues 80 to 98; and Nα4, residues 101 to 112). Another three α-helices (Cα1, 134 to 140; Cα2, 168 to 172; and Cα3, 174 to 179) packed against a five-stranded antiparallel β-sheet (β1, residues 115 to 118; β2, residues 127 to 132; β3, residues 146 to 149; β4, residues 152 to 160; and β5, residues 184 to 190) form the C-terminal “grip” domain, which has an α/β fold.
Superimposition of the FCoV Nsp8 C-terminal “grip” domain onto the same regions of SARS-CoV Nsp8I and Nsp8II indicates that the C-terminal “grip” domains are quite conserved (Fig. 2d), with RMSD values (for 79 Cα atoms) of 1.9 and 1.1 Å, respectively. However, the Nsp8 N-terminal shaft domains of the three molecules (SARS-CoV Nsp8I and II, and FCoV Nsp8) display totally different orientations (Fig. 2d). Similar to the situation seen for half of the Nsp8 molecules (Nsp8II or “kinked golf club”) in the hexadecameric SARS-CoV Nsp7+8 complex (31), the Nsp8 molecules in both copies of our heterotrimeric FCoV Nsp7+8 complex possess a bend (displaying poorly defined electron density) between helices Nα2 and Nα3, hinting at the flexibility of this region. It is likely that the bend provides rotational freedom for the N-terminal shaft domain to swing around the C-terminal “grip” domain, thereby resulting in the change of the orientation of the N-terminal shaft domain.
Interactions between components of the heterotrimer.
In the FCoV Nsp7+8 heterotrimeric complex, Nsp8, Nsp7I, and Nsp7II interact predominantly via hydrophobic interactions, which are augmented by a few intermolecular hydrogen bonds and salt bridges. The interaction interfaces between Nsp8 and Nsp7I, Nsp8 and Nsp7II, and Nsp7I and Nsp7II feature a total buried surface area of 3,500, 1,250, and 1,310 Å2, respectively. Most of the residues involved in the interactions are quite conserved among coronaviruses (Fig. 3).
The interaction between Nsp8 and Nsp7I involves two parts. The first interface is located at Nα3 of the Nsp8 N-terminal shaft domain. Residues of Nsp7I helices H1 (Met6, Val12, and Leu16) and H4 (Leu71, Tyr75, and Phe76) form a hydrophobic core with residues of Nsp8 helix Nα3 (Met87, Leu91, Phe92, Met94, and Leu98); a 3.16-Å hydrogen bond is observed between Nsp7I Lys2 Nζ and Nsp8 Leu98 O. The second hydrophobic interface is mainly composed of residues on H3 (Leu49, Leu53, Ile56, Ala57, and Leu60), as well as H4 (Ile72) of Nsp7I, and residues on Nα4 (Ile106 and Ile107) as well as β1 (Leu115 and Phe116) of Nsp8. The side chain of Nsp8 Arg111 (Nε) forms hydrogen bonds and salt bridges with the side chains of both Nsp7I Glu73 (Oε2, 3.14 Å) and Glu77 (Oε1, 3.25 Å). The two interfaces are also observed in the hexadecameric complex of SARS-CoV Nsp7+8 (31) and its truncated heterodimeric form (16).
The hydrophobic interface between Nsp7II and Nsp8 is mainly formed between helix H1 of Nsp7II (Val11 and Leu14) and helix Nα2 (Leu59, Met62, and Ala66) of the Nsp8 N-terminal shaft domain, indicating that the binding of Nsp7II has influence on the orientation of this domain. In addition, Phe76 on H4 of Nsp7II and Ile83 and Met87 on Nα3 of Nsp8 also contribute to the hydrophobic interactions between these two polypeptides.
The Nsp7I-Nsp7II interface is formed mostly by hydrophobic interactions between residues on helices H1 and H2 of Nsp7I (Cys8, Val11, Val12, Leu14, and Leu41) and residues on helices H3 and H4 of Nsp7II (Leu49, Leu53, Ile56, Leu60, and Ile72). In addition, Ser18 (Oγ), Trp29 (Nε1), and Asn37 (Nδ2) of Nsp7I form hydrogen bonds with Glu73 (Oε1, 2.46 Å), Asp67 (Oδ1, 2.70 Å), and Cys66 (O, 2.82 Å) of Nsp7II, respectively.
The two copies of the Nsp7+8 heterotrimer present in the asymmetric unit of the crystal form a heterohexamer through interaction between the N-terminal shaft domains of the Nsp8 molecules. To investigate whether the heterohexamer is formed by crystal packing or does also exist in solution, we performed a series of biochemical and biophysical experiments.
Solution state of Nsp7, Nsp8, and the Nsp7+8 complex.
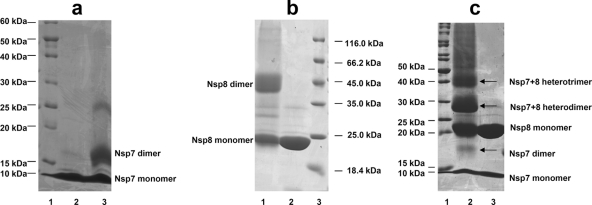
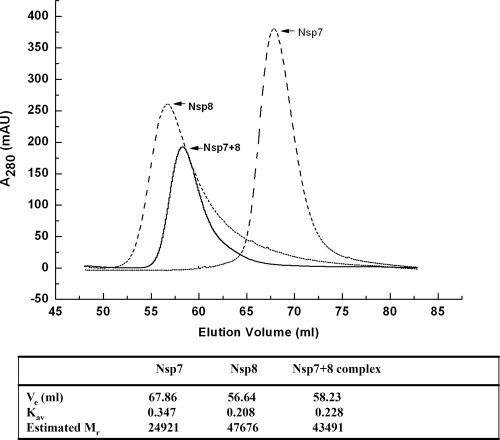
In addition to the question regarding which quaternary structure of FCoV Nsp7+8 exists in solution (heterotrimer or heterohexamer), the multimeric states of isolated coronavirus Nsp7 and Nsp8 are also poorly described and remain controversial (13, 25, 31). To study the quaternary structures of Nsp7, Nsp8, and the Nsp7+8 complex in solution, we first performed a chemical cross-linking analysis (Fig. 4). After treatment with 0.01% glutaraldehyde, both isolated Nsp7 and isolated Nsp8 displayed cross-linked homodimers (18 kDa for Nsp7 and 43 kDa for Nsp8) in addition to their monomers (Fig. 4a and b). When the Nsp7+8 complex was cross-linked by 0.05% glutaraldehyde, we detected the following species: Nsp7 monomer (9 kDa), Nsp7 dimer (18 kDa), Nsp8 monomer (21.5 kDa), Nsp7/Nsp8 heterodimer (30.5 kDa), and the Nsp7+8 2:1 heterotrimer (39.5 kDa). Higher-order multimers were not detected (Fig. 4c). We then also used SEC to determine the solution states of Nsp7, Nsp8, and the Nsp7+8 complex. As shown in Fig. 5, the Nsp7+8 complex eluted with a retention volume of 58.23 ml, representing an estimated MM of 43.5 kDa, which corresponded reasonably well to the MM indicated by cross-linking (39.5 kDa). Nsp7 and Nsp8 had retention volumes of 67.86 ml and 56.64 ml, respectively. The estimated MMs (24.9 kDa for Nsp7 and 47.7 kDa for Nsp8) calculated from the retention volumes also agreed well with the MMs of their cross-linked dimers.
Fig 4.
SDS-PAGE analyses of chemically cross-linked Nsp7, Nsp8, and the Nsp7+8 complex. (a) Nsp7, lane 1, MM marker proteins; lane 2, untreated Nsp7; lane 3, Nsp7 cross-linked by 0.01% glutaraldehyde. The monomer (9 kDa) and the dimer (18 kDa) are detected. The weak band at ∼25 kDa is possibly due to a nonspecifically cross-linked Nsp7 trimer. (b) Nsp8, lane 1, Nsp8 cross-linked by 0.01% glutaraldehyde (the monomer [21.5 kDa] and the dimer [43 kDa] are detected); lane 2, untreated Nsp8; lane 3, MM marker proteins. (c) Nsp7+8 complex, lane 1, MM marker proteins; lane 2, Nsp7+8 complex cross-linked by 0.05% glutaraldehyde; lane 3, untreated Nsp7+8 complex. The Nsp7 monomer (9 kDa), Nsp7 dimer (18 kDa), Nsp8 monomer (21.5 kDa), heterodimer (30.5 kDa), and heterotrimer (39.5 kDa) are all observed.
Fig 5.
SEC of Nsp7, Nsp8, and the Nsp7+8 complex. An estimation of protein solution state based on SEC data is shown in the table. Ve is the elution volume, and Kav is the gel-phase distribution coefficient. Calibration of the column determined the relationship between Kav and MM as Kav = −0.49265 log MM + 2.51301. Estimated Mr values from the SEC analyses also indicate that Nsp7 and Nsp8 alone are homodimers in solution but that their complex is a 2:1 heterotrimer.
In order to investigate the quaternary structures of the FCoV Nsp7+8 complex and of isolated Nsp7 further, we applied SAXS using synchrotron radiation. The linear Guinier plot of the SAXS profile for the Nsp7+8 complex yielded a radius of gyration (Rg) of 30.0 ± 0.5 Å, which agrees well with the Rg of 30.3 Å derived from the atomic structure of the heterotrimer. In contrast, the calculated Rg for the heterohexamer (dimer of heterotrimers) found in the asymmetric unit of the crystal is 35.0 Å. The maximum particle size Dmax derived from the experimental data is 105 ± 5 Å, which agrees well with the value of 115 Å for the heterotrimer, whereas the Dmax for the asymmetric heterohexamer is ∼130 Å. As shown in Fig. 6, a direct comparison of the experimental scattering profile with that calculated from the atomic structure for the trimer yields a good fit with discrepancy χ = 1.3, in contrast to the much worse fit with χ = 6.0 for the hexamer (the discrepancy χ should be equal to 1 for a perfect fit).
Fig 6.
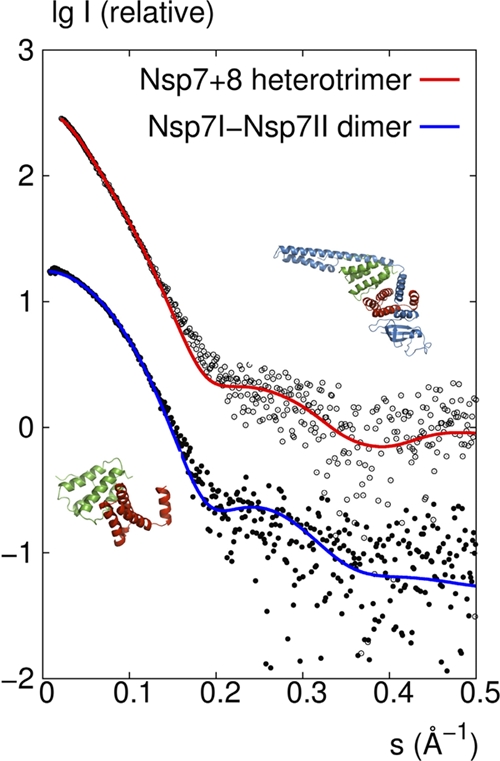
SAXS investigations of the FCoV Nsp7+8 complex and Nsp7 in solution. Comparison of the experimental SAXS profile (empty circles) with a CRYSOL-calculated scattering curve (red) of the heterotrimer model consisting of Nsp7I, Nsp7II, and Nsp8 derived from the crystal structure and comparison of the SAXS data (filled circles) with the calculated scattering curve (blue) of the dimer model consisting of Nsp7I and Nsp7II, as found in the crystal structure of the heterotrimer.
The MM of Nsp7 estimated from the Guinier analysis of the SAXS data was 19 kDa. This value is twice the molecular mass (9 kDa) calculated from the sequence of a monomer, suggesting that the protein forms dimers in solution. There is a good agreement between the experimental SAXS curve and the theoretical curve calculated by CRYSOL from a dimeric model containing the two conformations, Nsp7I and Nsp7II, observed in the crystal structure of the Nsp7+8 heterotrimer (discrepancy χ = 1.3; Fig. 6).
Taking into account all of the evidence from chemical cross-linking, SEC, and small-angle X-ray scattering, it is clear that the Nsp7+8 complex is heterotrimeric in solution, whereas the isolated Nsp7 and Nsp8 form homodimers.
RdRp activity of Nsp8.
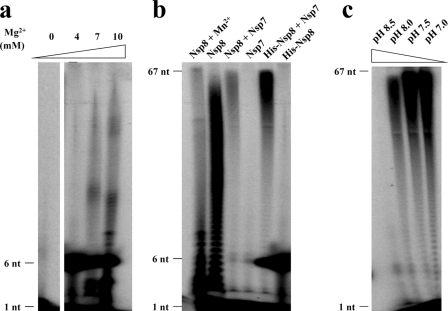
Imbert et al. (12) reported that the N-terminally His6-tagged Nsp8 of SARS-CoV is a noncanonical RdRp capable of synthesizing short oligoribonucleotides (<6 residues), which might be used as primers by the canonical RdRp, Nsp12. In our study, we initially detected a similar activity for the HCoV-229E Nsp7-10 polyprotein and for N-terminally His6-tagged Nsp8 of FCoV (Fig. 7 and Fig. 8a); both of these proteins were able to mainly synthesize short RNA strands of ∼6 nucleotides. The polymerase activity of FCoV Nsp8 also required the presence of magnesium ions, since activity was not detectable without Mg2+. Surprisingly, however, upon increasing the Mg2+ concentration, limited amounts of longer RNA products could also be observed in case of N-terminally His6-tagged Nsp8 (Fig. 8a). We subsequently tested the RNA synthesis activity of the FCoV Nsp8 construct lacking the His6 tag while carrying four additional residues (GPLG) at its N terminus; this protein was the one used for cocrystallization with Nsp7. Interestingly, compared to His6-tagged Nsp8, GPLG-Nsp8 showed a much more pronounced ability to synthesize longer RNA, although it still could hardly synthesize RNA of template length (Fig. 8b).
Fig 7.
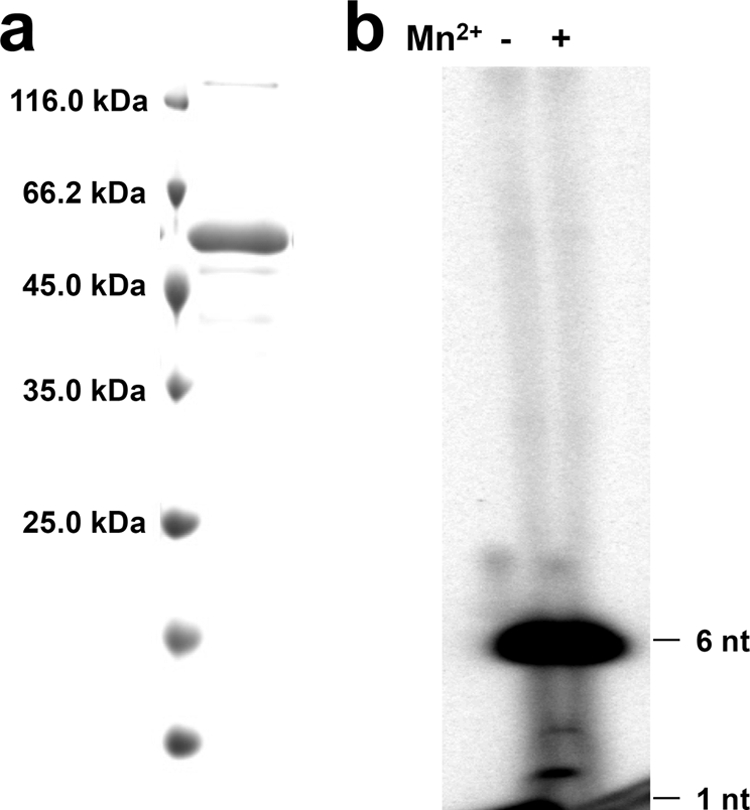
The HCoV-229E Nsp7-10 polyprotein is a de novo RdRp in vitro. (a) Purified HCoV-229E Nsp7-10 polyprotein (52.5 kDa) analyzed by SDS–15% PAGE. (b) RdRp activity of HCoV-229E Nsp7-10 polyprotein in the presence or absence of Mn2+.
Fig 8.
De novo RdRp activities of FCoV Nsp8-containing variants. (a) Mg2+ concentration influence on the RdRp processivity of His-Nsp8 (N-terminally His6 tagged). (b) Side-by-side comparison of the polymerase activities of the Nsp8, Nsp8+7 complex, His6-Nsp8, and His6-Nsp8+7 complex. Nsp7 was used as a negative control; Nsp8 with 4 mM Mn2+ was also tested. (c) Influence of pH on Nsp7+8 complex polymerase activity. RNA oligonucleotide representing stem-loop 1 of the 3′UTR of the FCoV strain FIPV WSU-79/1146 genome was used as a template in all RdRp assays.
Next, we tested these two Nsp8 variants in complex with Nsp7. Compared to the two Nsp8 variants alone, we noticed that both of the complexes exhibited much higher processivity and were able to synthesize template-length RNA (67 nucleotides; Fig. 8b). This observation provides clear evidence for an important role of Nsp7 in the RNA polymerase activity; apparently, it helps Nsp8 reduce abortion during replication and transcription and thus significantly increases its processivity. We also studied the influence of pH on the activity of the Nsp7+8 complex and found optimum activity at neutral pH, while at pH 8.5, the polymerase activity was almost abolished (Fig. 8c). Different from the report on SARS-CoV Nsp8 (12), manganese ions did not enhance the polymerase activity of the HCoV-229E Nsp7-10 polyprotein (Fig. 7b) or the FCoV Nsp7+8 complex (data not shown). On the contrary, the addition of 4 mM Mn2+ even had a negative effect on the polymerase activity (Fig. 8b). Possibly, Mn2+ competes with Mg2+ for binding to the coordinating residues.
Oligoribonucleotides representing stem-loop 1 (SL1) and the complete 3′UTR of SARS-CoV were also used as templates for testing the polymerase activities of the N-terminally His6-tagged FCoV Nsp8 and the Nsp7+8 complex (data not shown). The identity between the SL1 sequences of SARS-CoV and FCoV is ca. 50%. We did not observe significant changes in the processivity compared to using FCoV SL1 as a template, implying that Nsp8 does not exhibit strong (if any) preference for template sequences.
DISCUSSION
Unlike the SARS-CoV Nsp7+8 hexadecamer complex, in which eight Nsp8 interact with eight Nsp7, the Nsp7+8 complex of FCoV is a heterotrimer formed by two Nsp7 molecules and one Nsp8. In our study, the Nsp7 and the Nsp8 have been mixed at different ratios, but the resulting complex always eluted with a retention volume of ∼58.2 ml from the size exclusion column, corresponding to the heterotrimer. Chemical cross-linking and SAXS also indicate that the 2:1 stoichiometry of Nsp7:Nsp8 observed in our crystal structure exists in solution as well.
The crystal structure shows that the N-terminal shaft domain of FCoV Nsp8 is rich in positively charged amino acid residues on one side (Fig. 1f), which could be responsible for RNA binding. In the SARS-CoV Nsp7+8 complex (31), a similar charge distribution was observed; replacing Arg and Lys residues in this region by Ala significantly decreased its nucleic-acid-binding affinity (31). Previous studies in our group, using one-dimensional NMR and circular dichroism spectroscopy, showed that the N-terminal shaft domain of isolated Nsp8 of SARS-CoV is partially disordered in solution, in agreement with the conclusions of Rao and coworkers (16, 31). The same domain of FCoV Nsp8 is well defined in our complex structure, with the exception of the bend between helices Nα2 and Nα3. Interestingly, the orientation of the FCoV Nsp8 shaft domain is distinct from that of its counterpart in SARS-CoV. The latter makes the same interactions with Nsp7 as FCoV Nsp8 does with Nsp7I, but the second Nsp7 molecule (with its distinct conformation) is lacking in the hexadecameric SARS-CoV complex. It is quite possible that the interaction with two Nsp7 molecules, especially the binding of Nsp7II, not only improves folding of the Nsp8 N-terminal shaft domain but also has an influence on its orientation.
We show here that, like SARS-CoV, FCoV and HCoV 229E possess a primer-independent RNA polymerase, Nsp8, in addition to the canonical, primer-dependent RNA polymerase Nsp12. Since Nsp8 of SARS-CoV was reported to produce oligoribonucleotides with fewer than six residues, it was postulated that the protein acts as a primase and the primers synthesized by Nsp8 would be subsequently utilized by the primer-dependent polymerase Nsp12 for further elongation (12). Distinct from previous observations with SARS-CoV, our results demonstrate that both the Nsp8 and the Nsp7+8 complex of FCoV are able to synthesize much longer transcripts (up to template length). Interestingly, while the present study was under review, te Velthuis et al. showed that Nsp8 of SARS-CoV with authentic N terminus is capable of extending primed RNA templates, implying the possibility that Nsp8 and the Nsp7+8 complex of SARS-CoV may also synthesize longer products (29). We conclude that Nsp8 is not simply a primase that merely provides primers for Nsp12 but that it also executes its own polymerase function and associated roles in coronavirus replication and transcription. In addition, considering the fact that the FCoV Nsp7+8 complex exhibits a much higher processivity than does Nsp8 alone in de novo RNA synthesis, we consider Nsp7's role to be more than just mortar to stabilize the FCoV Nsp7+8 complex structure. We postulate that Nsp7 could enhance the RNA-binding affinity of the Nsp8 N-terminal shaft domain by stabilizing its orientation and folding and thereby increasing the polymerase processivity of Nsp8.
In the present study, we have also tested the RNA synthesis function of the precursor polyprotein Nsp7-10 of HCoV 229E. Sawicki et al. had proposed that the MHV-A59 precursor polyprotein Nsp4-10 might possess independent functions before being proteolytically processed (23). Deming et al. further supported this idea by demonstrating that rearrangement of the Nsp7 and Nsp8 coding sequences in MHV-A59 was not permissive for virus replication (6). Here we demonstrate that the purified Nsp7-10 polyprotein of HCoV 229E, a presumable replicase precursor, can function as RdRp in vitro, even though it is only able to produce short RNA oligonucleotides. In comparison, the mature FCoV Nsp8 and the Nsp7+8 complex exhibit much stronger polymerase processivity, suggesting that polyprotein processing and replication/transcription complex formation are crucial for the polymerase activity of Nsp8, well in agreement with the observation that disruption of the Nsp7-Nsp8 cleavage site of MHV-A59 is lethal for viral replication (6). In addition, the distinct polymerase processivity of different Nsp8 variants implies that at different stages of replication and transcription, different Nsp8-containing variants may be required as de novo RdRps. The proteolytic processing of the replicase polyproteins and the assembly of the replication complex might be critical for regulating virus replication and transcription, for instance, for switching from negative-strand RNA synthesis to positive-strand RNA synthesis (1). Nsp8 or Nsp8-containing polyprotein might act as primase at the initial step of coronavirus RNA synthesis, but after being cleaved and assembled into the Nsp7+8 complex, the latter might be responsible for further elongation of the primers.
As mentioned above, we noticed that FCoV GPLG-Nsp8 showed much higher polymerase processivity than the N-terminally His6-tagged Nsp8 and the HCoV-229E Nsp7-10 polyprotein, indicating that additional residues at the Nsp8 N terminus might have significant influence on the elongation step during RNA synthesis. It might be argued that the increased processivity is due to the four extra residues (GPLG) remaining at the N terminus of FCoV Nsp8 from the cloning procedure, although such a gain of function would be extremely unlikely for these four amino acid residues devoid of functional groups in their side chains. In addition, the recent report by te Velthuis et al. (29) on a primer extension activity of SARS-CoV Nsp8 with authentic N terminus supports our view that the primer-independent polymerase activities of FCoV Nsp8 and the Nsp7+8 complex are not due to a gain of function caused by these four extra residues. Rather, a hexahistidine tag or large additional sequences, such as in HCoV-229E Nsp7-10, seem to cause a loss of function in terms of polymerase processivity. We do not have a good explanation for the apparent influence of residues at the N terminus of Nsp8, since a structure of free Nsp8 is not available. We are planning more structural and functional studies to address this issue.
The fact that the Nsp7-10 polyprotein of HCoV 229E, which is very unlikely to form the same quaternary structure as either the SARS-CoV or the FCoV Nsp7+8 complex (hexadecamer and heterotrimer, respectively), shows polymerase activity, suggests that the Nsp8 de novo RNA synthesis activity probably does not depend on its multimeric state. This is supported by the observation of polymerase activity for the dimeric FCoV Nsp8 and the heterotrimeric Nsp7+8 complex.
Unfortunately, the molecular mechanism of the RNA polymerase activity of Nsp8 still cannot be derived from the available Nsp7+8 complex structures, since these do not allow the localization of the active site(s), in spite of a large body of mutational data (12, 31). Canonical primases and RNA polymerases usually feature an (Asp/Glu)-X-(Asp/Glu) motif at their active site, which binds to Mg2+ or Mn2+ (3). FCoV Nsp8 comprises a Glu-X-Glu sequence within its helix Nα3, but the two acidic side chains are oriented toward different sides of the helix. However, we cannot exclude the possibility that the active conformation of Nsp8 could be quite different when in complex with RNA, since preliminary results from our fluorescence resonance energy transfer measurements demonstrate that fluorophor-labeled Nsp8 of FCoV undergoes significant conformational changes upon addition of RNA (unpublished data). Therefore, it is important to perform more biochemical and structural studies on the Nsp8-RNA interaction in order to reveal the mechanism of this noncanonical RNA-dependent RNA polymerase.
ACKNOWLEDGMENTS
We thank John Ziebuhr for providing the plasmid of HCoV-229E Nsp7-10 and Bruno Coutard and Bruno Canard for providing plasmids encoding FCoV Nsp7 and Nsp8. We are grateful to Ulrike Haas, Sarah Willkomm, and Winfried Wünsche for assistance with polymerase activity assays and to Yuri Kusov and Jeroen R. Mesters for helpful discussion. We also acknowledge access to the beamline BL14.2 of the BESSY II storage ring (Berlin, Germany) via the Joint Berlin MX-Laboratory sponsored by the Helmholtz Zentrum Berlin für Materialien und Energie, the Freie Universität Berlin, the Humboldt-Universität zu Berlin, the Max-Delbrück Centrum, and the Leibniz-Institut für Molekulare Pharmakologie.
This study was partly supported by the European Commission through its projects VIZIER (contract LSHG-CT-2004-511960 [for work on FCoV proteins]) and subsequently by its SILVER project (contract HEALTH-F3-2010-260644 [for work on HCoV-229E proteins]). R.H. thanks the DFG Cluster of Excellence “Inflammation at Interfaces” (EXC306) and the Fonds der Chemischen Industrie for continuous support. R.H. is also supported by the Chinese Academy of Sciences (CAS) through a CAS Visiting Professorship for Senior International Scientists (grant 2010T1S6). D.I.S. acknowledges support from BMBF research grant SYNC-LIFE (contract 05K10YEA).
Footnotes
Published ahead of print 8 February 2012
REFERENCES
- 1. Brayton PR, Lai MM, Patton CD, Stohlman SA. 1982. Characterization of two RNA polymerase activities induced by mouse hepatitis virus. J. Virol. 42:847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bricogne G, et al. 2010. BUSTER, version 2.11.0. Global Phasing, Ltd., Cambridge, United Kingdom [Google Scholar]
- 3. Castro C, et al. 2007. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc. Natl. Acad. Sci. U. S. A. 104:4267–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Y, et al. 2009. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 106:3484–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Decroly E, et al. 2011. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 7:e1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deming DJ, Graham RL, Denison MR, Baric RS. 2007. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J. Virol. 81:10280–10291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doublie S. 1997. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276:523–530 [PubMed] [Google Scholar]
- 8. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of COOT. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guinier A. 1939. La diffraction des rayons X aux trés petits angles: application a l'étude de phenomenes ultramicroscopiques. Ann. Phys. (Paris) 12:161–237 [Google Scholar]
- 10. Hattne J, Lamzin VS. 2008. Pattern-recognition-based detection of planar objects in three-dimensional electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 64:834–842 [DOI] [PubMed] [Google Scholar]
- 11. Hilgenfeld R, Tan J, Chen S, Shen X, Jiang H. 2008. Structural proteomics of emerging viruses: the examples of SARS-CoV and other coronaviruses, p 361–433 In Sussman J, Silman I. (ed), Structural proteomics and its impact on the life sciences. WorldScientific, Singapore [Google Scholar]
- 12. Imbert I, et al. 2006. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 25:4933–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson MA, Jaudzems K, Wüthrich K. 2010. NMR structure of the SARS-CoV nonstructural protein 7 in solution at pH 6.5. J. Mol. Biol. 402:619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kabsch W. 2010. XDS. Acta Crystallogr. D Biol. Crystallogr. 66:125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. 2003. PRIMUS: a Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36:1277–1282 [Google Scholar]
- 16. Li S, et al. 2010. New nsp8 isoform suggests mechanism for tuning viral RNA synthesis. Protein Cell 1:198–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 18. Peti W, et al. 2005. Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7. J. Virol. 79:12905–12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ponnusamy R. 2010. Crystallographic and biochemical investigations on coronavirus replication proteins: nonstructural proteins 8 and 9. Ph.D. thesis University of Lübeck, Lübeck, Germany [Google Scholar]
- 20. Roessle MW, et al. 2007. Upgrade of the small-angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory, Hamburg. J. Appl. Crystallogr. 40(Suppl):190–194 [Google Scholar]
- 21. Round AR, et al. 2008. Automated sample-changing robot for solution scattering experiments at the EMBL Hamburg SAXS station X33. J. Appl. Crystallogr. 41:913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sawicki SG, Sawicki DL, Siddell SG. 2007. A contemporary view of coronavirus transcription. J. Virol. 81:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sawicki SG, et al. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 1:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sheldrick GM. 2010. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 66:479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sutton G, et al. 2004. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure 12:341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Svergun DI. 1992. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25:495–503 [Google Scholar]
- 27. Svergun DI, Barberato C, Koch MHJ. 1995. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28:768–773 [Google Scholar]
- 28. te Velthuis AJ, Arnold JJ, Cameron CE, van den Worm SH, Snijder EJ. 2010. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 38:203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. te Velthuis AJ, van den Worm SH, Snijder EJ. 29 October 2012. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. [Epub ahead of print.] doi:10.1093/nar/gkr893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Hemert MJ, et al. 2008. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 4:e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhai Y, et al. 2005. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 12:980–986 [DOI] [PMC free article] [PubMed] [Google Scholar]